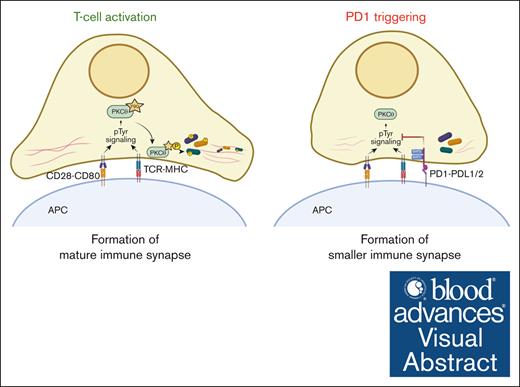
PD1 inhibits PKCθ synaptic localization and phosphorylation of its cytoskeleton targets, leading to formation of smaller immune synapses.
Reduced Vimentin phosphorylation on S39 is a putative marker for PD1 engagement in T cells in a PDL1+ tumor microenvironment.
Visual Abstract
The inhibitory surface receptor programmed cell death protein 1 (PD1) is a major target for antibody–based cancer immunotherapies. Nevertheless, a substantial number of patients fail to respond to the treatment or experience adverse effects. An improved understanding of intracellular pathways targeted by PD1 is thus needed to develop better predictive and prognostic biomarkers. Here, via unbiased phosphoproteome analysis of primary human T cells, we demonstrate that PD1 triggering inhibited the phosphorylation and physical association with protein kinase Cθ (PKCθ) of a variety of cytoskeleton-related proteins. PD1 blocked activation and recruitment of PKCθ to the forming immune synapse (IS) in a Src homology-2 domain–containing phosphatase-1/2 (SHP1/SHP2)-dependent manner. Consequently, PD1 engagement led to impaired synaptic phosphorylation of cytoskeleton-related proteins and formation of smaller IS. T-cell receptor induced phosphorylation of the PKCθ substrate and binding partner vimentin was long-lasting and it could be durably inhibited by PD1 triggering. Vimentin phosphorylation in intratumoral T cells also inversely correlated with the levels of the PD1 ligand, PDL1, in human lung carcinoma. Thus, PKCθ and its substrate vimentin represent important targets of PD1-mediated T-cell inhibition, and low levels of vimentin phosphorylation may serve as a biomarker for the activation of the PD1 pathway.
Introduction
T cells are crucial for effective anticancer immune responses. Their activation requires triggering of the T-cell receptor (TCR) by major histocompatibility complex (MHC)–bound antigen, together with a costimulatory signal provided by the CD28 coreceptor.1,2 This initiates a complex signaling cascade that involves the membrane-proximal tyrosine kinases Lck and ZAP70 and the serine/threonine (S/T) kinase PKCθ (protein kinase Cθ), and culminates in the induction of major transcriptional changes.1-3 Upon activation, the T cell and the antigen-presenting cells also form a stable, mature immune synapse (IS), required for polarized secretion of inflammatory cytokines and restricted killing of target cells.4-6
In the tumor microenvironment, various strategies are at work to dampen T-cell activation, allowing tumor immune escape.7 The receptor programmed cell death protein 1 (PD1), upregulated on activated T cells, is 1 of the major coinhibitors of T-cell activation.8 PD1 can be triggered by 1 of its ligands, PD ligand 1 (PDL1) or PDL2, expressed by immune and nonimmune cells, including infected and cancer cells.9 Ligand-dependent engagement of PD1 leads to T-cell inhibition.10-12 Therefore, the PD1/PDL1 axis has gained much attention as a drug target in antibody–based tumor immunotherapies. Therapeutic blockade of PD1 or PDL1 has become a first- or second-line therapy in the treatment of several cancers, such as non–small cell lung cancer (NSCLC), melanoma, Hodgkin lymphoma or advanced renal carcinoma.13-16 Despite significant advancements in these therapies, PD1 targeting fails to provide the desired outcome in a substantial number of patients. It is estimated that targeting PD1/PDL1 with PD1-specific (eg, nivolumab) or PDL1-specific (eg, atezolizumab) monoclonal antibodies, achieves response rates between 10% and 25% in bladder, gastric or ovarian cancer.17 A better understanding of the PD1–mediated intracellular mechanisms leading to T-cell inhibition would help to develop predictive biomarkers, enabling more precise stratification of patients expected to benefit from PD1-based immunotherapies.
The tyrosine phosphatases Src homology 2 domain-containing phosphatase 1 (SHP1) and SHP2 are signaling proteins that mediate PD1–dependent T-cell inhibition.18-20 PD1 triggering leads, in presence of a simultaneous stimulatory trigger, to the Lck-mediated phosphorylation of tyrosine residues Y223 and Y248 of PD1, which in turn provide binding sites with partially overlapping binding specificity for SHP1 and SHP2, respectively.19-23 The recruitment of SHP1/SHP2 to PD1 is thought to activate the phosphatases, causing dephosphorylation of tyrosine residues of membrane-proximal proteins required for T-cell activation, such as ZAP70 and CD28.18,21,24 However, T-cell activation relies not only on tyrosine phosphorylation of membrane-proximal proteins, but also on changes within the serine and threonine (S/T) phosphoproteome,25 which have received less attention in the context of PD1–mediated T-cell inhibition.
Here, we show that PD1 triggering leads to dephosphorylation of cytoskeleton-related proteins that are substrates and/or binding partners of the S/T kinase PKCθ. We additionally identify PKCθ itself as a key target of PD1-induced, SHP1/SHP2-mediated tyrosine dephosphorylation and PD1-induced inhibition of IS formation. Furthermore, we demonstrate that the phosphorylation status of the cytoskeleton protein and PKCθ target vimentin is a bona fide indicator of PD1 engagement. Finally, we provide evidence for an inverse correlation between the status of vimentin phosphorylation on S39 in T cells and the extent of PDL1 expression in human lung adenocarcinoma tissues. These findings identify a new aspect of PD1 signaling with potential relevance for biomarker development.
Materials and methods
Details on the cells, constructs, reagents, and assays used for this study are provided in the supplemental Material and Methods.
T-cell stimulation
For beads-mediated stimulation, Dynabeads M450Epoxy were coupled according to manufacturer’s instructions with anti-CD3 ab (OKT3) at 5%, anti-CD28 ab (CD28.2) at 10% and TACI-Fc or PDL1-Fc at 85% of beads capacity to obtain stimulatory (S) or inhibitory (I) beads, respectively. TACI-Fc is a recombinant receptor whose ligands BAFF and APRIL are not expressed on T cells and served as a control protein. Cells were stimulated at a 1:3 (cell:bead) ratio for the indicated time points. If required, we prestimulated CD4 and CD8 T cells to increase PD1 surface expression with cross-linked anti-CD3 and anti-CD28 abs at 1 μg/mL for 48 hours; we then washed and incubated the cells for 24 hours in 10% fetal calf serum/RPMI before beads-mediated stimulus. For stimulation of Jurkat cells or human primary T cells with Raji cells, the latter were incubated for 30 minutes with staphylococcal enterotoxin E (SEE) or with a mixture of SEA, SEB, and SEE at indicated concentrations. Raji cells were then mixed with Jurkat cells at a 1:1 ratio or with primary T cells at a 1:2 ratio for the time of stimulation. Cells were stimulated with cross-linked anti-CD3 and anti-CD28 abs or PMA (phorbol 12-myristate 13-acetate, 20 ng/mL) and ionomycin (1 μM) for the indicated times and concentrations. Where indicated, cells were pretreated with the PKCθ inhibitor (2,4-diamino-5-nitropyrimidine) or the kinase C inhibitor BIM8 (bisindolylmaleimide VIII acetate) at indicated concentrations for 60 minutes at 37°C.
Labeling and conjugate formation
Jurkat and Raji cells were labeled with CMTMR (1 μM) or CFSE (10 nM) ± SEE, respectively, for 30 minutes at 37°C, then pelleted, resuspended at 106 cells/mL in 10% fetal calf serum/RPMI-1640, mixed at a 1:1 ratio and incubated for 30 minutes (unless indicated otherwise) at 37°C to allow conjugate formation.
Immunocytochemistry staining and confocal microscopy
Cells were layered on polylysine coverslips, centrifuged (1 minute, 450 g), incubated on ice for 5 minutes, fixed with 4% paraformaldehyde (PFA) for 15 minutes on ice and quenched with 50 mM NH4Cl for 10 minutes. Cells were treated with 0.05% saponin/0.2% bovine serum albumin in phosphate-buffered saline for 15 minutes. Fc receptors were blocked using Fc receptor blocking solution. Immunostainings were performed with indicated antibodies and samples were mounted in medium with DAPI to stain nuclei. Images were acquired using a Zeiss LSM880 confocal microscope with EC Plan Neofluar 40x/1.3 NA and EC Plan Neofluar 63x/1.25 NA oil immersion objectives. For quantification of confocal images, the total fluorescence intensity was measured in whole cells or IS regions of the focal plane using Fiji software. Red-green-blue (RGB) intensity plot was obtained using ImageJ plugin RGB Profiler.
Image stream
Analysis was conducted using an ImageStream X MKII (ISx) system. Up to 40 000 cell events per sample were collected at 60× magnification.
Lung adenocarcinoma samples
Ten lung adenocarcinomas cases (5 PDL1low and 5 PDL1high) were collected upon receiving consent at the Centre Hospitalier Universitaire Vaudois and anonymized, followed by 5-plex staining of CD3, PD1, pan-cytokeratin (PanCK), pS39Vimentin, and PDL1.
Western blots
Cells were lysed in SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) protein sample buffer and boiled at 96°C for 10 minutes, followed by analysis on high-resolution SDS-PAGE and immunoblot as described.26
Phosphoproteome analysis
Cells were cultured in stable isotope labeling by amino acids in cell culture (SILAC) medium for more than 2 weeks, stimulated and lysed in 8M urea and digested essentially as described.27 Phosphopeptide enrichment was performed as described.28 Details on liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis and data processing are described in the supplemental Material and Methods.
PKCθ interactome
PKCθ immunoprecipitates were analyzed by Coomassie blue staining, gel lanes were excised into 6 equal regions and in-gel digested with trypsin (Promega) as described.29 LC-MS/MS analysis was carried out on a Q-exactive Plus orbitrap mass spectrometer (for details see supplemental Material and Methods).
Blood samples from healthy, informed, and consenting human donors were obtained from Interregional Blood Transfusion SRC Ltd (Epalinges, Switzerland). Lung adenocarcinomas cases (5 PDL1low and 5 PDL1high) were collected upon consent at the Centre Hospitalier Universitaire Vaudois and were anonymized.
Results
PD1 inhibits S/T phosphorylation of cytoskeleton-related proteins in primary human T cells
As an experimental system to identify PD1-specific changes in the phosphoproteome, we used primary human T cells in which PD1 expression was induced by prestimulation with cross-linked stimulatory anti-CD3/CD28 antibodies (supplemental Figure 1A). PD1 expression was typically at the highest 48 hours after stimulation, as previously reported.30 To assess the functionality of PD1, T cells were then incubated with dynabeads, coupled with anti-CD3 and anti-CD28 antibodies,12 together with recombinant PDL1-Fc (for inhibitory beads), or TACI-Fc protein (for stimulatory beads). Compared with stimulatory beads, inhibitory beads efficiently impaired extracellular signal-regulated kinase 1/2 phosphorylation (supplemental Figure 1B) and IL-2 secretion in CD4 and CD8 T cells (supplemental Figure 1C-D).
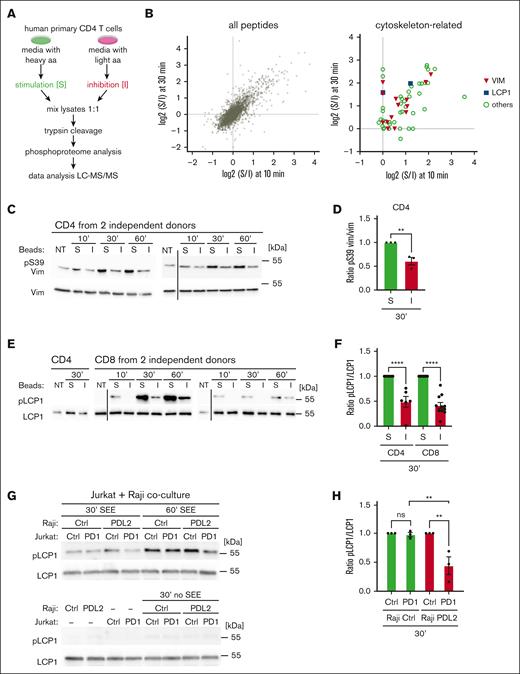
Next, we treated PD1+ primary human CD4 T cells with stimulatory vs inhibitory beads for 10 and 30 minutes and monitored differences in the total phosphoproteome (Figure 1A). For this, cells were grown under conditions of SILAC, and labeling efficiency (not shown) and T-cell responsiveness were verified prior to the experiment (supplemental Figure 1E). Using mass spectrometry, we identified and quantified over 6000 phosphorylation sites across the 2 time points (Figure 1B, left panel; supplemental Table 1; all phosphosites). Of these, 278 sites showed significant changes, when comparing inhibitory to stimulatory conditions (supplemental Table 1; significant phosphosites). Of note, we identified a PD1-induced decrease in phosphorylation of 60 sites within prominent cytoskeleton components or regulators (Figure 1B, right panel; supplemental Table 2). These included the intermediate filament protein vimentin, previously identified as a PKCθ substrate,31 the actin-bundling protein L-plastin (also called lymphocyte cytosolic protein, LCP1),32,33 previously reported to undergo phosphorylation upon TCR/CD28 costimulation,34 and other actin cytoskeleton regulators (Figure 1B, right panel; supplemental Table 2). This suggests that PD1 promotes T-cell exhaustion, at least in part, through effects on cytoskeleton rearrangements that are important for IS formation.5,6,35
PD1 triggering induced a reduction in S/T phosphorylation of cytoskeleton-related proteins. (A) Scheme of the SILAC labeling and mass spectrometry approach used to identify PD1-mediated changes in the phosphoproteome of human primary CD4 T cells. Cells were labeled with heavy or light SILAC media for 2 weeks and treated for 10 or 30 minutes with beads coated with anti-CD3 and anti-CD28 antibodies in combination with TACI-Fc (stimulatory beads) or PDL1-Fc (inhibitory beads). After mixing, lysates were digested with trypsin and phosphorylated peptides were identified by LC-MS/MS analysis. (B) Graphs summarizing phosphorylation events that differed between stimulatory (S) and inhibitory (I) conditions. Left graph: all identified sites. Dots in the upper right quarter represent sites for which phosphorylation decreased upon PD1 triggering at both, the 10- and 30-minute time points. Right graph: only cytoskeleton-related proteins, including vimentin and LCP1. (C) Western blot analysis monitoring phosphorylation of S39 (pS39) of vimentin (Vim) in human primary CD4 T cells from 2 independent donors, left untreated (NT) or triggered for the indicated time points with S or I beads. Black vertical line indicates removal of an irrelevant lane. (D) Quantification of the experiments performed as in panel C: pS39 Vim levels were normalized to total Vim levels for the 30-minute condition for 3 independent experiments. (E) Western blot analysis monitoring phosphorylation of S5 of L-plastin (pLCP1) in human primary CD4 and CD8 T cells from 2 independent donors. Cells were left untreated (NT) or triggered for the indicated time points with S or I beads. Black vertical line indicates the cut of the image performed to remove 5-minute time point, which was not relevant for this figure. (F) Quantification of experiments performed as in panel E: pS5 LCP1 levels in CD4 (upper panel) and CD8 (lower panel) were normalized to total LCP1 levels for the 30-minute condition, for 6 and 11 independent experiments, respectively. (G) Western blot analysis monitoring phosphorylation of S5 of L-plastin (pLCP1) in Jurkat Ctrl cells or in Jurkat cells expressing WT PD1 (PD1) triggered for the indicated time points with Raji Ctrl cells or Raji PDL2 in presence or absence of SEE at 6 ng/mL. (H) Quantification of experiments performed as in panel G: pS5 LCP1 level in Jurkat cells was normalized to total LCP1 level for the 30-minute condition for 3 independent experiments. Graphs in panels D,F,H show mean value ± SEM. Statistical analysis for graphs in panels D,F was performed using a 2-tailed, unpaired t test. Statistical analysis for graph in panel H was performed using 1-way ANOVA with Šídák's multiple comparisons test. ∗∗∗∗P < .0001; ∗∗∗P < .001; ∗∗P < .01. ANOVA, analysis of variance; SEM, standard error of the mean.
PD1 triggering induced a reduction in S/T phosphorylation of cytoskeleton-related proteins. (A) Scheme of the SILAC labeling and mass spectrometry approach used to identify PD1-mediated changes in the phosphoproteome of human primary CD4 T cells. Cells were labeled with heavy or light SILAC media for 2 weeks and treated for 10 or 30 minutes with beads coated with anti-CD3 and anti-CD28 antibodies in combination with TACI-Fc (stimulatory beads) or PDL1-Fc (inhibitory beads). After mixing, lysates were digested with trypsin and phosphorylated peptides were identified by LC-MS/MS analysis. (B) Graphs summarizing phosphorylation events that differed between stimulatory (S) and inhibitory (I) conditions. Left graph: all identified sites. Dots in the upper right quarter represent sites for which phosphorylation decreased upon PD1 triggering at both, the 10- and 30-minute time points. Right graph: only cytoskeleton-related proteins, including vimentin and LCP1. (C) Western blot analysis monitoring phosphorylation of S39 (pS39) of vimentin (Vim) in human primary CD4 T cells from 2 independent donors, left untreated (NT) or triggered for the indicated time points with S or I beads. Black vertical line indicates removal of an irrelevant lane. (D) Quantification of the experiments performed as in panel C: pS39 Vim levels were normalized to total Vim levels for the 30-minute condition for 3 independent experiments. (E) Western blot analysis monitoring phosphorylation of S5 of L-plastin (pLCP1) in human primary CD4 and CD8 T cells from 2 independent donors. Cells were left untreated (NT) or triggered for the indicated time points with S or I beads. Black vertical line indicates the cut of the image performed to remove 5-minute time point, which was not relevant for this figure. (F) Quantification of experiments performed as in panel E: pS5 LCP1 levels in CD4 (upper panel) and CD8 (lower panel) were normalized to total LCP1 levels for the 30-minute condition, for 6 and 11 independent experiments, respectively. (G) Western blot analysis monitoring phosphorylation of S5 of L-plastin (pLCP1) in Jurkat Ctrl cells or in Jurkat cells expressing WT PD1 (PD1) triggered for the indicated time points with Raji Ctrl cells or Raji PDL2 in presence or absence of SEE at 6 ng/mL. (H) Quantification of experiments performed as in panel G: pS5 LCP1 level in Jurkat cells was normalized to total LCP1 level for the 30-minute condition for 3 independent experiments. Graphs in panels D,F,H show mean value ± SEM. Statistical analysis for graphs in panels D,F was performed using a 2-tailed, unpaired t test. Statistical analysis for graph in panel H was performed using 1-way ANOVA with Šídák's multiple comparisons test. ∗∗∗∗P < .0001; ∗∗∗P < .001; ∗∗P < .01. ANOVA, analysis of variance; SEM, standard error of the mean.
PD1 engagement inhibits serine phosphorylation of vimentin and L-plastin
Next, we validated our phosphoproteome data using commercial antiphospho-Serine (pS) antibodies, available for the PD1–targeted phosphorylation sites identified here, pS39 and pS72 of vimentin and pS5 of LCP1. These revealed efficient induction of vimentin and LCP1 phosphorylation upon various conditions of primary human CD4 T-cell activation (supplemental Figure 2A). Although resting cells exhibited some constitutive vimentin phosphorylation at S39 and S72, phosphorylation at S39 was enhanced by stimulatory beads, while inhibitory beads induced a decrease of both, pS39 and pS72, compared with stimulatory beads (Figure 1C-D; supplemental Figure 2B-C). Moreover, we observed a strong PD1-mediated inhibition of LCP1 S5 phosphorylation in CD4 and CD8 T cells (Figure 1E-F). We confirmed our findings using PD1–transduced wild-type Jurkat T cells (Jurkat-PD1 WT), which were cocultured with mock-transduced or PDL1- or PDL2-transduced Raji cells, in the presence or absence of the superantigen SEE (supplemental Figure 3A-B).21,36 In this setting, PD1 engagement strongly decreased SEE-dependent IL-2 secretion (supplemental Figure 3C) and LCP1 S5 phosphorylation (Figure 1G-H). No such effect was seen on Jurkat cells lacking PD1 expression. Thus, PD1 triggering inhibits S/T phosphorylation of multiple cytoskeleton-related proteins in human primary T cells and Jurkat T cells.
PD1-induced inhibition of LCP1 phosphorylation depends on SHP1 and SHP2
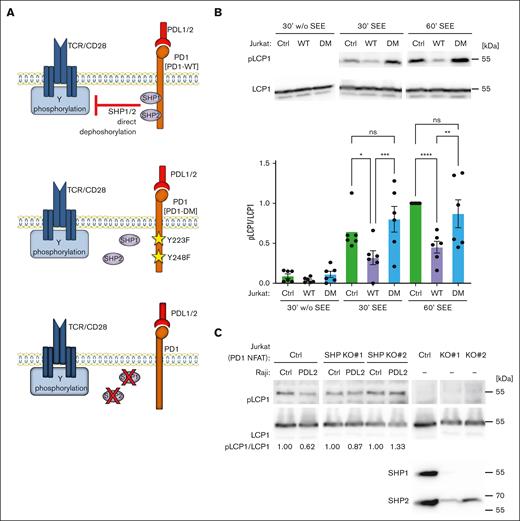
The tyrosine phosphatases SHP1 and SHP2 contribute to PD1–mediated T-cell inhibition, even if their individual contributions remain somewhat controversial.19-21,37-40 To assess if these phosphatases play a role in the PD1–induced dephosphorylation of cytoskeleton proteins, we developed 2 strategies (Figure 2A). First, we generated Jurkat cell lines expressing either PD1 WT or a PD1 double mutant (PD1 DM) for both SHP1 and SHP2 binding sites, Y223F and Y248F (Figure 2A, middle panel; supplemental Figure 3B). Second, we stably silenced SHP1 and SHP2 expression in a commercially available Jurkat cell line expressing PD1 and an NFAT reporter (Jurkat-PD1 NFAT cells) (Figure 2A, lower panel). For both strategies, we verified surface levels of PD1, CD3 and CD28 (supplemental Figures 3B and 4). Triggering of Jurkat cells with Raji PDL2 cells inhibited LCP1 S5 phosphorylation in Jurkat-PD1 WT, but not Jurkat-PD1 DM or Jurkat control (Ctrl) cells, suggesting that PD1-mediated recruitment of SHP1 and SHP2 was necessary for LCP1 dephosphorylation (Figure 2B). Similarly, CRISPR/Cas9–mediated silencing of both, SHP1 and SHP2 prevented PD1-induced dephosphorylation of LCP1 in Jurkat-PD1 NFAT cell lines (Figure 2C). Thus, PD1 triggering inhibits LCP1 phosphorylation in a SHP1/SHP2-dependent manner.
PD1-triggered inhibition of LCP1 phosphorylation is SHP1/SHP2-dependent. (A) Overview of the Jurkat cell lines used to assess the relevance of SHP1/2 phosphatases in the PD1-induced dephosphorylation of cytoskeleton proteins. Upper section: Ctrl cells expressing WT PD1. Middle section: Jurkat cells expressing PD1 DM, Y223F/Y248F, unable to bind SHP1/SHP2. Lower section: CRISPR/Cas9-mediated silencing of SHP1 and SHP2 in Jurkat-PD1-NFAT cells. (B) Upper panel: western blot monitoring phosphorylation of S5 of L-plastin (pLCP1) in the indicated cell lines. Jurkat-PD1 WT and Jurkat-PD1 DM cells were cocultured for the indicated times with PDL2–expressing Raji cells (Raji PDL2) in presence or absence of SEE at 6 ng/mL. Lower panel: quantification of experiments, pS5 LCP1 was normalized to total LCP1 for the 60-minute condition for 6 independent experiments. (C) Western blot monitoring phosphorylation of S5 of L-plastin (pLCP1) in 2 independent Jurkat-PD1-NFAT cell lines stably transduced with SHP1- and SHP2–specific CRISPR constructs and cocultured for 30 minutes with Raji Ctrl cells or Raji PDL2 in presence of SEE at 6 ng/mL. Representative data from 4 independent experiments.
PD1-triggered inhibition of LCP1 phosphorylation is SHP1/SHP2-dependent. (A) Overview of the Jurkat cell lines used to assess the relevance of SHP1/2 phosphatases in the PD1-induced dephosphorylation of cytoskeleton proteins. Upper section: Ctrl cells expressing WT PD1. Middle section: Jurkat cells expressing PD1 DM, Y223F/Y248F, unable to bind SHP1/SHP2. Lower section: CRISPR/Cas9-mediated silencing of SHP1 and SHP2 in Jurkat-PD1-NFAT cells. (B) Upper panel: western blot monitoring phosphorylation of S5 of L-plastin (pLCP1) in the indicated cell lines. Jurkat-PD1 WT and Jurkat-PD1 DM cells were cocultured for the indicated times with PDL2–expressing Raji cells (Raji PDL2) in presence or absence of SEE at 6 ng/mL. Lower panel: quantification of experiments, pS5 LCP1 was normalized to total LCP1 for the 60-minute condition for 6 independent experiments. (C) Western blot monitoring phosphorylation of S5 of L-plastin (pLCP1) in 2 independent Jurkat-PD1-NFAT cell lines stably transduced with SHP1- and SHP2–specific CRISPR constructs and cocultured for 30 minutes with Raji Ctrl cells or Raji PDL2 in presence of SEE at 6 ng/mL. Representative data from 4 independent experiments.
PD1 inhibits PKCθ-mediated binding and phosphorylation of cytoskeleton-related proteins
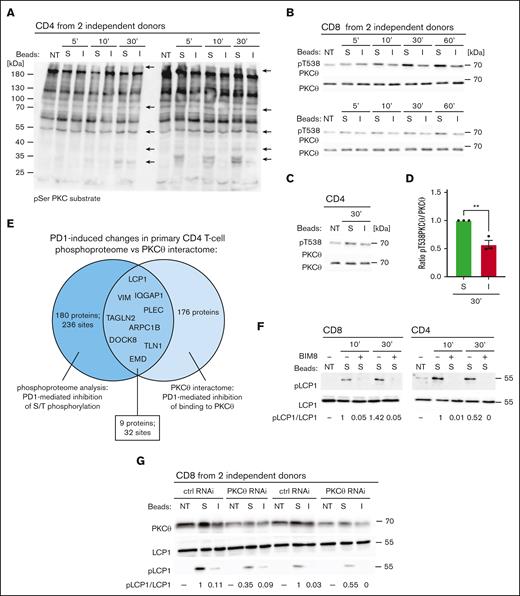
PKC family members regulate phosphorylation and activation of various cytoskeletal compounds,41-43 including vimentin.31 These findings, together with a previous report describing PD1-mediated inhibition of PKCθ threonine phosphorylation,24 allows us to hypothesize that PKCθ might be a major mediator of the observed PD1-induced reduction in the S/T phosphoproteome. We, thus, assessed the total phosphorylation levels of PKCθ substrates in primary CD4 T cells treated with stimulatory or inhibitory beads by Western blotting, using an anti-pS PKC substrate antibody (Figure 3A). This revealed a PD1-induced decrease of phosphorylation of multiple proteins. Next, we tested the status of PKCθ phosphorylation at T538, a surrogate marker for PKCθ kinase activation.44 In unstimulated T cells, we observed a basal level of T538 phosphorylation of PKCθ. Addition of stimulatory beads led to a further increase in phosphorylation for at least 60 minutes, and this was inhibited by PD1 triggering in both CD8 and CD4 T cells (Figure 3B-D). Next, we performed an MS-based analysis of PKCθ binding partners in CD4 T cells triggered with stimulatory or inhibitory beads for 10 minutes. PKCθ was immunoprecipitated (IP) with comparable efficiency in both conditions (supplemental Table 3). When using a cutoff of at least 4 spectral counts/protein in 1 of the conditions for each identified target, we identified 176 proteins, including 33 cytoskeleton-related proteins, whose interaction with PKCθ was severely diminished upon PD1 triggering (supplemental Table 3; cytoskeleton-related proteins). Among these were 9 proteins, including vimentin and LCP1, who showed a diminished interaction with PKCθ and a correlating decrease in S/T phosphorylation upon PD1 triggering (Figure 3E; supplemental Tables 1 and 3). Finally, we monitored the effects of pretreatment with the pan-PKC inhibitor BIM8 or of PKCθ silencing on the status of LCP1 pS5 in stimulated primary T cells. Pretreatment with BIM8 completely blocked stimulation–induced LCP1 S5 phosphorylation in CD4 and CD8 T cells (Figure 3F). Similarly, pretreatment of Jurkat-PD1 NFAT and Jurkat-PD1 WT cells with BIM8 or a PKCθ inhibitor blocked stimulation–induced LCP1 S5 phosphorylation (supplemental Figure 5). We also observed a marked decrease in LCP1 S5 phosphorylation in stimulated, PKCθ-silenced primary T cells cells compared with control cells (Figure 3G). Thus, LCP1 is a direct or indirect target of PKCθ-mediated phosphorylation.
PD1 triggering inhibits binding and phosphorylation of proteins by PKCθ. (A) Western blot monitoring phosphorylation of PKC substrates on serine residues embedded within the pentapeptide motif K/R-X-pS-hydrophobic aa-K/R in lysates of human primary CD4 T cells from 2 independent donors. Cells were left untreated (NT) or triggered for the indicated time points with stimulatory (S) or inhibitory (I) beads. Arrows indicate protein bands that show reduced phosphorylation upon treatment with inhibitory beads. (B,C) Western blot monitoring PKCθ phosphorylation at T538 and total PKCθ levels in lysates of human primary CD8 from 2 independent donors (B) and CD4 (C) T cells. Cells were left NT or triggered for the indicated time points with S or I beads. (D) Quantification of experiments performed as in panels B,C: pT538 PKCθ levels in human primary T cells were normalized to total PKCθ levels for the 30-minute condition for 3 independent experiments. Mean values ± SEM. Statistical analysis was performed with a 2-tailed, unpaired t test. ∗∗P < .01. (E) Venn diagram graphically representing results of the PKCθ interactome and phosphoproteome analyses: 171 proteins underwent a significant PD1-mediated reduction in serine or threonine phosphorylation (dark blue circle), 253 proteins bound less well or did not bind to PKCθ upon PD1 triggering (light blue circle). The overlapping section contains 9 cytoskeleton proteins that show a significant decrease in serine phosphorylation and decreased binding to PKCθ upon PD1 triggering compared with the stimulating condition. (F) Western blot monitoring the effect of pretreatment with the pan-PKC inhibitor BIM8 on the phosphorylation of S5 of L-plastin (pLCP1) in human primary CD4 and CD8 T cells. Cells were pretreated as indicated for 1 hour, followed by triggering with S beads for the indicated times. Two representative blots of 3 independent experiments are shown. Densitometry analysis: the relative ratio of pLCP1 to total LCP in the samples is indicated below. (G) Western blot monitoring phosphorylation of S5 of L-plastin (pLCP1) in human primary CD8 T cells from 2 independent donors, nucleoporated with PKCθ-specific or Ctrl siRNAs. Seventy-two hours after nucleoporation, cells were triggered for 30 minutes with S or I beads. The relative ratio of pLCP1 to total LCP in the samples is indicated below.
PD1 triggering inhibits binding and phosphorylation of proteins by PKCθ. (A) Western blot monitoring phosphorylation of PKC substrates on serine residues embedded within the pentapeptide motif K/R-X-pS-hydrophobic aa-K/R in lysates of human primary CD4 T cells from 2 independent donors. Cells were left untreated (NT) or triggered for the indicated time points with stimulatory (S) or inhibitory (I) beads. Arrows indicate protein bands that show reduced phosphorylation upon treatment with inhibitory beads. (B,C) Western blot monitoring PKCθ phosphorylation at T538 and total PKCθ levels in lysates of human primary CD8 from 2 independent donors (B) and CD4 (C) T cells. Cells were left NT or triggered for the indicated time points with S or I beads. (D) Quantification of experiments performed as in panels B,C: pT538 PKCθ levels in human primary T cells were normalized to total PKCθ levels for the 30-minute condition for 3 independent experiments. Mean values ± SEM. Statistical analysis was performed with a 2-tailed, unpaired t test. ∗∗P < .01. (E) Venn diagram graphically representing results of the PKCθ interactome and phosphoproteome analyses: 171 proteins underwent a significant PD1-mediated reduction in serine or threonine phosphorylation (dark blue circle), 253 proteins bound less well or did not bind to PKCθ upon PD1 triggering (light blue circle). The overlapping section contains 9 cytoskeleton proteins that show a significant decrease in serine phosphorylation and decreased binding to PKCθ upon PD1 triggering compared with the stimulating condition. (F) Western blot monitoring the effect of pretreatment with the pan-PKC inhibitor BIM8 on the phosphorylation of S5 of L-plastin (pLCP1) in human primary CD4 and CD8 T cells. Cells were pretreated as indicated for 1 hour, followed by triggering with S beads for the indicated times. Two representative blots of 3 independent experiments are shown. Densitometry analysis: the relative ratio of pLCP1 to total LCP in the samples is indicated below. (G) Western blot monitoring phosphorylation of S5 of L-plastin (pLCP1) in human primary CD8 T cells from 2 independent donors, nucleoporated with PKCθ-specific or Ctrl siRNAs. Seventy-two hours after nucleoporation, cells were triggered for 30 minutes with S or I beads. The relative ratio of pLCP1 to total LCP in the samples is indicated below.
Collectively, these findings suggest that PD1 triggering inhibits the activation of PKCθ, as well as the PKCθ-dependent physical recruitment and phosphorylation of cytoskeleton-related proteins, including vimentin and LCP1.
PD1 inhibits PKCθ localization to the forming IS
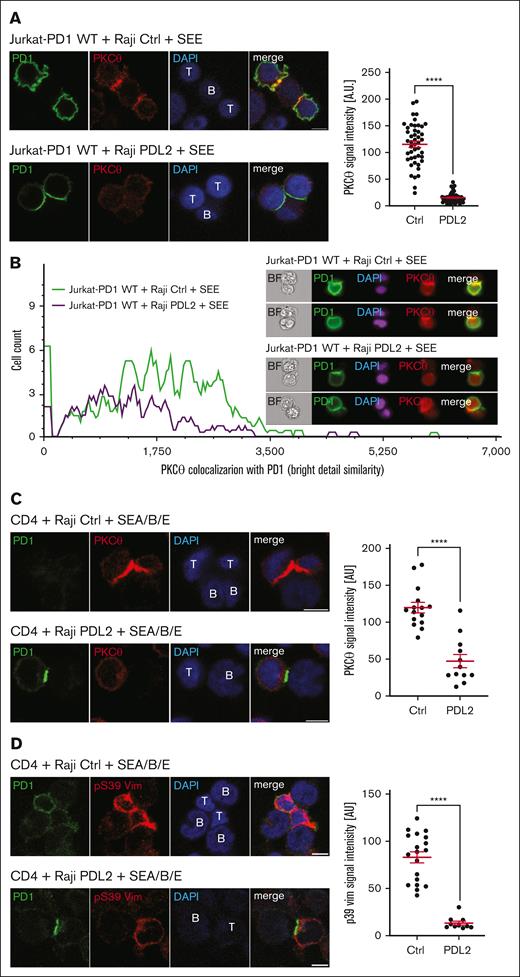
Next, we investigated the consequences of PD1 triggering on the known IS recruitment of PKCθ.45 We observed a clear translocation of PKCθ to the forming IS in Jurkat-PD1 WT cells upon coculture with Raji Ctrl cells (Figure 4A, upper panels), but not with Raji PDL2 cells, where PKCθ remained dispersed, with little or no enrichment at the IS (Figure 4A, lower panels). Importantly, PD1 triggering also led to strong accumulation of PD1 at the contact zone, as described.19 We next quantified the effect of PD1 on PKCθ recruitment to the IS using ImageStream (Figure 4B) gating on clusters of at least 2 cells, including 1 positive for PD1 staining (supplemental Figure 6). Quantification of the PKCθ and PD1 signal overlap within the synapse region confirmed that the PKCθ signal was reduced when Jurkat-PD1 cells were triggered with PDL2–expressing Raji cells (Raji PDL2), whereas PD1 accumulated within the IS (Figure 4B).
PD1 triggering affects PKCθ recruitment and vimentin phosphorylation at the IS. (A) Left: immunofluorescent labeling of PD1 and PKCθ upon 60 minutes of coculture of Jurkat-PD1 WT cells with Raji Ctrl cells (upper panel) or PDL2 expressing Raji cells (lower panel), preincubated with 6 ng/mL of SEE and mixed with Jurkat-PD1 WT cells at a 1:1 ratio. Right: quantification of the PKCθ signal intensity in Jurkat cells within proximity of the T and B cell interface from 1 experiment. Representative results of 4 independent experiments are shown. Each dot represents a value from 1 Jurkat cell, 46 and 56 cells were quantified, respectively. (B) ImageStream-based quantification of the colocalization between PD1 and PKCθ in Jurkat-PD1 WT cells cocultured for 60 minutes with Raji Ctrl or Raji PDL2 cells, preincubated with 6 ng/mL of SEE and mixed with Jurkat-PD1 WT cells at a 1:1 ratio. Upon fixation and permeabilization, samples were stained for PKCθ, PD1 and nuclei (DAPI) and analyzed by ImageStream flow cytometry. Left: histogram measuring the extent of signal overlap between PKCθ and PD1, with PD1 signal in the IS used as a mask; right: examples of images acquired. Representative results of 2 independent experiments are shown. (C) Left: immunofluorescent labeling of PD1 and PKCθ upon 30 minutes of coculture of human primary CD4 T cells with Raji Ctrl cells (upper panel) or Raji PDL2 cells (lower panel). Raji cells were preincubated with 1 ng/mL of SEA/SEB/SEE and mixed with CD4 cells at a 1:2 (Raji:T cells) ratio. Images shown are from a single Z plane, which likely accounts for the concentration of staining in the T cell in the absence of PDL2 ligation, whereas the signal is weaker in the Raji PDL2 condition (bottom panel). Right: quantification of the PKCθ signal intensity in T cells within proximity of the T cell–B cell interface from 1 experiment. Representative results of 3 independent experiments are shown. Each dot represents a value from 1 primary T cell, 15 and 12 cells were quantified, respectively. (D) Left: immunofluorescent labeling of PD1 and pS39 Vim upon 60 minutes of coculture of human primary CD4 T cells with Raji Ctrl cells (upper panel) or Raji PDL2 cells (lower panel). Raji cells were preincubated with 6 ng/mL of SEE and mixed with Jurkat-PD1 WT cells at a 1:2 (Raji:T cells) ratio. Right: quantification of the total pVim39 signal intensity in T cells from 1 experiment. Representative results of 3 independent experiments are shown. Each dot represents a value from 1 primary T cell, 19 and 10 cells were quantified, respectively. Scale bar: 5 μm (panels A,C-D). Graphs in panels A,C-D show mean value ± SEM. Statistical analysis was performed using a 2-tailed, unpaired t-test. ∗∗∗∗P < .0001.
PD1 triggering affects PKCθ recruitment and vimentin phosphorylation at the IS. (A) Left: immunofluorescent labeling of PD1 and PKCθ upon 60 minutes of coculture of Jurkat-PD1 WT cells with Raji Ctrl cells (upper panel) or PDL2 expressing Raji cells (lower panel), preincubated with 6 ng/mL of SEE and mixed with Jurkat-PD1 WT cells at a 1:1 ratio. Right: quantification of the PKCθ signal intensity in Jurkat cells within proximity of the T and B cell interface from 1 experiment. Representative results of 4 independent experiments are shown. Each dot represents a value from 1 Jurkat cell, 46 and 56 cells were quantified, respectively. (B) ImageStream-based quantification of the colocalization between PD1 and PKCθ in Jurkat-PD1 WT cells cocultured for 60 minutes with Raji Ctrl or Raji PDL2 cells, preincubated with 6 ng/mL of SEE and mixed with Jurkat-PD1 WT cells at a 1:1 ratio. Upon fixation and permeabilization, samples were stained for PKCθ, PD1 and nuclei (DAPI) and analyzed by ImageStream flow cytometry. Left: histogram measuring the extent of signal overlap between PKCθ and PD1, with PD1 signal in the IS used as a mask; right: examples of images acquired. Representative results of 2 independent experiments are shown. (C) Left: immunofluorescent labeling of PD1 and PKCθ upon 30 minutes of coculture of human primary CD4 T cells with Raji Ctrl cells (upper panel) or Raji PDL2 cells (lower panel). Raji cells were preincubated with 1 ng/mL of SEA/SEB/SEE and mixed with CD4 cells at a 1:2 (Raji:T cells) ratio. Images shown are from a single Z plane, which likely accounts for the concentration of staining in the T cell in the absence of PDL2 ligation, whereas the signal is weaker in the Raji PDL2 condition (bottom panel). Right: quantification of the PKCθ signal intensity in T cells within proximity of the T cell–B cell interface from 1 experiment. Representative results of 3 independent experiments are shown. Each dot represents a value from 1 primary T cell, 15 and 12 cells were quantified, respectively. (D) Left: immunofluorescent labeling of PD1 and pS39 Vim upon 60 minutes of coculture of human primary CD4 T cells with Raji Ctrl cells (upper panel) or Raji PDL2 cells (lower panel). Raji cells were preincubated with 6 ng/mL of SEE and mixed with Jurkat-PD1 WT cells at a 1:2 (Raji:T cells) ratio. Right: quantification of the total pVim39 signal intensity in T cells from 1 experiment. Representative results of 3 independent experiments are shown. Each dot represents a value from 1 primary T cell, 19 and 10 cells were quantified, respectively. Scale bar: 5 μm (panels A,C-D). Graphs in panels A,C-D show mean value ± SEM. Statistical analysis was performed using a 2-tailed, unpaired t-test. ∗∗∗∗P < .0001.
We also assessed the effect of PD1 engagement on the subcellular localization of PD1 and PKCθ in primary human CD4 T cells upon coculture with Raji Ctrl vs Raji PDL2 cells presenting the superantigens SEA, SEB, and SEE. Coculture with Raji PDL2 cells led to a reduced IL-2 response in this system (supplemental Figure 7) and resulted in accumulation of PD1, but not of PKCθ at the IS (Figure 4C). Moreover, superantigen stimulation induced a pS39 vimentin signal at the IS, which was almost totally abrogated by PD1 engagement (Figure 4D). ImageStream analysis confirmed PD1-induced inhibition of Vimentin phosphorylation (supplemental Figure 8). Thus, PD1 triggering leads to enrichment of PD1 at the IS, and prevents PKCθ recruitment and substrate phosphorylation at the IS.
PD1 inhibits IS recruitment and Y90 phosphorylation of PKCθ in a SHP1/2-dependent manner
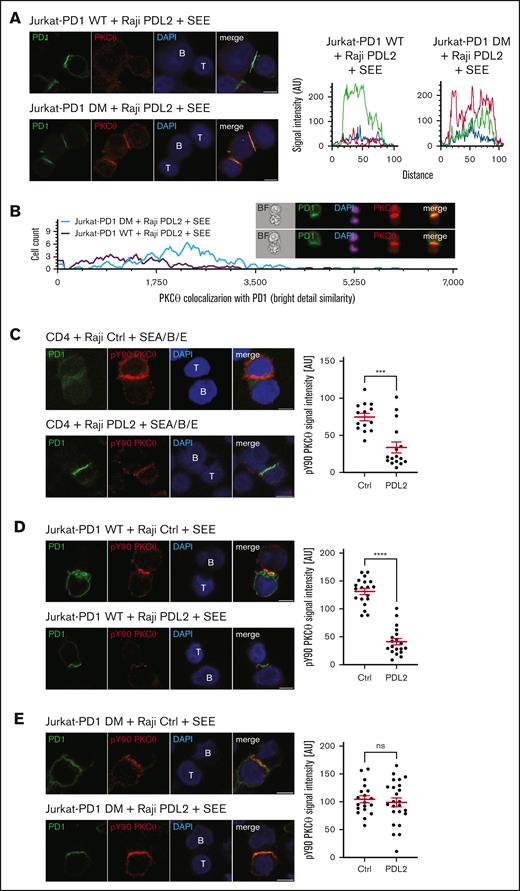
To assess the relevance of PD1–induced SHP1/2 activation for IS recruitment of PKCθ, we compared the pattern of PKCθ localization in Jurkat cells expressing WT PD1 or its SHP1/SHP2 binding–deficient DM version, upon triggering with Raji PDL2 cells (Figure 5A). In Jurkat-PD1 WT cells, PKCθ was dispersed throughout the cell without any clear enrichment pattern, as observed before (Figure 4A). In contrast, in Jurkat-PD1 DM cells triggered with Raji PDL2 cells, PKCθ was concentrated within the Jurkat-Raji interface and colocalized with PD1 DM, which still enriched within the same interface (Figure 5A, RGB profile histogram). Triggering of Jurkat-PD1 WT or DM cells in the absence of PD1 engagement resulted in comparable enrichment of PKCθ within the IS (supplemental Figure 9).
PD1-induced inhibition of PKCθ localization to the IS is SHP1/SHP2-dependent. (A) Left: immunofluorescent labeling of PD1 and PKCθ of Jurkat-PD1 WT cells (upper panel) and Jurkat-PD1 DM cells (lower panel) upon 60 minute of coculture with Raji PDL2 cells, preincubated with 6 ng/mL of SEE and mixed with Jurkat cells at a 1:1 ratio. Right: representative histogram of the colocalization profile prepared with RGB profiler plugin of ImageJ. Representative images of 3 independent experiments are shown. (B) ImageStream-based quantification of the colocalization between PD1 and PKCθ in Jurkat-PD1 WT cells and Jurkat-PD1 DM cells cocultured for 60 minutes with Raji PDL2 cells. Raji PDL2 cells were preincubated with 6 ng/mL of SEE and mixed with Jurkat cells at 1:1 ratio. Upon fixation and permeabilization, samples were stained for PKCθ, PD1, and nuclei (DAPI) and analyzed by Amnis Image Stream imaging flow cytometry. Left: histogram measuring the extent of signal overlap between PKCθ and PD1 with PD1 signal in the IS used as a mask; right: examples of images acquired. Representative results of 2 independent experiments are shown. (C) Left: immunofluorescent labeling of PD1 and pY90 PKCθ of human primary CD4 T cells upon 60 minute of coculture with Raji Ctrl cells (upper panel) or Raji PDL2 cells (lower panel). Raji cells were preincubated with 1 ng/mL of SEA/SEB/SEE and mixed with CD4 cells at a 1:2 (Raji:T cells) ratio. Right panel: quantification of total pY90 PKCθ signal intensity in T cells from 1 experiment. Representative results of 3 independent experiments are shown. Each dot represents the value from 1 primary T cell, 14 and 16 cells were quantified, respectively. (D) Left: immunofluorescent labeling of PD1 and pY90 PKCθ of Jurkat-PD1 WT cells upon 30 minutes of coculture with Raji Ctrl cells (upper panel) or Raji PDL2 cells (lower panel). Raji cells were preincubated with 6 ng/mL of SEE and mixed with Jurkat-PD1 WT cells at 1:1 ratio. Right panel: quantification of total pY90 PKCθ signal intensity in Jurkat-PD1 WT cells. Representative results of 3 independent experiments are shown. Each dot represents a value from 1 Jurkat cell, 19 and 19 cells were quantified, respectively. (E) Left: immunofluorescent labeling of PD1 and pY90 PKCθ of Jurkat-PD1 DM cells upon 60 minute of coculture with Raji Ctrl cells (upper panel) or Raji PDL2 cells (lower panel). Raji cells were preincubated with 6 ng/mL of SEE and mixed with Jurkat-PD1 DM cells at 1:1 ratio. Right panel: quantification of total pY90 PKCθ signal intensity in Jurkat-PD1 DM cells. Representative results of 3 independent experiments are shown. Each dot represents a value from 1 Jurkat cell, 19 and 24 cells were quantified, respectively. Scale bar, 5 μm. Graphs in panels C-E show mean value ± SEM. Statistical analysis was performed using a 2-tailed, unpaired t test. ∗∗∗∗P < .001; ∗∗∗P < .001; ns, not significant.
PD1-induced inhibition of PKCθ localization to the IS is SHP1/SHP2-dependent. (A) Left: immunofluorescent labeling of PD1 and PKCθ of Jurkat-PD1 WT cells (upper panel) and Jurkat-PD1 DM cells (lower panel) upon 60 minute of coculture with Raji PDL2 cells, preincubated with 6 ng/mL of SEE and mixed with Jurkat cells at a 1:1 ratio. Right: representative histogram of the colocalization profile prepared with RGB profiler plugin of ImageJ. Representative images of 3 independent experiments are shown. (B) ImageStream-based quantification of the colocalization between PD1 and PKCθ in Jurkat-PD1 WT cells and Jurkat-PD1 DM cells cocultured for 60 minutes with Raji PDL2 cells. Raji PDL2 cells were preincubated with 6 ng/mL of SEE and mixed with Jurkat cells at 1:1 ratio. Upon fixation and permeabilization, samples were stained for PKCθ, PD1, and nuclei (DAPI) and analyzed by Amnis Image Stream imaging flow cytometry. Left: histogram measuring the extent of signal overlap between PKCθ and PD1 with PD1 signal in the IS used as a mask; right: examples of images acquired. Representative results of 2 independent experiments are shown. (C) Left: immunofluorescent labeling of PD1 and pY90 PKCθ of human primary CD4 T cells upon 60 minute of coculture with Raji Ctrl cells (upper panel) or Raji PDL2 cells (lower panel). Raji cells were preincubated with 1 ng/mL of SEA/SEB/SEE and mixed with CD4 cells at a 1:2 (Raji:T cells) ratio. Right panel: quantification of total pY90 PKCθ signal intensity in T cells from 1 experiment. Representative results of 3 independent experiments are shown. Each dot represents the value from 1 primary T cell, 14 and 16 cells were quantified, respectively. (D) Left: immunofluorescent labeling of PD1 and pY90 PKCθ of Jurkat-PD1 WT cells upon 30 minutes of coculture with Raji Ctrl cells (upper panel) or Raji PDL2 cells (lower panel). Raji cells were preincubated with 6 ng/mL of SEE and mixed with Jurkat-PD1 WT cells at 1:1 ratio. Right panel: quantification of total pY90 PKCθ signal intensity in Jurkat-PD1 WT cells. Representative results of 3 independent experiments are shown. Each dot represents a value from 1 Jurkat cell, 19 and 19 cells were quantified, respectively. (E) Left: immunofluorescent labeling of PD1 and pY90 PKCθ of Jurkat-PD1 DM cells upon 60 minute of coculture with Raji Ctrl cells (upper panel) or Raji PDL2 cells (lower panel). Raji cells were preincubated with 6 ng/mL of SEE and mixed with Jurkat-PD1 DM cells at 1:1 ratio. Right panel: quantification of total pY90 PKCθ signal intensity in Jurkat-PD1 DM cells. Representative results of 3 independent experiments are shown. Each dot represents a value from 1 Jurkat cell, 19 and 24 cells were quantified, respectively. Scale bar, 5 μm. Graphs in panels C-E show mean value ± SEM. Statistical analysis was performed using a 2-tailed, unpaired t test. ∗∗∗∗P < .001; ∗∗∗P < .001; ns, not significant.
We also assessed the overlap of PKCθ with PD1 signals in the IS using ImageStream (Figure 5B). As evident from the histogram analysis, the intensity of the PKCθ signal colocalized with PD1 was considerably higher in Jurkat-PD1 DM cells than in Jurkat-PD1 WT cells upon triggering with Raji-PDL2 cells (Figure 5B, plot for Jurkat-PD1 WT cells is the same as shown in Figure 4B). Thus, signaling through the PD1 SHP1/SHP2 binding sites is required to inhibit recruitment of PKCθ to the IS.
As Lck-mediated phosphorylation of Y90 of PKCθ is required for its targeting to the IS,46,47 we next monitored its phosphorylation status under stimulatory vs inhibitory conditions using a PCKθ pY90-specific antibody. Superantigen-mediated stimulation of CD4 T cells with Raji Ctrl cells led to a strong, IS-localized pY90 PKCθ signal (Figure 5C), which was greatly reduced upon PD1 engagement. Moreover, PKCθ Y90 phosphorylation was inhibited by WT PD1, but not by PD1 DM (Figure 5D-E). Indeed, for Jurkat-PD1 DM cells, the localization and intensity of PKCθ Y90 phosphorylation was comparable between stimulatory and inhibitory conditions (Figure 5E). Altogether, these data suggest that the initial, PD1- and SHP1/SHP2-dependent Y90 dephosphorylation of PKCθ represents a crucial link to the subsequent alterations in S/T-phosphorylation events at the IS.
PD1 engagement inhibits formation of the IS
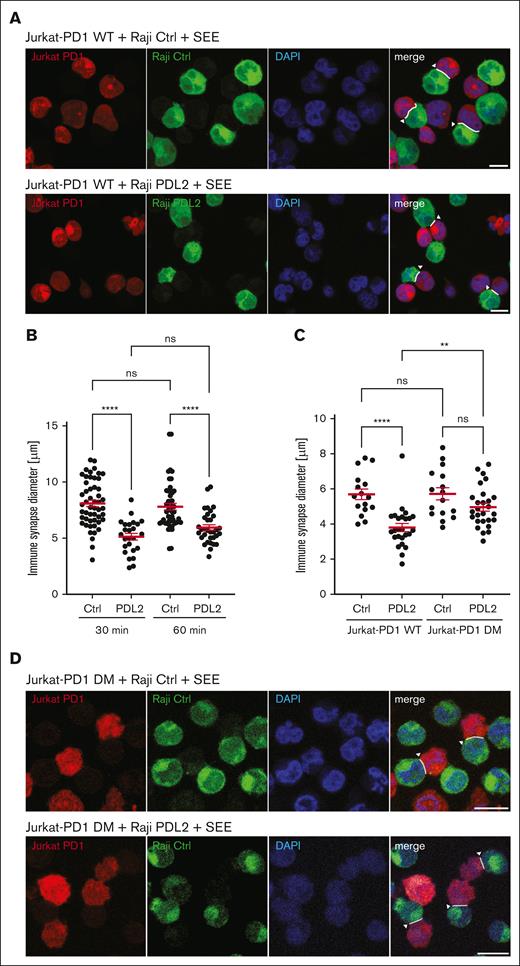
We reasoned that the observed PD1-mediated inhibition of localized phosphorylation of cytoskeleton proteins should alter cytoskeleton plasticity and translate to an inhibition of the formation of the IS. Therefore, we measured the size of IS upon coculture of Jurkat-PD1-NFAT cells with Raji Ctrl or Raji PDL2 cells for 30 or 60 minutes. PD1 triggering with PDL2 led to formation of IS that were significantly reduced in size (Figure 6A-B). Importantly, no significant effect on IS size was observed when Jurkat-PD1 DM cells were triggered with Raji PDL2 cells (Figure 6C-D). We also assessed whether PD1 engagement affected the frequency of conjugate formation between Jurkat and SEE–loaded Raji cells by flow cytometry (data not shown). However, the low concentration of SEE required to allow efficient PD1–mediated T-cell inhibition was not sufficient to trigger robust formation of conjugates stable enough for analysis by flow cytometry. Together with our biochemical data, these findings suggest that PD1 inhibits T-cell activation, at least in part, by preventing the synaptic translocation and activation of PKCθ and its targeting of cytoskeleton substrates required for the extension of the size of the IS.
PD1 triggering decreases IS size through inhibition of PKCθ recruitment. (A) Assessment of the effect of PD1 triggering on the size (maximal diameter of the contact zone) of the IS. Jurkat-PD1-NFAT cells (labeled with CMTMR) were incubated with Raji Ctrl and Raji PDL2 cells (labeled with CFSE in presence of 6 ng/mL SEE), at a 1:1 cell ratio for 30 minutes at 37°C to facilitate IS formation. Image data from representative experiment, highlighting quantified IS by white arrowheads. Representative images of 2 independent experiments are shown. (B-C) Quantification of the size of the IS formed as described in panel A, using Jurkat-PD1-WT or DM cells (B) or Jurkat-PD1-WT cells, at different time points of incubation with Raji Ctrl and Raji PDL2 cells (C). Representative results of 2 and 3 independent experiments, respectively, are shown. Each dot represents a value from 1 Jurkat cell, (B) 51, 26, 50 and 33 cells; (C)16, 25, 16 and 27 cells were quantified, respectively. (D) Assessment of the effect of PD1 triggering on the size (maximal diameter of the contact zone) of the IS. Jurkat-PD1-WT and Jurkat-PD1-DM cells (labeled with CMTMR) were incubated with Raji PDL2 cells (labeled with CFSE in presence of 6 ng/mL SEE), at a 1:1 cell ratio for 30 minutes at 37°C to facilitate IS formation. Image data from representative experiments, highlighting quantified IS by white arrowheads. Representative images of 3 independent experiments are shown. Scale bar, 10 μm. Graphs in panels B-C show mean value ± SEM. Statistical analysis was performed using 1-way ANOVA with Šídák’s multiple comparisons test. ∗∗∗∗P < .0001; ∗∗P < .01; ns, not significant.
PD1 triggering decreases IS size through inhibition of PKCθ recruitment. (A) Assessment of the effect of PD1 triggering on the size (maximal diameter of the contact zone) of the IS. Jurkat-PD1-NFAT cells (labeled with CMTMR) were incubated with Raji Ctrl and Raji PDL2 cells (labeled with CFSE in presence of 6 ng/mL SEE), at a 1:1 cell ratio for 30 minutes at 37°C to facilitate IS formation. Image data from representative experiment, highlighting quantified IS by white arrowheads. Representative images of 2 independent experiments are shown. (B-C) Quantification of the size of the IS formed as described in panel A, using Jurkat-PD1-WT or DM cells (B) or Jurkat-PD1-WT cells, at different time points of incubation with Raji Ctrl and Raji PDL2 cells (C). Representative results of 2 and 3 independent experiments, respectively, are shown. Each dot represents a value from 1 Jurkat cell, (B) 51, 26, 50 and 33 cells; (C)16, 25, 16 and 27 cells were quantified, respectively. (D) Assessment of the effect of PD1 triggering on the size (maximal diameter of the contact zone) of the IS. Jurkat-PD1-WT and Jurkat-PD1-DM cells (labeled with CMTMR) were incubated with Raji PDL2 cells (labeled with CFSE in presence of 6 ng/mL SEE), at a 1:1 cell ratio for 30 minutes at 37°C to facilitate IS formation. Image data from representative experiments, highlighting quantified IS by white arrowheads. Representative images of 3 independent experiments are shown. Scale bar, 10 μm. Graphs in panels B-C show mean value ± SEM. Statistical analysis was performed using 1-way ANOVA with Šídák’s multiple comparisons test. ∗∗∗∗P < .0001; ∗∗P < .01; ns, not significant.
Presence of PDL1 negatively correlates with vimentin phosphorylation in T cells in human lung adenocarcinoma
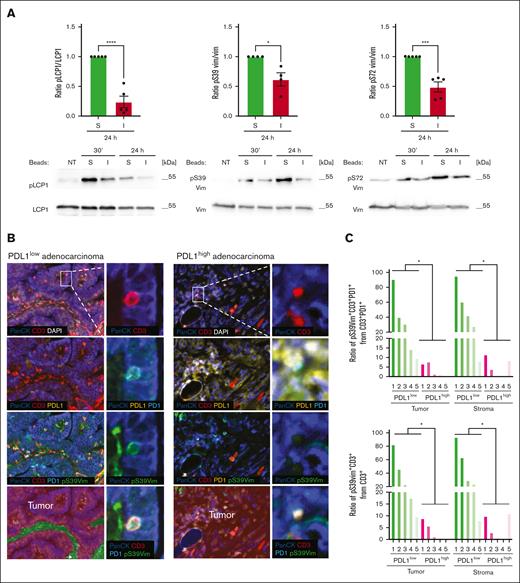
While most of the described PD1-induced changes in T-cell signaling are phospho-tyrosine-based and thus rather short lasting,21 the stimulation-induced phosphorylation of LCP1 and vimentin and their PD1-mediated dephosphorylation were still detectable 24 hours after T-cell triggering (Figure 7A). This prompted us to investigate a potential inverse correlation between vimentin phosphorylation in T cells and PDL1 expression on cancer cells in lung adenocarcinoma—a subtype of NSCLC responsive to targeting the PD1-PDL1 axis.48,49 We stained a total of 10 human biopsy samples, characterized by either low or high PDL1 expression in cancer cells, for PanCK (present in tumor cells), CD3, PD1 and vimentin pS39. In PDL1low tumor samples, a significant proportion of the CD3+PD1+ T cells stained positive for vimentin pS39. In contrast, this proportion was low or undetectable in PDL1high tumors (Figure 7B-C, left panel). Similar observations were made when all CD3+ T cells were analyzed (Figure 7C, right panel). For overall numbers of CD3+ or CD3+PD1+ cells in the tumor or stroma regions, we did not notice a consistent correlation of T-cell numbers with PDL1 expression in the tumors (supplemental Figure 10). These data suggest that low or absent pSer39 vimentin staining might provide a useful readout to monitor functional PD1 engagement in tumor-infiltrating T cells.
PD1 induces long-lasting dephosphorylation of cytoskeleton proteins in vitro and in human lung adenocarcinoma tissues. (A) Western blot analysis monitoring phosphorylation of S5 of L-plastin (pS5 LCP1), S39 of vimentin (pS39 Vim), and S72 of vimentin (pS72 Vim) in human primary CD4 T cells. Cells were left untreated (NT) or triggered for the indicated time points with stimulatory (S) or inhibitory (I) beads. Quantification of the signal at 24 hours was performed by normalization of the phosphorylation signal to the signal of nonphosphorylated total proteins and calculated for 4 (pS39 Vim) and 5 independent experiments (pS5 LCP1 and pS72 Vim). Mean value ± SEM. Statistical analysis was performed using a 2-tailed, unpaired t test. ∗P < .05; ∗∗∗P < .001; ∗∗∗∗P < .0001. (B) Immunohistochemistry staining of human lung adenocarcinoma tissue samples characterized by low or high PDL1 expression. Samples where fluorescently labeled for PanCK, PDL1, PD1, CD3 and pS39 Vim. Nuclei were visualized with DAPI. Insets show intratumoral CD3+ lymphocytes, positive for PD1, coexpressing pS39 Vim in the case of PDL1low adenocarcinoma, and negative for pS39 Vim in PDL1high adenocarcinoma. Images were acquired using Vectra Polaris imaging system. Images were segmented into “tumor” or “stroma” sections using the inForm software and based on PanCK and DAPI signals. (C) Quantification of the ratio of pS39Vim+ cells amongst CD3+PD1+ cells (left panel) or CD3+ cells (right panel) in tumor and stroma regions performed in 5 cases of PDL1low and in 5 cases of PDL1high human lung adenocarcinomas. Number of imaging fields collected per each case for PDL1low: 1-21; 2-16; 3-19; case 4-15; case 5-18. Number of imaging fields collected per each case for PDL1high: 1-19; 2-22; 3-13; case 4-20; case 5-11. Number of tumor cells analysed per each case for PDL1low: 1-30 853; 2-81 983; 3-60 988; 4-14 298; 5- 35 166. Number of tumor cells analysed per each case for PDL1high: 1-5 900; 2-89 931; 3-27 363; 4-89 448; 5-20 981. Number of stroma cells analysed per each case for PDL1low: 1-39 006; 2-31 193; 3-19 132; 4-32 378; 5-36 296. Number of stroma cells analysed per each case for PDL1high: 1-112 832; 2-15 749; 3-10 887; 4-2; 5-51 525. Statistical analysis was performed using a 2-tailed, unpaired t test. ∗P < .05.
PD1 induces long-lasting dephosphorylation of cytoskeleton proteins in vitro and in human lung adenocarcinoma tissues. (A) Western blot analysis monitoring phosphorylation of S5 of L-plastin (pS5 LCP1), S39 of vimentin (pS39 Vim), and S72 of vimentin (pS72 Vim) in human primary CD4 T cells. Cells were left untreated (NT) or triggered for the indicated time points with stimulatory (S) or inhibitory (I) beads. Quantification of the signal at 24 hours was performed by normalization of the phosphorylation signal to the signal of nonphosphorylated total proteins and calculated for 4 (pS39 Vim) and 5 independent experiments (pS5 LCP1 and pS72 Vim). Mean value ± SEM. Statistical analysis was performed using a 2-tailed, unpaired t test. ∗P < .05; ∗∗∗P < .001; ∗∗∗∗P < .0001. (B) Immunohistochemistry staining of human lung adenocarcinoma tissue samples characterized by low or high PDL1 expression. Samples where fluorescently labeled for PanCK, PDL1, PD1, CD3 and pS39 Vim. Nuclei were visualized with DAPI. Insets show intratumoral CD3+ lymphocytes, positive for PD1, coexpressing pS39 Vim in the case of PDL1low adenocarcinoma, and negative for pS39 Vim in PDL1high adenocarcinoma. Images were acquired using Vectra Polaris imaging system. Images were segmented into “tumor” or “stroma” sections using the inForm software and based on PanCK and DAPI signals. (C) Quantification of the ratio of pS39Vim+ cells amongst CD3+PD1+ cells (left panel) or CD3+ cells (right panel) in tumor and stroma regions performed in 5 cases of PDL1low and in 5 cases of PDL1high human lung adenocarcinomas. Number of imaging fields collected per each case for PDL1low: 1-21; 2-16; 3-19; case 4-15; case 5-18. Number of imaging fields collected per each case for PDL1high: 1-19; 2-22; 3-13; case 4-20; case 5-11. Number of tumor cells analysed per each case for PDL1low: 1-30 853; 2-81 983; 3-60 988; 4-14 298; 5- 35 166. Number of tumor cells analysed per each case for PDL1high: 1-5 900; 2-89 931; 3-27 363; 4-89 448; 5-20 981. Number of stroma cells analysed per each case for PDL1low: 1-39 006; 2-31 193; 3-19 132; 4-32 378; 5-36 296. Number of stroma cells analysed per each case for PDL1high: 1-112 832; 2-15 749; 3-10 887; 4-2; 5-51 525. Statistical analysis was performed using a 2-tailed, unpaired t test. ∗P < .05.
Discussion
Here, we provide several lines of evidence for a crucial role of PD1–mediated PKCθ inhibition in the control of TCR/CD28-mediated IS formation and T-cell activation. We identified decreased phosphorylation of multiple cytoskeleton-related proteins as a major outcome of PD1–induced T-cell inhibition. Second, we identified PKCθ as the PD1 target and master kinase required for the recruitment and phosphorylation of 9 of these proteins. Third, we provided a link between transient PD1-induced changes in the Y-phosphoproteome and long-term changes in S/T-phosphorylation of cytoskeleton-related proteins and IS formation. Finally, we propose PD1-controlled phosphorylation of vimentin as a potential marker for assessing T-cell effector functions in human lung cancer samples.
Importantly, our discoveries are based on an unbiased phospho-proteomics approach, performed on human primary CD4 T cells in the background of suboptimal CD3/CD28 stimulation, which likely allowed for optimal detection of PD1–mediated inhibitory effects. Inclusion of CD28 triggering was paramount for our study, because CD28 signaling is required for full T-cell activation, including PKCθ activation and actin cytoskeleton remodeling.50 In addition, CD28 signaling is important for both, PD1–induced T-cell inhibition and effective rescue of T-cell exhaustion via PD1-targeting therapies.21,51,52 Our study, therefore, provided mechanistic insights into PKCθ-dependent cytoskeleton changes that go beyond previous phospho-proteomics studies, which used Jurkat cells stimulated with anti-CD3 alone, likely not engaging PKCθ efficiently.53 The latter revealed mainly CD3-driven alterations in tyrosine phosphorylation of cytoskeleton proteins and no obvious effects of PD1 on an in silico predicted PKCθ substrate profile.
We also identified a requirement for SHP1 and SHP2 in the PD1-mediated inhibition of Y-phosphorylation and IS recruitment of PKCθ, and the phosphorylation of the PKCθ substrate LCP1. These findings support a model in which PD1 inhibits cytoskeleton rearrangements, directly or indirectly, by SHP1/SHP2-dependent dephosphorylation of PKCθ and its cytoskeleton targets (supplemental Figure 11). Of note, PD1 accumulation at the IS did not depend on its immunoreceptor tyrosine-based inhibitory motif and immunoreceptor tyrosine-based switch motif, it must, thus, rely on signaling events that are independent of SHP1/SHP2 binding. The latter may indeed be responsible for the recently reported, SHP1/2-independent effects of PD1 on the actin cytoskeleton,54 which likely synergizes with the PD1-mediated, SHP1/2-and PKCθ-dependent inhibition of IS formation that we report here.
Among the cytoskeleton proteins whose serine phosphorylation was decreased upon PD1 engagement, we validated LCP1 S5, vimentin S39 and S72 using commercial antibodies. The actin-bundling protein LCP1 is known to be phosphorylated on S5 in activated T cells, which affects the formation of high affinity clusters of the adhesion molecule LFA1 within the IS.34 Our data suggest a role for PKCθ in LCP1 phosphorylation and identify S5 of LCP1 as a target of PD1-mediated inhibition. Vimentin has been previously proposed to be a substrate and binding partner of PKCθ. Indeed, both proteins colocalize in a molecular superstructure formed at the distal pole of regulatory T cells, opposite the IS, likely as a mechanism to augment the suppressive activity of regulatory T cells.31 Our data extend these findings by identifying vimentin as a PKCθ binding partner and PD1 target in the IS of effector T cells. Moreover, we identified the PKCθ-dependent vimentin phosphorylation sites S39 and S72 and provided proof-of-concept evidence for vimentin S39 phosphorylation as a biomarker indicating functional engagement of PD1 in tumor and stroma samples of patients with NSCLC. Although several lines of evidence support a correlation between PDL1 expression on tumors and response to therapy,55-57 not all PDL1 positive tumors respond to targeting the PD1/PDL1 axis. PDL1 expression can be induced through, tumor-intrinsic oncogenic pathways58,59; it is not necessarily indicative of the intratumoral presence or fitness of T cells. Thus, a suitable marker predicting T-cell effector functions in combination with PDL1 expression might allow for better selection of patients who would benefit from therapeutic PD1/PDL1-targeting. Future studies should focus on measurement of vimentin S39 phosphorylation in PDL1+ tumors collected from responders and nonresponders to PD1/PDL1-targeting therapies.
Our research lays the ground for future investigations into the role of PD1–induced cytoskeleton changes, which could yield precious mechanistic insight and identify further targets with therapeutic promise or utility as biomarkers for PD1-targeting immunotherapies.
Acknowledgments
The authors thank Nagham Alouche for assistance with flow cytometry–based experiments, Pascal Schneider for recombinant TACI-Fc, Giuseppe Pantaleo for providing the Jurkat-PD1 NFAT cell line, Marcus Long and Shiro Shibayama for valuable discussions, advice, and critical review of the manuscript, Florence Morgenthaler for her assistance in confocal imaging, and Emilie Lingre for her help with multiplex immunofluorescence stainings.
L.d.L. and D.V. received funding from Swiss National Science Foundation grant 310030_172954. This study was supported by grants from Swiss Cancer Research (KFS-4095-02-2017-R; M.T.), the ISREC Foundation (S.P.), the Emma Muschamp foundation (M.T.), and a collaboration agreement with Ono Pharmaceutical (M.T.).
Authorship
Contribution: D.C. and M.T. developed the study concept, interpreted data, and prepared the manuscript; D.C., S.P., R.S., C.D., and M.G. performed experiments; M.T., D.C., and M.Q. designed the proteomic studies; M.Q. and D.C. performed and analyzed the proteomic studies; D.C. and K.B. designed the ImageStream experiments; K.B. performed and analyzed the ImageStream experiments; D.V., K.I., and L.d.L. designed the immunohistochemistry (IHC) experiments; D.V. and K.I. performed the IHC experiments; K.I., D.V., and L.d.L. analyzed the IHC experiments; and D.C. designed all other experiments, analyzed data, and prepared figures.
Conflict-of-interest disclosure: This study was supported by a collaboration agreement between M.T. and ONO Pharmaceutical, Japan. The remaining authors declare no competing financial interests.
Correspondence: Margot Thome, Department of Immunobiology, University of Lausanne, Chemin des Boveresses 155, CH-1066, Epalinges, Switzerland; email: margot.thomemiazza@unil.ch.
References
Author notes
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD044028.
Data may be accessed at http://www.ebi.ac.uk/pride (username: reviewer_pxd044028@ebi.ac.uk, password: paxjQmcF).
The other original data that support the findings of this study are available on reasonable request from the corresponding author, Margot Thome (margot.thomemiazza@unil.ch).
The full-text version of this article contains a data supplement.