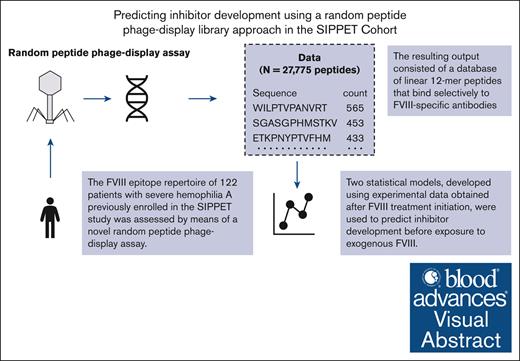
A novel random peptide phage display assay can be used to predict future inhibitor development before exposure to exogenous FVIII.
Visual Abstract
Inhibitor development is the most severe complication of hemophilia A (HA) care and is associated with increased morbidity and mortality. This study aimed to use a novel immunoglobulin G epitope mapping method to explore the factor VIII (FVIII)–specific epitope profile in the SIPPET cohort population and to develop an epitope mapping–based inhibitor prediction model. The population consisted of 122 previously untreated patients with severe HA who were followed up for 50 days of exposure to FVIII or 3 years, whichever occurred first. Sampling was performed before FVIII treatment and at the end of the follow-up. The outcome was inhibitor development. The FVIII epitope repertoire was assessed by means of a novel random peptide phage-display assay. A least absolute shrinkage and selection operator (LASSO) regression model and a random forest model were fitted on posttreatment sample data and validated in pretreatment sample data. The predictive performance of these models was assessed by the C-statistic and a calibration plot. We identified 27 775 peptides putatively directed against FVIII, which were used as input for the statistical models. The C-statistic of the LASSO and random forest models were good at 0.78 (95% confidence interval [CI], 0.69-0.86) and 0.80 (95% CI, 0.72-0.89). Model calibration of both models was moderately good. Two statistical models, developed on data from a novel random peptide phage display assay, were used to predict inhibitor development before exposure to exogenous FVIII. These models can be used to set up diagnostic tests that predict the risk of inhibitor development before starting treatment with FVIII.
Introduction
Recent advances in the treatment of patients with hemophilia A (HA) have greatly improved clinical outcomes and quality of life. Nevertheless, one of the greatest treatment complications in severe HA is still the development of anti–factor VIII (anti-FVIII) alloantibodies that neutralize FVIII (also called inhibitors). At least one-third of patients treated with FVIII replacement therapy develop an inhibitor during the first 20 to 30 days of exposure to FVIII (also called exposure days or EDs),1 making treatment with FVIII ineffective. This in turn leads to increased morbidity and mortality among these patients.1
This complication is the result of a multicausal immune response involving both patient- and treatment-related factors.1 The type of FVIII product is one of the most important risk factors for inhibitor development, with the SIPPET randomized clinical trial showing that patients treated with recombinant FVIII (rFVIII) have an almost twofold higher risk of developing an inhibitor than those treated with plasma-derived FVIII (pdFVIII) products.2 The pathophysiological mechanisms behind this increased immunogenicity remains unknown. Some plausible biological explanations have been postulated, such as the different posttranslational modifications caused by the use of different cell lines during the manufacturing process of rFVIII products and the protective role played by von Willebrand factor (VWF) in pdFVIII products.3
Mature FVIII consists of 6 major domains (A1, A2, B, A3, C1, and C2) and 3 acidic linking regions (a1, a2, and a3); A1-a1-A2-a2-B-a3-A3-C1-C2. The VWF-FVIII complex forms through a high-affinity interaction between the FVIII light chain and the VWF D′D3 domains.4 FVIII is activated by limited proteolysis through thrombin cleavage of 3 peptide bonds at Arg391 (a1-A2 junction), Arg759 (a2-B junction), and Arg1708 (a3-A3 junction).5 After thrombin cleavage, activated FVIII (without the B domain) is released from VWF and binds to phosphatidylserine on the extracellular surface of activated platelets.6,7
The anti-FVIII humoral immune response is highly polyclonal and consists primarily of immunoglobulin G (IgG) antibodies recognizing variable multiple epitopes among patients and even in the same patient over time.8 Several studies have examined the immunogenicity of FVIII and the mechanisms underlying inhibitor development during treatment with FVIII.3,9,10 The role of FVIII B-cell epitopes in inhibitor development has been previously investigated using different techniques. Specific regions in the A2 (region encompassing Arg484-Ile50811), A3 (Gln1778-Asp184012), C1 (Lys2065-Trp221213 and residues 2063-207114), and C2 (residues Glu2181-Val224315 as well as other residues16-18) FVIII domains were shown to be target domains for FVIII alloantibodies interaction using several methods including low resolution immunoprecipitation, western blotting, and antibody neutralization assays,8,19 as well as high resolution methods such as hydrogen-deuterium exchange mass spectrometry,18 crystallographic studies,20 surface plasmon resonance–based methods,17 and phage display.21-24
In recent years, quantitative immunoproteomics has developed rapidly, offering high-throughput analyses at relatively low cost. The aim of this study was to use a novel high-throughput epitope mapping technique based on a random peptide phage-display method to explore the overall FVIII epitope profile and to develop an epitope–based inhibitor prediction model.
Methods
Patient population
Study samples were obtained from patients enrolled in the SIPPET trial, which was designed to investigate the immunogenicity of different FVIII products in patients with severe HA who were previously untreated with any FVIII concentrates (PUPs) or minimally treated with blood components.2 Samples from 122 patients were used for this study. These patients were treated with 8 different FVIII products (4 pdFVIII products and 4 rFVIII products). Inhibitor development was measured using the Bethesda assay with Nijmegen modification.25 Thirty-nine of 122 individuals developed an inhibitor.
One sample of citrated plasma was collected at baseline before exposure to FVIII (pretreatment) and on sample at the end of the study (posttreatment). As previously described,2 in inhibitor-positive patients, the end of the study was the time of inhibitor development. In inhibitor-negative patients, the study ended when the patient reached 50 EDs or after 3 years of follow-up, whichever occurred first.
Approval for this study was obtained from the medical ethics committee at each study center, and informed consent was obtained from all parents/guardians of patients.
MVA
Assay set-up
The total IgG epitope repertoire was assessed using mimotope-variation analysis (MVA), a phage display–based method (Protobios, Tallinn, Estonia) as described previously.26 A combinatorial library of randomized linear 12-mer peptides fused to the pIII minor coat protein of M13 phages (Ph.D.-12, New England Biolabs, Ipswich, MA) was used according to the manufacturer’s protocol. Two μL of plasma was incubated with 5 μL of phage library (∼5 × 1010 phage particles; overnight at 4°C). The human IgG–captured phages were pulled down by protein G-coated magnetic beads (NEB, S1506S). Phage DNA was extracted, enriched and samples were barcoded by polymerase chain reaction amplification. Pooled samples were analyzed by Illumina sequencing (50-bp single end read, Genohub). The resulting DNA sequences were in silico translated to 12 amino acid (aa) long peptide sequences. To correct for differences in sequencing depth among the samples, the total count of each unique peptide per sample was normalized in its counts per 3 million. The resulting output consisted of a database of 12-mer peptides with varying degrees of apparent affinity for IgG antibodies. In the context of the assay, apparent affinity was defined as the frequency with which a 12-mer peptide was detected (ie, the peptide count). These peptides are often referred to in the literature as “mimotopes,” due to the fact that they may mimic the structure of an epitope.
Two versions of the assay were performed, the standard MVA assay (described above) and a competition assay. In the MVA competition assay, the same FVIII products that were used to treat the patient (Alphanate [Grifols], Fanhdi [Grifols], Emoclot [Kedrion Biopharma], Factane [LFB], Advate [Baxalta], Kogenate FS [Bayer AG], ReFacto AF [Pfizer], or Recombinate [Baxalta]) were also used to precondition study samples before competition analyses. In detail, respective FVIII products (final concentration, 3 uM) were incubated with 2 μL of plasma for 2 hours at room temperature before proceeding with the MVA assay as described above.
Removal of TUPs
One issue in conducting phage display experiments is the presence of so-called target-unrelated peptides (TUPs). These are false-positive results caused by selection-related TUPs, which are peptides binding to materials and reagents used in the assay (for example, plastic surfaces and albumin), or propagation-related TUPs, caused by faster propagation of some phage clones, resulting in a higher peptide count for some peptides. To minimize the effect of these TUPs, we removed all peptides that were predicted to be TUPs using the SAROTUP software tool.27 Using this tool, known TUPs were filtered out exploiting the TUPscan and the mimosearch algorithms. Peptides with a high likelihood (P > .8) to bind to polystyrene, as assessed by the PSBinder algorithm, were also filtered out.
Quality control using intra-assay and inter-assay replicates
To improve the assay signal to noise ratio, we focused on the most abundant peptides in the data set. To establish an optimal abundance threshold, we relied on technical replicates of a control sample within the same MVA plate (intra-assay) and across the different assay plates (inter-assay). Each technical replicate was compared with its intra-assay and inter-assay littermates using each possible filtering threshold. As similarity metric, we accounted for the proportion of peptides in the data set found in only 1 replicate at each possible threshold.
Identification of FVIII mimotopes
To find peptides identified using the MVA competition assay that bind selectively to FVIII-specific antibodies (FVIII mimotopes), the posttreatment sample was analyzed twice, once using the standard MVA assay and once using the MVA competition assay. FVIII mimotopes were defined as peptides that were present in the posttreatment sample in which the standard MVA assay was performed but not in the posttreatment sample in which the MVA competition assay was performed. Consequently, the abundance of each peptide in the standard MVA assay vs the MVA competition assay was compared using the Fisher exact test. Adjustment for multiple testing was done using the Bonferroni method. An adjusted P value < .05 was considered statistically significant. Only peptides significantly underrepresented in the MVA competition assay samples when compared with the standard MVA assay samples were considered to be FVIII mimotopes and used for further analyses.
Clustering workflow
Each FVIII epitope can be conceptualized as being represented by multiple peptide sequences, each containing the antibody-binding motif. Therefore, the Hammock algorithm was used to cluster peptides based on sequence similarity before further analyses.28 A complete linkage clustering algorithm was used for the initial clustering step. Cluster iterative merging was based on 3 iterations, maximum alignment length was set at 150% of that of the input peptide, and 5% of the initial clusters were used as seeds for cluster merging. Applying the algorithm resulted in clusters of highly similar peptides. For each cluster, a consensus motif was generated based on the multiple sequence alignment of the sequences. Each consensus motif can be interpreted as representing an epitope motif. Highly conserved residues (>60%) were denoted with an uppercase symbol, whereas moderately conserved residues (30%-60%) were denoted with a lower case symbol. Columns in the multiple sequence alignment in which no single residue had a prevalence of >30% were denoted with “x.” The total peptide count of each cluster was calculated as the sum of the count of each peptide included in a cluster. Clusters with an epitope motif that contained <4 conserved residues were filtered out from the data set.
Alignment of epitope motifs to FVIII
Epitope motifs from the remaining clusters were then aligned to the linear amino acid sequence of FVIII (UniProt database ID P00451). Local pairwise alignment using the Smith-Waterman algorithm was used (R package; “Biostrings” version 2.40.2). The degree to which a given residue on FVIII was surface accessible was calculated using the GETAREA algorithm29 using as input the crystal structure of a B-domain–deleted FVIII molecule30 (PDB ID 3CDZ). Based on the literature,31 a relative solvent accessibility of ≥20% was used as a cutoff for defining whether a residue was buried.
Statistical analyses
Descriptive statistics
For the descriptive analyses, data were summarized using the mean and standard deviation, or median and interquartile range, or as proportions.
Prediction modeling
To find biomarkers that were able to predict inhibitor development before the start of FVIII therapy, 2 statistical prediction models were fitted to the data. Both models were trained on data generated from the posttreatment samples and were validated on data generated from the pretreatment samples.
Firstly, a logistic regression model using L1 regularization (R package; “glmnet” version 4.1.7), also called least absolute shrinkage and selection operator (LASSO) logistic regression, was evaluated using all clusters as the input. Leave-one-out cross-validation was used to select the optimal value for the regularization parameter. All clusters were used as the input for the model, the variables were centered and scaled before model fitting. Secondly. A random forest model (R package; “randomForest” version 4.7-1.1) was evaluated using all clusters as the input. Values for the number of trees in the model and the number of variables at each split were selected by fitting models with different values of these parameters and then selecting the parameter value that minimized the out-of-bag error rate.
Evaluation of predictive performance
Predictive performance of these models was evaluated in 2 ways. Firstly, we assessed the degree to which a model could discriminate between patients with and without inhibitors, using a receiver operating characteristic (ROC) curve. Secondly, we evaluated the degree to which the predicted cumulative incidence of inhibitor development matched the observed cumulative incidence using a calibration plot.
Selection of important clusters
To identify the clusters that were most important for inhibitor development, we first ranked the importance of each variable in the random forest model for model prediction by permutation feature importance. We then selected all the clusters with a feature importance score in the 90th percentile. From this set, we then selected all the clusters that were also present (ie, with a nonzero model coefficient) in the final LASSO regression model. We then generated descriptive statistics for this final set of clusters.
Approval for this study was obtained from the medical ethics committee at each study center, and informed consent was obtained from all parents/guardians of patients.
Results
MVA
Of the 122 previously untreated patients with hemophilia selected from the SIPPET study cohort for this analysis, 39 patients developed an inhibitor during follow-up (Table 1). Inhibitor-positive patients were slightly more likely to have a null mutation in the F8 gene and slightly more likely to use a recombinant FVIII product than inhibitor-negative patients, although these changes were not significant (Table 1).
Patient characteristics
| . | Inhibitor-negative (n = 83) . | Inhibitor-positive (n = 39) . |
|---|---|---|
| Age at first treatment, mean (SD), mo | 20.7 (17.6) | 17.6 (13.0) |
| Family history of inhibitor development | ||
| No | 67 (80.7%) | 31 (79.5%) |
| Yes | 12 (14.5%) | 4 (10.3%) |
| Unknown | 4 (4.8%) | 4 (10.3%) |
| F8 gene mutation (null vs non-null) | ||
| Non-null mutation | 15 (18.1%) | 1 (2.6%) |
| Null mutation | 67 (80.7%) | 35 (89.7%) |
| Unknown | 1 (1.2%) | 3 (7.7%) |
| F8 gene mutation (detailed) | ||
| Frameshift mutation | 10 (12.0%) | 7 (17.9%) |
| Intron 1 inversion | 4 (4.8%) | 0 (0%) |
| Intron 22 inversion | 32 (38.6%) | 20 (51.3%) |
| Large deletion | 5 (6.0%) | 2 (5.1%) |
| Nonsense mutation | 16 (19.3%) | 6 (15.4%) |
| Missense mutation | 8 (9.6%) | 0 (0%) |
| Splice site mutation | 4 (4.8%) | 1 (2.6%) |
| Non-null mutation, type unknown | 3 (3.6%) | 0 (0%) |
| Unknown | 1 (1.2%) | 3 (7.7%) |
| FVIII product (type) | ||
| Recombinant FVIII product | 39 (47.0%) | 22 (56.4%) |
| Plasma-derived FVIII product | 44 (53.0%) | 17 (43.6%) |
| . | Inhibitor-negative (n = 83) . | Inhibitor-positive (n = 39) . |
|---|---|---|
| Age at first treatment, mean (SD), mo | 20.7 (17.6) | 17.6 (13.0) |
| Family history of inhibitor development | ||
| No | 67 (80.7%) | 31 (79.5%) |
| Yes | 12 (14.5%) | 4 (10.3%) |
| Unknown | 4 (4.8%) | 4 (10.3%) |
| F8 gene mutation (null vs non-null) | ||
| Non-null mutation | 15 (18.1%) | 1 (2.6%) |
| Null mutation | 67 (80.7%) | 35 (89.7%) |
| Unknown | 1 (1.2%) | 3 (7.7%) |
| F8 gene mutation (detailed) | ||
| Frameshift mutation | 10 (12.0%) | 7 (17.9%) |
| Intron 1 inversion | 4 (4.8%) | 0 (0%) |
| Intron 22 inversion | 32 (38.6%) | 20 (51.3%) |
| Large deletion | 5 (6.0%) | 2 (5.1%) |
| Nonsense mutation | 16 (19.3%) | 6 (15.4%) |
| Missense mutation | 8 (9.6%) | 0 (0%) |
| Splice site mutation | 4 (4.8%) | 1 (2.6%) |
| Non-null mutation, type unknown | 3 (3.6%) | 0 (0%) |
| Unknown | 1 (1.2%) | 3 (7.7%) |
| FVIII product (type) | ||
| Recombinant FVIII product | 39 (47.0%) | 22 (56.4%) |
| Plasma-derived FVIII product | 44 (53.0%) | 17 (43.6%) |
SD, standard deviation.
The mean number of unique peptides generated from each patients’ posttreatment samples was 356 365. After removing potential TUPs, the mean number of unique peptides generated for each patient decreased to 313 340. As shown in supplemental Figure 1, both intra-assay and inter-assay reproducibility started dropping when considering peptides with abundance <250 reads. Therefore, only peptides found with abundance >250 in at least 1 sample were considered for further analyses. This yielded 27 775 unique peptides that were identified as being FVIII mimotopes, with a median number of 266 (range, 4-1101) peptides per patient. These 27 775 peptides were then clustered (as described in “Methods”), which resulted in 223 clusters.
Location of clusters on FVIII
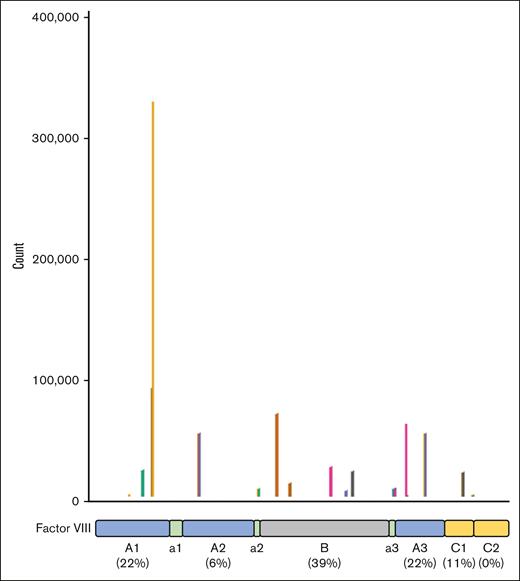
Using pairwise local alignment, 18 of 223 clusters were mapped with acceptable alignment to the linear sequence of FVIII. Most of these clusters were mapped against the B domain (39%; Figure 1). Of the 10 clusters that were mapped to parts of the linear sequence for which information on surface-accessibility was available, 9 were aligned to positions on FVIII that were partially surface accessible (Table 2). All B-domain–aligned peptide clusters had nonzero mean peptide counts in patients using a B-domain–deleted product. Furthermore, of the 7 peptide clusters that were aligned to the B domain, 4 peptide clusters had a higher mean peptide count among patients receiving treatment with full-length FVIII than among patients receiving B-domain–deleted FVIII. (Table 3).
Alignment of epitope motifs on the linear sequence of FVIII. The plot shows the alignment of 18 epitope motifs on the linear sequence of FVIII. The x-axis represents the linear sequence of FVIII, from position 20 to 2351. The y-axis shows a count for each position of the FVIII sequence. The count for each position was defined as the weighted sum of each cluster whose epitope motif was mapped to that position, using the peptide count of each cluster as weights. For example, if 2 clusters with a peptide count of 20 and 10, respectively, were mapped to a given position, then the total score for that position would be cluster 1 ∗ 20 + cluster 2 ∗ 10 = 30. The number of epitope motifs mapped to each domain, as a proportion of all 18 aligned epitope motifs, is shown at the bottom of the figure.
Alignment of epitope motifs on the linear sequence of FVIII. The plot shows the alignment of 18 epitope motifs on the linear sequence of FVIII. The x-axis represents the linear sequence of FVIII, from position 20 to 2351. The y-axis shows a count for each position of the FVIII sequence. The count for each position was defined as the weighted sum of each cluster whose epitope motif was mapped to that position, using the peptide count of each cluster as weights. For example, if 2 clusters with a peptide count of 20 and 10, respectively, were mapped to a given position, then the total score for that position would be cluster 1 ∗ 20 + cluster 2 ∗ 10 = 30. The number of epitope motifs mapped to each domain, as a proportion of all 18 aligned epitope motifs, is shown at the bottom of the figure.
Linear alignment of mimotope clusters on FVIII
| Mimotope core motif sequence . | FVII sequence . | Alignment . | Residue number, start . | Residue number, end . | Domain . | No. of surface-accessible residues∗ . |
|---|---|---|---|---|---|---|
| nxRRPfflnsg | LNSG | LNSG | 187 | 190 | A1 | 0 |
| glggLi | LPGLI | L?GLI | 261 | 265 | A1 | 3 |
| dPxqtll | QTLL | QTLL | 316 | 319 | A1 | 2 |
| GLGqLL | LGQFL | LGQ?L | 322 | 326 | A1 | 4 |
| nqkms | NQIMS | NQ?MS | 583 | 587 | A2 | 5 |
| pdtppSxp | PPSMP | PPS?P | 925 | 929 | B | NA† |
| txxKtxIxTxt | TNRKTHI | T??KT?I | 1028 | 1034 | B | NA† |
| Ppdixspp | PPDAQNP | PPD???P | 1105 | 1111 | B | NA† |
| KVFRxp | KQFRLP | K?FR?P | 1335 | 1340 | B | NA† |
| VFRlpxtxt | FRLP | FRLP | 1337 | 1340 | B | NA† |
| TxltRtls | LTRVL | LTR?L | 1423 | 1427 | B | NA† |
| qNLsl | NLSL | NLSL | 1461 | 1464 | B | NA† |
| ydkadnerarlg | YDEDENQSPR | YD???N???R | 1699 | 1708 | other | NA† |
| dRxeLNmxxxl | RGELN | R?ELN | 1768 | 1772 | A3 | 3 |
| lNEvLv | LNEHL | LNE?L | 1771 | 1775 | A3 | 3 |
| hTnln | HTNTLN | HTN-LN | 1878 | 1883 | A3 | 4 |
| kxDiLaxl | KVDLLA | K?D?LA | 2091 | 2096 | C1 | 3 |
| KxDssGP | DSSG | DSSG | 2150 | 2153 | C1 | 3 |
| Mimotope core motif sequence . | FVII sequence . | Alignment . | Residue number, start . | Residue number, end . | Domain . | No. of surface-accessible residues∗ . |
|---|---|---|---|---|---|---|
| nxRRPfflnsg | LNSG | LNSG | 187 | 190 | A1 | 0 |
| glggLi | LPGLI | L?GLI | 261 | 265 | A1 | 3 |
| dPxqtll | QTLL | QTLL | 316 | 319 | A1 | 2 |
| GLGqLL | LGQFL | LGQ?L | 322 | 326 | A1 | 4 |
| nqkms | NQIMS | NQ?MS | 583 | 587 | A2 | 5 |
| pdtppSxp | PPSMP | PPS?P | 925 | 929 | B | NA† |
| txxKtxIxTxt | TNRKTHI | T??KT?I | 1028 | 1034 | B | NA† |
| Ppdixspp | PPDAQNP | PPD???P | 1105 | 1111 | B | NA† |
| KVFRxp | KQFRLP | K?FR?P | 1335 | 1340 | B | NA† |
| VFRlpxtxt | FRLP | FRLP | 1337 | 1340 | B | NA† |
| TxltRtls | LTRVL | LTR?L | 1423 | 1427 | B | NA† |
| qNLsl | NLSL | NLSL | 1461 | 1464 | B | NA† |
| ydkadnerarlg | YDEDENQSPR | YD???N???R | 1699 | 1708 | other | NA† |
| dRxeLNmxxxl | RGELN | R?ELN | 1768 | 1772 | A3 | 3 |
| lNEvLv | LNEHL | LNE?L | 1771 | 1775 | A3 | 3 |
| hTnln | HTNTLN | HTN-LN | 1878 | 1883 | A3 | 4 |
| kxDiLaxl | KVDLLA | K?D?LA | 2091 | 2096 | C1 | 3 |
| KxDssGP | DSSG | DSSG | 2150 | 2153 | C1 | 3 |
SD, standard deviation.
The number of surface-accessible residues for each alignment was calculated using the GETAREA algorithm, as described in “Methods.”
No information on surface-accessibility of B domain and some adjacent residues.
Mean peptide count (SD) of peptide clusters that were aligned to the B domain, stratified by use of B-domain–deleted or full-length FVIII
| . | B-domain–deleted FVIII (n = 7) . | Full-length FVIII (n = 115) . |
|---|---|---|
| qNLsl | 175 (56.0) | 349 (543) |
| KVFRxp | 405 (627) | 383 (948) |
| Ppdixspp | 63.6 (89.0) | 59.2 (288) |
| TxltRtls | 119 (248) | 1 160 (10 800) |
| txxKtxIxTxt | 51.1 (63.3) | 304 (1790) |
| VFRlpxtxt | 157 (391) | 395 (1870) |
| pdtppSxp | 27.7 (27.4) | 291 (716) |
| . | B-domain–deleted FVIII (n = 7) . | Full-length FVIII (n = 115) . |
|---|---|---|
| qNLsl | 175 (56.0) | 349 (543) |
| KVFRxp | 405 (627) | 383 (948) |
| Ppdixspp | 63.6 (89.0) | 59.2 (288) |
| TxltRtls | 119 (248) | 1 160 (10 800) |
| txxKtxIxTxt | 51.1 (63.3) | 304 (1790) |
| VFRlpxtxt | 157 (391) | 395 (1870) |
| pdtppSxp | 27.7 (27.4) | 291 (716) |
SD, standard deviation.
Predictive value of clusters
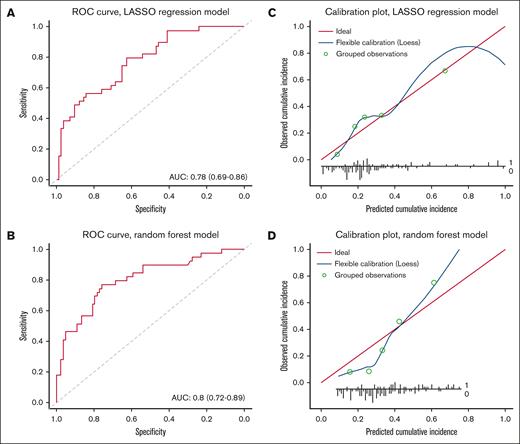
Next, we constructed 2 statistical prediction models to assess the degree to which the presence of these clusters in patients’ samples were able to predict inhibitor development. First, a LASSO logistic regression model was fitted to all 223 clusters in the posttreatment patient samples. The fitted model was then used to predict inhibitor development using the pretreatment patient samples. The C-statistic was 0.78 (95% confidence interval, 0.69-0.86; Figure 2A). Model calibration was good because the cumulative incidence of inhibitor development predicted by the model was roughly in line with the observed incidence across the entire risk range (Figure 2C). Next, a random forest model was fitted to all 223 clusters in the posttreatment patient samples. The fitted model was then used to predict inhibitor development using the pretreatment patient samples. The C-statistic was 0.80 (95% confidence interval, 0.72-0.89; Figure 2B). Model calibration was moderate, due to the model somewhat overpredicting the observed cumulative incidence of inhibitor development in the lower-risk range (Figure 2D).
Evaluation of the degree to which the logistic regression model and random forest classifier model can predict inhibitor development. Model discrimination (ie, the degree to which a model assigns a higher risk to an inhibitor-positive patient vs an inhibitor-negative patient) was assessed by plotting ROC curves and by calculating the area under the curve (AUC). The AUC varies between 0.5 (no discrimination) to 1 (perfect discrimination). (A-B) The ROC curves of the LASSO logistic regression model and the random forest model are shown in (A) and (B), respectively. Model calibration (ie, the degree to which the predicted cumulative incidence of inhibitor development matched the observed cumulative incidence) was assessed using a calibration plot. For each quintile of predicted cumulative incidence, we plotted the mean predicted cumulative incidence of inhibitor development in a group against the observed cumulative incidence of inhibitor development in that group. In addition, we plotted a LOESS (locally estimated scatterplot smoothing) line in the same figure to assess model calibration across the full risk range. Ideally, all points should lie exactly on the diagonal line (which represents perfect agreement between predicted and observed values). (C-D) The calibrations plots of the LASSO logistic regression model and the random forest model are shown in (C) and (D), respectively.
Evaluation of the degree to which the logistic regression model and random forest classifier model can predict inhibitor development. Model discrimination (ie, the degree to which a model assigns a higher risk to an inhibitor-positive patient vs an inhibitor-negative patient) was assessed by plotting ROC curves and by calculating the area under the curve (AUC). The AUC varies between 0.5 (no discrimination) to 1 (perfect discrimination). (A-B) The ROC curves of the LASSO logistic regression model and the random forest model are shown in (A) and (B), respectively. Model calibration (ie, the degree to which the predicted cumulative incidence of inhibitor development matched the observed cumulative incidence) was assessed using a calibration plot. For each quintile of predicted cumulative incidence, we plotted the mean predicted cumulative incidence of inhibitor development in a group against the observed cumulative incidence of inhibitor development in that group. In addition, we plotted a LOESS (locally estimated scatterplot smoothing) line in the same figure to assess model calibration across the full risk range. Ideally, all points should lie exactly on the diagonal line (which represents perfect agreement between predicted and observed values). (C-D) The calibrations plots of the LASSO logistic regression model and the random forest model are shown in (C) and (D), respectively.
There were 12 clusters that had a feature importance score in the 90th percentile in the random forest model and were also part of the final LASSO logistic regression model (Table 4). Of these 12 clusters, only 2 mapped with good alignment to the linear sequence of FVIII. These 2 clusters were mapped to the A2 and A3 domain (Table 4). Ten of 12 clusters had a higher peptide count in inhibitor-positive patients than in inhibitor-negative patients (Table 4). No clear differences were seen in the mean peptide counts of the clusters when measured in patients treated with plasma-derived FVIII vs patients treated with rFVIII. (Table 5).
Peptide clusters that were used as predictors in both the logistic regression model and the random forest classifier model
| Core motif . | Mean peptide count in INH– group . | Mean peptide count in INH+ group . | Fold change . | FVIII domain . | Number of unique peptides in cluster (%) . | Peptide count of cluster (%) . |
|---|---|---|---|---|---|---|
| kxPxstw | 133 | 1826 | 13.70 | - | 24 (0.09%) | 55 764 (0.16%) |
| hntMels | 38 | 119 | 3.10 | - | 11 (0.04%) | 20 460 (0.06%) |
| hTnln | 151 | 454 | 3.01 | A3 | 48 (0.17%) | 66 660 (0.19%) |
| nqkms | 140 | 365 | 2.60 | A2 | 31 (0.11%) | 66 660 (0.19%) |
| dxxYxlxm | 438 | 1114 | 2.54 | - | 34 (0.12%) | 63 368 (0.18%) |
| Yvntxxxt | 193 | 475 | 2.46 | - | 11 (0.04%) | 25 499 (0.07%) |
| LtqM | 159 | 302 | 1.90 | - | 31 (0.11%) | 61 253 (0.17%) |
| pQyxnxxk | 454 | 738 | 1.63 | - | 51 (0.18%) | 52 743 (0.15%) |
| sxnKP | 325 | 483 | 1.49 | - | 53 (0.19%) | 52 537 (0.15%) |
| WDVpPxxxxt | 301 | 445 | 1.48 | - | 21 (0.08%) | 33 105 (0.09%) |
| KxxHyxk | 459 | 459 | 1.00 | - | 15 (0.05%) | 39 173 (0.11%) |
| qTAkfh | 41 | 40 | 0.96 | - | 41 (0.15%) | 48 928 (0.14%) |
| Core motif . | Mean peptide count in INH– group . | Mean peptide count in INH+ group . | Fold change . | FVIII domain . | Number of unique peptides in cluster (%) . | Peptide count of cluster (%) . |
|---|---|---|---|---|---|---|
| kxPxstw | 133 | 1826 | 13.70 | - | 24 (0.09%) | 55 764 (0.16%) |
| hntMels | 38 | 119 | 3.10 | - | 11 (0.04%) | 20 460 (0.06%) |
| hTnln | 151 | 454 | 3.01 | A3 | 48 (0.17%) | 66 660 (0.19%) |
| nqkms | 140 | 365 | 2.60 | A2 | 31 (0.11%) | 66 660 (0.19%) |
| dxxYxlxm | 438 | 1114 | 2.54 | - | 34 (0.12%) | 63 368 (0.18%) |
| Yvntxxxt | 193 | 475 | 2.46 | - | 11 (0.04%) | 25 499 (0.07%) |
| LtqM | 159 | 302 | 1.90 | - | 31 (0.11%) | 61 253 (0.17%) |
| pQyxnxxk | 454 | 738 | 1.63 | - | 51 (0.18%) | 52 743 (0.15%) |
| sxnKP | 325 | 483 | 1.49 | - | 53 (0.19%) | 52 537 (0.15%) |
| WDVpPxxxxt | 301 | 445 | 1.48 | - | 21 (0.08%) | 33 105 (0.09%) |
| KxxHyxk | 459 | 459 | 1.00 | - | 15 (0.05%) | 39 173 (0.11%) |
| qTAkfh | 41 | 40 | 0.96 | - | 41 (0.15%) | 48 928 (0.14%) |
Total number of unique peptides were 27 775. Total peptide count was 35 452 858. The “-” indicates lack of good alignment on linear sequence of FVIII.
Mean count of peptide clusters, stratified by inhibitor status, F8 gene mutation, and FVIII product type
| . | Inhibitor status . | F8 Gene mutation∗ . | FVIII product type . | |||||
|---|---|---|---|---|---|---|---|---|
| Inhibitor-negative (n = 83) . | Inhibitor-positive, total (n = 39) . | Inhibitor-positive, low-titer (n = 15) . | Inhibitor-positive, high-titer (n = 24) . | Non-null mutation (n = 16) . | Null mutation (n = 102) . | rFVIII product (n = 61) . | pdFVIII product (n = 61) . | |
| Mean peptide count (SD) | ||||||||
| kxPxstw | 133 (119) | 1830 (9810) | 4360 (15800) | 240 (337) | 145 (119) | 779 (6070) | 1190 (7850) | 164 (196) |
| hntMels | 38.5 (44.8) | 119 (266) | 256 (398) | 33.4 (28.7) | 88.5 (264) | 61.6 (139) | 91.8 (212) | 36.6 (61.5) |
| hTnln | 151 (372) | 454 (1540) | 850 (2370) | 206 (564) | 295 (590) | 247 (985) | 350 (1260) | 145 (345) |
| nqkms | 140 (199) | 365 (1240) | 168 (139) | 489 (1570) | 159 (215) | 223 (784) | 267 (996) | 157 (223) |
| dxxYxlxm | 438 (1590) | 1110 (3850) | 451 (840) | 1530 (4850) | 332 (440) | 717 (2770) | 889 (3140) | 420 (1750) |
| Yvntxxxt | 193 (290) | 475 (891) | 448 (523) | 492 (1070) | 185 (167) | 299 (615) | 286 (532) | 280 (608) |
| LtqM | 159 (483) | 302 (1000) | 159 (316) | 391 (1250) | 61.3 (106) | 234 (752) | 187 (797) | 222 (573) |
| pQyxnxxk | 454 (1170) | 738 (1890) | 109 (157) | 1130 (2340) | 176 (189) | 614 (1560) | 683 (1540) | 407 (1320) |
| sxnKP | 325 (674) | 483 (858) | 717 (995) | 337 (746) | 208 (293) | 413 (794) | 459 (888) | 293 (543) |
| WDVpPxxxxt | 301 (464) | 445 (1130) | 277 (277) | 549 (1430) | 331 (468) | 358 (791) | 330 (520) | 364 (917) |
| KxxHyxk | 459 (3120) | 459 (1880) | 99.5 (155) | 684 (2380) | 65.6 (164) | 532 (3030) | 617 (3630) | 301 (1510) |
| qTAkfh | 41.5 (103) | 39.7 (82.9) | 39.3 (97.9) | 40.0 (74.3) | 88.9 (164) | 34.3 (81.9) | 44.8 (104) | 37.0 (88.6) |
| . | Inhibitor status . | F8 Gene mutation∗ . | FVIII product type . | |||||
|---|---|---|---|---|---|---|---|---|
| Inhibitor-negative (n = 83) . | Inhibitor-positive, total (n = 39) . | Inhibitor-positive, low-titer (n = 15) . | Inhibitor-positive, high-titer (n = 24) . | Non-null mutation (n = 16) . | Null mutation (n = 102) . | rFVIII product (n = 61) . | pdFVIII product (n = 61) . | |
| Mean peptide count (SD) | ||||||||
| kxPxstw | 133 (119) | 1830 (9810) | 4360 (15800) | 240 (337) | 145 (119) | 779 (6070) | 1190 (7850) | 164 (196) |
| hntMels | 38.5 (44.8) | 119 (266) | 256 (398) | 33.4 (28.7) | 88.5 (264) | 61.6 (139) | 91.8 (212) | 36.6 (61.5) |
| hTnln | 151 (372) | 454 (1540) | 850 (2370) | 206 (564) | 295 (590) | 247 (985) | 350 (1260) | 145 (345) |
| nqkms | 140 (199) | 365 (1240) | 168 (139) | 489 (1570) | 159 (215) | 223 (784) | 267 (996) | 157 (223) |
| dxxYxlxm | 438 (1590) | 1110 (3850) | 451 (840) | 1530 (4850) | 332 (440) | 717 (2770) | 889 (3140) | 420 (1750) |
| Yvntxxxt | 193 (290) | 475 (891) | 448 (523) | 492 (1070) | 185 (167) | 299 (615) | 286 (532) | 280 (608) |
| LtqM | 159 (483) | 302 (1000) | 159 (316) | 391 (1250) | 61.3 (106) | 234 (752) | 187 (797) | 222 (573) |
| pQyxnxxk | 454 (1170) | 738 (1890) | 109 (157) | 1130 (2340) | 176 (189) | 614 (1560) | 683 (1540) | 407 (1320) |
| sxnKP | 325 (674) | 483 (858) | 717 (995) | 337 (746) | 208 (293) | 413 (794) | 459 (888) | 293 (543) |
| WDVpPxxxxt | 301 (464) | 445 (1130) | 277 (277) | 549 (1430) | 331 (468) | 358 (791) | 330 (520) | 364 (917) |
| KxxHyxk | 459 (3120) | 459 (1880) | 99.5 (155) | 684 (2380) | 65.6 (164) | 532 (3030) | 617 (3630) | 301 (1510) |
| qTAkfh | 41.5 (103) | 39.7 (82.9) | 39.3 (97.9) | 40.0 (74.3) | 88.9 (164) | 34.3 (81.9) | 44.8 (104) | 37.0 (88.6) |
SD, standard deviation.
Four patients were excluded from this analysis as their F8 gene mutation was unknown.
Discussion
We assessed the FVIII-specific epitope profile of 122 previously untreated patients with HA, using a novel random peptide phage-display assay. Our results show that the apparent FVIII-specific antibody response is highly polyclonal, with many different epitope motifs. Among the 18 epitope motifs that were mapped to the linear sequence of FVIII, most of the mimotopes aligned with A1, A3, and B domain sequences. Using information on the presence of these epitope motifs in patient samples, 2 statistical prediction models (developed on posttreatment samples and validated in pretreatment samples) were found to be predictive for inhibitor development.
Seven of the 18 epitope motifs (39%) with good alignment to FVIII were mapped to the B domain. It is important to note that the alignments were not confirmed in vitro (eg, by antibody-binding assays using FVIII proteins with mutations at the relevant residues). All B-domain–aligned peptide clusters had nonzero mean peptide counts in patients using a B-domain–deleted product. This provides evidence against the hypothesis that the motifs of these peptide clusters truly represent targets of antibodies that are highly specific to a linear epitope on the B domain. That being said, these results are based on very small numbers because there were only 7 patients using a B-domain–deleted FVIII product (all ReFacto).
Previous studies have suggested that antibodies against the B domain might be predominantly of the nonneutralizing type,32-34 because the B domain is not essential for the role of FVIII in blood clotting and is cleaved off after FVIII is activated. We could not verify this in our data set because almost all epitope motifs that were most important for inhibitor prediction (Table 4) did not map well to the linear sequence of FVIII.
In the past, several studies have tried to develop models to predict inhibitor development. Most of these models were based on either clinical parameters (eg, the age of FVIII treatment initiation, the type of FVIII product used, or the intensity of the first treatment moment with FVIII) or genetic parameters (eg, family history of inhibitor development, the type of F8 gene mutation, HLA type, or gene polymorphisms in immunoregulatory genes such as IL10 or CTLA4).35 Previously, a study in the SIPPET cohort also assessed the predictive value of the presence of nonneutralizing antibodies detected before treatment as part of a larger clinical prediction model for inhibitor development.36 None of the aforementioned prediction models were able to accurately predict inhibitor development. In addition, some prediction models were only implementable after starting treatment with FVIII, due to the inclusion of treatment-related predictors (eg, information on treatment intensity can only be obtained after a couple of days of exposure to FVIII). This limits the applicability of these models as one would ideally want to have an idea about the risk of inhibitor development before FVIII treatment is initiated so that certain types of treatment modalities (eg, exposure to exogenous FVIII) can be avoided. The models in this publication only use information on the pretreatment epitope repertoire of the patient and can therefore be used before FVIII treatment initiation.
The presence of peptides that specifically bind to anti-FVIII antibodies in samples taken before treatment with FVIII might seem unexpected at first glance. However, several studies have reported the presence of nonneutralizing anti-FVIII antibodies in healthy controls.37 In addition, a previous study using pretreatment samples of the current cohort reported that ∼10% of patients had measurable anti-FVIII antibodies.38 This suggests that natural autoreactivity against endogenous FVIII is relatively common in patients as well as healthy controls. Another hypothesis could be that the detected antibodies were not initially directed against FVIII but were the result of previous exposure to a pathogen (eg, a bacteria or virus) that contained a similar epitope. This cross-reactivity of the antibody response has been previously reported in several autoimmune disorders.39 Our results indicate that the presence of anti-FVIII antibodies before treatment with FVIII might be a risk factor for inhibitor development in a subset of patients.
This approach has some limitations. Firstly, it has been shown that only a handful of contact residues within an epitope make a significant contribution to antibody binding.40 In this study, we tried to identify these residues by clustering highly similar FVIII mimotopes and generating a consensus motif. Using alanine walk mutational analysis, the study by Kahle et al24 showed that there was reasonable agreement between a given consensus motif and the crucial binding residues of an epitope. Therefore, the consensus motifs derived from the multiple sequence alignment of each cluster of peptide sequences can, in theory, be considered to be potential epitope motifs. However, the accuracy of this approach is unknown, and further verification is needed to identify the exact residues involved in binding to an antibody.
Secondly, the final epitope motifs were mapped to FVIII by aligning the motifs to the linear sequence of FVIII. However, it has been reported that the majority of B-cell epitopes are conformational41,42 (although the exact proportion of B-cell epitopes purported to be conformational is unknown). In this case, clustering based on sequence similarity might yield the correct conformational epitope motif, but the linear alignment procedure will produce faulty alignment. An alternative approach would involve mapping the epitope motifs to the 3-dimensional structure of FVIII, using an in silico approach. However, a recent study that assessed a set of B-cell epitope prediction algorithms against a benchmark data set reported that all algorithms performed relatively poorly at mapping a potential epitope to the right location on an antigen.43 That being said, knowing the correct location of these putative B-cell epitopes is not needed if the goal is only to predict inhibitor development.
We removed all peptides that were predicted to be target unrelated (based on software exploiting publicly available repositories27) from the final peptide database. However, the residual impact of TUPs that were not removed from the database on the results is difficult to quantify. In addition, some peptides can bind to both elements of assay as well as an IgG antibody (ie, they can be classified as both target-unrelated and target-related peptides). By removing these peptides, we might have inadvertently also removed some important peptides from the initial database.
From the output of the assay, only peptides with a count >250 were selected; this resulted in a much smaller data set. The cutoff was based on the intra-assay and inter-assay replicability (supplemental Figure 1). It is possible that many peptides that were the target of a FVIII-specific antibody were removed in this step.
Lastly, our analysis of the immune response did not include nonpeptidic epitopes (such as the glycans present on the surface of FVIII). One difference between rFVIII and pdFVIII is in their respective glycosylation patterns.44 Unfortunately, our approach does not allow for the assessment of the impact of differing glycosylation patterns on immunogenicity.
Conclusion
Two statistical models, developed on data from a novel random peptide phage display assay, were used to predict inhibitor development before exposure to exogenous FVIII. These models can be used to set up diagnostic tests that predict the risk of inhibitor development before starting treatment with FVIII.
Acknowledgments
Mimotope-variation analysis was performed by Protobios LLC.
This study was partially supported by the 2017 European Association for Haemophilia and Allied Disorders (EAHAD) grant (R.P.), 2017 Grifols investigator-sponsored research grant (F.P.), the Italian Ministry of Health–Bando Ricerca Corrente 2020 (F.P.), and a Horizon 2020 Program grant (grant agreement ID: 859974, project name: EDUC8).
Authorship
Contribution: S.H., G.B., L.M., R.L.R., and H.C. were involved in study design, manuscript development, and analysis; R.P., F.P., and F.R.R. were involved in study design, data collection, and manuscript development; and A.E.-B., M.E., V.R., P.E., and M.K. were involved in data collection and manuscript development.
Conflict-of-interest disclosure: R.P. has received speaker fees from Novo Nordisk for an educational workshop. F.P. reports honoraria for speaking at educational meetings from Grifols and Roche and was a member of advisory boards for Sanofi, Sobi, Takeda, Roche, and BioMarin. The remaining authors declare no competing financial interests.
Correspondence: Flora Peyvandi, Università degli Studi di Milano, Department of Pathophysiology and Transplantation, Via Francesco Sforza 35, 20122, Milan, Italy; email: flora.peyvandi@unimi.it.
References
Author notes
S.H. and G.B.contributed equally to this study.
Sequencing data generated and/or analyzed in this study are not publicly available due to containing sensitive clinical information but are available upon reasonable request from the corresponding author, Flora Peyvandi (flora.peyvandi@unimi.it).
The full-text version of this article contains a data supplement.