Machine learning optimized RHtyper for automated and accurate Rh blood group genotyping from WES data.
Visual Abstract
Rh phenotype matching reduces but does not eliminate alloimmunization in patients with sickle cell disease (SCD) due to RH genetic diversity that is not distinguishable by serological typing. RH genotype matching can potentially mitigate Rh alloimmunization but comprehensive and accessible genotyping methods are needed. We developed RHtyper as an automated algorithm to predict RH genotypes using whole-genome sequencing (WGS) data with high accuracy. Here, we adapted RHtyper for whole-exome sequencing (WES) data, which are more affordable but challenged by uneven sequencing coverage and exacerbated sequencing read misalignment, resulting in uncertain predictions for (1) RHD zygosity and hybrid alleles, (2) RHCE∗C vs. RHCE∗c alleles, (3) RHD c.1136C>T zygosity, and (4) RHCE c.48G>C zygosity. We optimized RHtyper to accurately predict RHD and RHCE genotypes using WES data by leveraging machine learning models and improved the concordance of WES with WGS predictions from 90.8% to 97.2% for RHD and 96.3% to 98.2% for RHCE among 396 patients in the Sickle Cell Clinical Research and Intervention Program. In a second validation cohort of 3030 cancer survivors (15.2% Black or African Americans) from the St. Jude Lifetime Cohort Study, the optimized RHtyper reached concordance rates between WES and WGS predications to 96.3% for RHD and 94.6% for RHCE. Machine learning improved the accuracy of RH predication using WES data. RHtyper has the potential, once implemented, to provide a precision medicine-based approach to facilitate RH genotype–matched transfusion and improve transfusion safety for patients with SCD. This study used data from clinical trials registered at ClinicalTrials.gov as #NCT02098863 and NCT00760656.
Introduction
Blood transfusion is an essential treatment for chronic anemia disorders, including sickle cell disease (SCD) and thalassemia. Exposure to donor red blood cell (RBC) antigens can lead to alloimmunization and increase the risk of hemolytic transfusion reactions with subsequent transfusions.1 Prophylactic matching for Rh (C, E or C/c, E/e) and K antigens lowers the risk of alloimmunization in patients with SCD and thalassemia but alloantibody formation against the Rh blood group remains a challenge because of the genetically diverse RH genes of Black patients and blood donors.2,3 The Rh blood group consists of 5 major antigens, D, C, c, E, and e, and is encoded by the highly homologous RHD and RHCE genes.4RHD and RHCE genes of individuals of African descent exhibit high diversity with single nucleotide polymorphisms (SNPs), insertions/deletions (indels), and structural variants. Approximately 450 RHD and 190 RHCE alleles have been identified, and >50 Rh variant antigens have been described serologically. We found in our practice that 48% to 49% of patients with SCD and 41% of Black blood donors in the United States have an RHD or RHCE variant (excluding altered alleles of RHD∗10.00 or RHD∗DAU0 and RHCE∗01.01 or RHCE∗ce48C),5,6 and 7% of D-positive patients with SCD have a partial D.7 In Brazil, 15% of patients with SCD and 8% of African Brazilian blood donors have both variant RHD and RHCE alleles.8 These variant RH alleles encode proteins associated with the loss of epitopes or the expression of neo-epitopes. Individuals with variant RH alleles are at risk of alloimmunization when exposed to conventional or variant Rh antigens differing from their own. Because serological antigen typing cannot distinguish the presence of most variant Rh antigens,2RH genotyping and consideration of RH genotype matching can potentially improve the resource allocation of valuable Black blood donors and avoid Rh alloimmunization.
Next-generation sequencing (NGS) data, such as whole-genome sequencing (WGS) and whole-exome sequencing (WES), offer comprehensive evaluation of the genome and have been used for RH genotyping.9-13 Genotyping RH using NGS data is challenging because RHD and RHCE are duplicated genes that share 97% sequence identity. Sequencing reads from highly homologous regions may map ambiguously, making it difficult to determine the true genomic origin of these reads. Therefore, analysis of NGS data from RH loci requires sophisticated bioinformatics tools that can differentiate between true genetic variants and sequencing artifacts. We previously developed RHtyper for automated and accurate detection of the complex RH genotypes of Black or African American individuals using WGS data.6 RHtyper relies on a Bayesian likelihood-based framework to infer RH genotypes directly after short-read sequence alignment. Both sequence consistency at each SNP/indel and phase consistency across adjacent SNPs/indels are considered to improve prediction accuracy. RHtyper also incorporates coverage profiling to determine RHD zygosity and hybrid alleles and can further define potential breakpoints of the hybrid RH alleles using the Circular Binary Segmentation algorithm. In a validation cohort of 57 patients with SCD, RHtyper achieved 100% accuracy for RHD and 98.2% accuracy for RHCE when compared with RH genotypes verified by multiple molecular methods. Upon application to the Sickle Cell Clinical Research and Intervention Program (SCCRIP) study cohort, RHtyper achieved high concordance rates of 98.3% with C serological typing (n = 360 patients) and 99.54% with D serological typing (n = 219 patients).
WES is a focused and cost-effective strategy to identify exonic variations but has limitations we sought to overcome with machine learning. Sequencing coverage of WES is uneven because of variable sequencing read enrichment by capturing oligonucleotides at different locations, leading to inaccurate prediction for copy numbers of exons and SNPs. WES data lacks most intronic sequence markers essential for aligning sequencing reads, resulting in misalignments among highly homologous exons. Here, we adapted RHtyper for WES data by leveraging machine learning to address uneven coverage and sequence misalignments and improved WES-based RH genotyping substantially.
Methods
Patients
Existing WES and WGS data from 396 patients with SCD enrolled in the SCCRIP study and 3030 cancer survivors enrolled in St. Jude Lifetime Cohort Study (SJLIFE) at St. Jude Children’s Research Hospital (SJCRH) were included in this study. Of the 396 patients with SCD, 56 had RH genotypes tested by the standard RH genotyping method, SNP-based and targeted molecular assays, and confirmed by Sanger sequencing and NGS as previously described6 and in supplemental Methods. They were used to further verify the WES-predicted genotypes. The SCCRIP is a lifetime longitudinal cohort study of patients with SCD, in which clinical information is prospectively collected and biologic samples are banked, including blood for genomics and proteomics studies (NCT02098863).14 The SJLIFE is a retrospective cohort study with prospective follow-up and ongoing accrual of oncology patients treated at SJCRH who were aged ≥18 years and ≥10 years after diagnosis from their malignancy (NCT00760656).15
WES, WGS, and serological typing
Genomic DNA was extracted from peripheral blood mononuclear cells using standard methods, and WGS and WES were performed at the HudsonAlpha Institute for Biotechnology and the SJCRH Hartwell Center for Bioinformatics and Biotechnology, as previously described.14,16 The paired-end reads were aligned against the human genome (hg38) using the Burrows-Wheeler Aligner software package.17 For patients in the SJLIFE cohort, serological typing of RhD only was performed.
Adjustment of RHtyper for WES data
The RH allele database was curated from the International Society of Blood Transfusion database and the now-retired National Center for Biotechnology Information-Blood Group Antigen Gene Mutation (BGMUT) database, as previously described.6 The consolidated database included 419 RHD and 130 RHCE alleles annotated for genotype determination. Variants were determined according to conventional RH messenger RNA sequences (RHD, L08429; RHCE, DQ322275), which differ from the reference genomic sequence (hg38) by 2 SNPs in the coding region (conventional RHD sequence, c.1136T, reference genomic sequence, c.1136C; conventional RHCE sequence, c.48G, reference genomic sequence, c.48C).
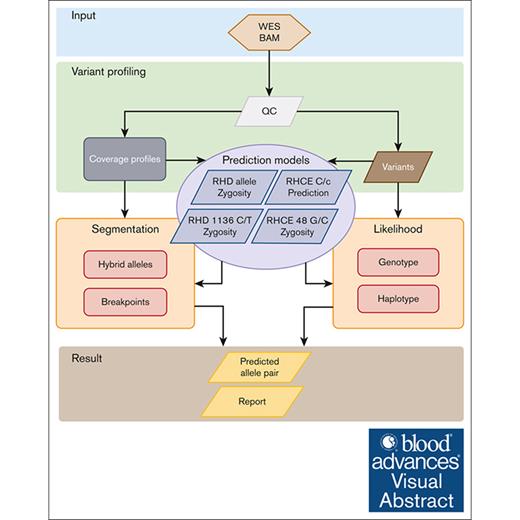
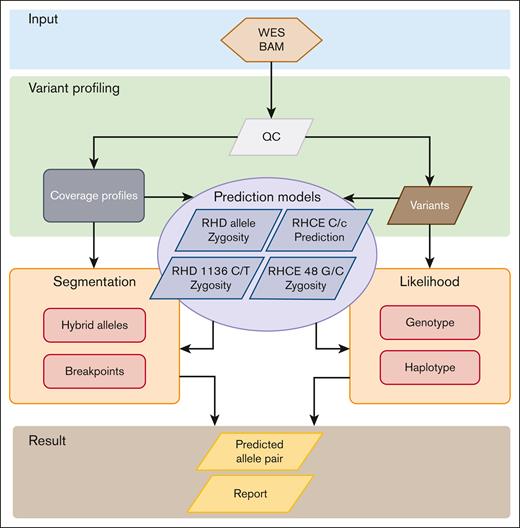
The WES-based RHtyper algorithm was developed according to the WGS-based RH genotyping approach6 with modification for WES data and by adding machine learning models to improve the prediction accuracy (Figure 1). Specifically, the WES-based RHtyper algorithm consists of 4 main steps: (1) variant profiling for SNPs/indels and coverage alterations; (2) predicting RHD zygosity and hybrid alleles, RHD c.1136C>T and RHCE c.48G>C, and the presence of RHCE∗C or RHCE∗c alleles using established machine learning models; (3) refining the hybrid allele and hybrid breakpoint predictions using segmentation; and (4) generating likelihood scores using genotypes and phased haplotype likelihoods to rank candidate allele pairs. Finally, the candidate allele pair with the highest likelihood score was reported as the predicted genotype.
Modification of RHtyper for WES data by adding machine learning. The WES-based RHtyper algorithm consists of 4 main steps: (1) variant profiling of SNPs/indels and coverage alterations. (2) Predicting RHD zygosity and hybrid alleles, RHCE∗C and RHCE∗c, and the zygosity of RHD c.1136C>T and RHCE c.48G>C using machine learning models. (3) Refining hybrid allele and breakpoint predictions using segmentation. (4) Generating likelihood scores using genotypes and phased haplotype likelihoods to rank candidate allele pairs. Finally, the candidate allele pair with the highest likelihood scores is considered as the predicted genotype. BAM, binary alignment map; QC, quality control.
Modification of RHtyper for WES data by adding machine learning. The WES-based RHtyper algorithm consists of 4 main steps: (1) variant profiling of SNPs/indels and coverage alterations. (2) Predicting RHD zygosity and hybrid alleles, RHCE∗C and RHCE∗c, and the zygosity of RHD c.1136C>T and RHCE c.48G>C using machine learning models. (3) Refining hybrid allele and breakpoint predictions using segmentation. (4) Generating likelihood scores using genotypes and phased haplotype likelihoods to rank candidate allele pairs. Finally, the candidate allele pair with the highest likelihood scores is considered as the predicted genotype. BAM, binary alignment map; QC, quality control.
RH variant calling and coverage profiling
Variants were called via the Samtools pileup method17 using WES reads that met predefined read criteria (base read quality ≥15, mapping read quality ≥10, and average read quality ≥15). Counts of A, T, G, and C nucleotides and indels were generated for each exonic position of RHD/RHCE genes. Exonic positions with variant allele-frequency >10% were classified as heterozygous sites. SNPs and indels were annotated subsequently with encoded amino acid changes. RHD/RHCE coverage profiling was performed as previously described, using WES data.6
Construction of machining learning models
The WGS-predicted genotypes served as control references.6 Informative features were selected using the Boruta algorithm (10.18637/jss.v036.i11), based on per-base coverage and variant allele frequency. The selected features were then incorporated to construct XGBoost models for model learning, using 75% of the WES data from the SCCRIP study. The modified RHtyper was next validated using the remaining 25% of the data from the SCCRIP study as well as a second patient cohort, SJLIFE.
This study was approved by the SJCRH institutional review board, and all participants or guardians provided written informed consent.
Results
Uneven sequencing coverage of WES data
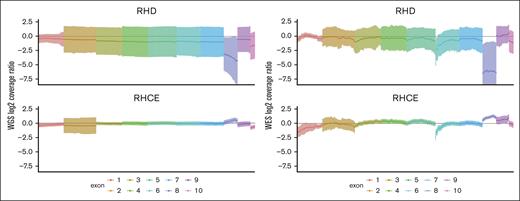
The average RH sequencing coverage for 396 patients with SCD in the SCCRIP cohort was 56.3× for WES compared with 35.7× for WGS. WES coverage demonstrated high regional unevenness, the normalized coverage per RHD exon ranged from –6.09 ± 5.09 to 0.21 ± 1.60 (mean ± standard deviation, “0” representing 2 copies), and the normalized coverage per RHCE exon ranged from –0.80 ± 0.78 to 1.01 ± 0.35 (Figure 2; supplemental Table 1). In contrast, the normalized WGS coverage fluctuated less, ranging from –3.58 ± 3.50 to –0.40 ± 0.85 per RHD exon, and from –0.70 ± 0.33 to 0.35 ± 0.41 per RHCE exon. Notably, RHD coverage varied more than RHCE regardless of sequencing method because RHD and RHCE have identical exon 8, and most sequencing reads from exon 8 align to RHCE, reducing RHD exon 8 coverage markedly. The unevenness of the WES coverage of RH genes affected the prediction of zygosity of alleles and SNPs.
Uneven sequencing coverage of RH genes by WES compared with WGS. Sequencing coverage is normalized by log2 transformation of the ratio between each exon and the average coverage of the sample. A value of “0” represents 2 copies. The exons are differentiated by color. Lines represent the mean sequencing coverage. Shadows represent the standard deviation.
Uneven sequencing coverage of RH genes by WES compared with WGS. Sequencing coverage is normalized by log2 transformation of the ratio between each exon and the average coverage of the sample. A value of “0” represents 2 copies. The exons are differentiated by color. Lines represent the mean sequencing coverage. Shadows represent the standard deviation.
Limitation of RHtyper using WES data
Because RHtyper was initially designed for WGS data analysis, we first modified the algorithm for WES data to not rely on intronic markers for identification. RHCE∗C can be predicted using WGS data with high confidence using a 109-base pair insertion in the RHCE∗C intron 2. Because this intronic region is not covered by WES, RHCE∗C was instead identified by increased coverage of RHD exon 2, because RHCE∗C and RHD exon 2 are identical, and the reads from RHCE∗C typically align with RHD.9 Notably, WES data cannot be used to identify alleles with only intronic variations.
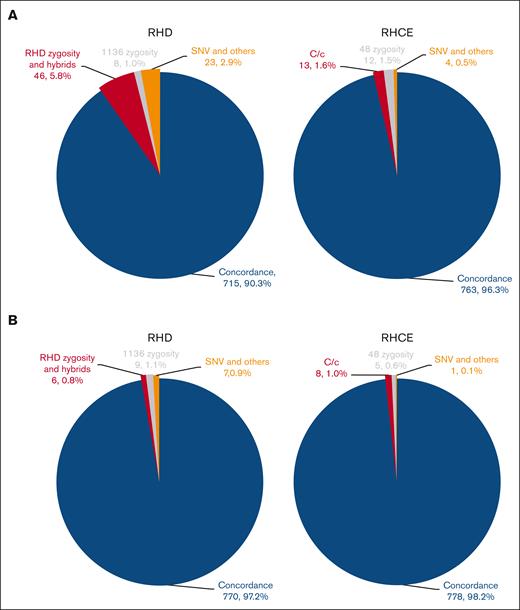
We next determined RH genotypes using WES data for 396 patients with SCD from the SCCRIP cohort, all of whom were Black or African American. The concordance rates between WES and previously reported WGS predictions6 were 90.3% (715/792 alleles) for RHD and 96.3% (763/792 alleles) for RHCE (Figure 3A). Problematic determinations included (1) RHD zygosity and hybrid alleles, (2) RHCE∗C vs RHCE∗c alleles, (3) RHD c.1136C>T zygosity, and (4) RHCE c.48G>C zygosity. RHD zygosity, hybrid alleles, and RHCE∗C were predicted based on sequencing coverage of the whole gene (ie, RHD zygosity) or certain exons of the gene (ie, RHCE exon 4 to 7 for RHD∗03N.01 or RHD∗DIIIa-CEVS(4-7)-D, RHD exon 2 for RHCE∗C), which was less accurate with WES data due to the fluctuated sequencing coverage. RHD c.1136C>T (p. Thr379Met), located in exon 8, is the most common missense RHD SNP in patients with SCD and is the characteristic SNP that defines the RHD DAU cluster.6,19 Because the reference genomic sequence of RHD represents RHD∗10.00 or RHD∗DAU0 with c.1136T, and the conventional RHD shares exon 8 with RHCE with c.1136C, almost all sequence reads from conventional RHD align with RHCE, resulting in reduced coverage of RHD exon 8. To circumvent the skewed coverage of exon 8, RHD c.1136 C>T zygosity was determined for the WGS data by dividing the reads containing the SNP by genome-wide average read coverage rather than position-specific read coverage. However, this approach was no longer reliable with WES data, given the highly variable exome-wide sequencing coverage. RHCE c.48G>C (p. Trp16Cys) resides in exon 1 and is the most common missense RHCE SNP found in patients with SCD.6 In addition, RHCE∗01.01 or RHCE∗ce48C is as common as the conventional RHCE∗01 or RHCE∗ce allele in Black individuls.5,6 The sequences of conventional RHD and RHCE exon 1 are highly homologous, differing only by 1 nucleotide: c.48C for RHD and c.48G for RHCE. Without the paired-mate sequencing reads that cover the surrounding introns to differentiate RHD from RHCE, sequence reads with RHCE c.48G>C frequently misaligned to RHD, resulting in an erroneous G/C fraction and subsequent incorrect allele prediction.
Concordance between WES and WGS predictions for the SCCRIP study. For the 396 patients with SCD enrolled in the SCCRIP study, machine learning increased the overall concordance rates between WES- and WGS-predicted RH genotypes from 90.3% for RHD and 96.3% for RHCE (A) to 97.2% for RHD, 98.2% for RHCE (B). The number and percentage of concordant alleles and various types of discordant alleles are shown in parentheses.
Concordance between WES and WGS predictions for the SCCRIP study. For the 396 patients with SCD enrolled in the SCCRIP study, machine learning increased the overall concordance rates between WES- and WGS-predicted RH genotypes from 90.3% for RHD and 96.3% for RHCE (A) to 97.2% for RHD, 98.2% for RHCE (B). The number and percentage of concordant alleles and various types of discordant alleles are shown in parentheses.
Modification and validation of RHtyper for WES data
Given the low concordance between WES and WGS predictions, we sought to improve RHtyper by incorporating machine learning specific to the problematic alleles and SNPs. The SCCRIP cohort was used for training and validating the machine learning models because (1) all patients in the SCCRIP cohort were of African descent with highly diverse RHD and RHCE genes6 and (2) despite Rh phenotype–matched blood transfusion, patients with SCD are still at high risk for Rh alloimmunization because of the genetic diversity of RH genes and will likely benefit the most from receiving RH genotype–matched blood transfusion.2,5,20 A total of 1547 informative features for RHD zygosity and hybrid alleles, 255 for RHCE∗C vs RHCE∗c allele differentiation, 240 for RHD c.1136C>T, and 253 for RHCE c.48G>C zygosity, were selected to build machine learning models (supplemental Figure 1). The RH genotypes predicted using the WGS data were used as reference genotypes because of their high accuracy.6 We randomly selected 75% of the WES data from the SCCRIP cohort for model training, and the remaining 25% for validation. Machine learning improved the concordance rates between WES and WGS predictions to 98.0% for RHD zygosity and hybrid alleles, 97.0% for RHCE∗C vs. RHCE∗c alleles, 97.0% for RHCE c.48G>C zygosity, and 96.0% for RHD c.1136C>T zygosity. The overall concordance rates for the SCCRIP cohort were 97.2% (770/792 alleles) for RHD and 98.2% (778/792 alleles) for RHCE (Figure 3B; supplemental Tables 2-3). The remaining discrepancies were due to, with substantially fewer numbers though, RHD zygosity and hybrid alleles (6 alleles 0.8% of total 792 RHD alleles), RHCE∗C vs RHCE∗c (8 alleles, 1% of total 792 RHCE alleles; 6 RHCE∗C misidentified as RHCE∗c, 0.8% of total RHCE alleles; 2 RHCE∗c as RHCE∗C, 0.2% of total RHCE alleles), RHCE c.48G>C zygosity (5 alleles, 0.6%), RHD c.1136C>T zygosity (9 alleles, 1.1%), and other SNP discrepancies (RHD, 7 alleles, 0.9%; RHCE, 1 allele, 0.1%).
In the SCCRIP cohort, the RH genotypes of 56 patients were also determined by standard RH genotyping methods of RH SNP array and targeted molecular assays, verified by Sanger sequencing or a second independent NGS as described previously6 and in supplemental Methods. Compared with the known genotypes, the modified RHtyper using WES data achieved 98.2% (110/112 alleles) accuracy for RHD and 94.6% (106/112 alleles) accuracy for RHCE alleles (Table 1). Notably, none of the erroneous predictions would have led to an increased risk of Rh alloimmunization. One erroneous prediction in which patient 1, with “RhC” (RHCE∗02/RHCE∗01.20.01 or RHCE∗Ce/RHCE∗ce733G), was misidentified by WES as “Rhc” (RHCE∗01/ RHCE∗01.20.02 or RHCE∗ce/RHCE∗48C733G) could have resulted in the dispensation of C-negative blood to a C-positive patient unnecessarily.
Discrepancies between known and WES-predicted genotypes in 56 patients with SCD
| Patient . | RH allele . | Known genotypes† . | WES-predicted genotypes . | Confirmation methods . |
|---|---|---|---|---|
| 1 | D | RHD∗01 | Same | ⎼ |
| D | Deletion | Same | ⎼ | |
| CE | RHCE∗02 (RHCE∗Ce) | RHCE∗01 (RHCE∗ce) | Serology | |
| CE | RHCE∗01.20.01 (RHCE∗ce733G) | RHCE∗01.20.02 (RHCE∗ce 48C, 733G) | Serology | |
| 2 | D | RHD | RHD∗10.00 (RHD∗DAU0) | Sanger sequencing |
| D | RHD | RHD∗10.00 or (RHD∗DAU0) | Sanger sequencing | |
| CE | RHCE∗02 or RHCE∗Ce | Same | ⎼ | |
| CE | RHCE∗01.20.01 (RHCE∗ce733G) | RHCE∗01.20.02 (RHCE∗ce 48C, 733G) | Sanger sequencing | |
| 3 | D | RHD∗01 | Same | ⎼ |
| D | Deletion | Same | ⎼ | |
| CE | RHCE∗01 (RHCE∗ce) | An extra RHCE c.105C>T identified | Sanger sequencing | |
| CE | RHCE∗01.20.02 (RHCE∗ce 48C, 733G) | Sanger sequencing | ||
| 4 | D | RHD∗01 | Same | ⎼ |
| D | Deletion | Same | ⎼ | |
| CE | RHCE∗01 (RHCE∗ce) | Same | ⎼ | |
| CE | RHCE∗01 (RHCE∗ce) | RHCE∗01.01 (RHCE∗ce48C) | Sanger sequencing |
| Patient . | RH allele . | Known genotypes† . | WES-predicted genotypes . | Confirmation methods . |
|---|---|---|---|---|
| 1 | D | RHD∗01 | Same | ⎼ |
| D | Deletion | Same | ⎼ | |
| CE | RHCE∗02 (RHCE∗Ce) | RHCE∗01 (RHCE∗ce) | Serology | |
| CE | RHCE∗01.20.01 (RHCE∗ce733G) | RHCE∗01.20.02 (RHCE∗ce 48C, 733G) | Serology | |
| 2 | D | RHD | RHD∗10.00 (RHD∗DAU0) | Sanger sequencing |
| D | RHD | RHD∗10.00 or (RHD∗DAU0) | Sanger sequencing | |
| CE | RHCE∗02 or RHCE∗Ce | Same | ⎼ | |
| CE | RHCE∗01.20.01 (RHCE∗ce733G) | RHCE∗01.20.02 (RHCE∗ce 48C, 733G) | Sanger sequencing | |
| 3 | D | RHD∗01 | Same | ⎼ |
| D | Deletion | Same | ⎼ | |
| CE | RHCE∗01 (RHCE∗ce) | An extra RHCE c.105C>T identified | Sanger sequencing | |
| CE | RHCE∗01.20.02 (RHCE∗ce 48C, 733G) | Sanger sequencing | ||
| 4 | D | RHD∗01 | Same | ⎼ |
| D | Deletion | Same | ⎼ | |
| CE | RHCE∗01 (RHCE∗ce) | Same | ⎼ | |
| CE | RHCE∗01 (RHCE∗ce) | RHCE∗01.01 (RHCE∗ce48C) | Sanger sequencing |
Determined by standard RH genotyping methods of RH SNP array and targeted molecular assays, verified by Sanger sequencing or a second independent NGS or serology.
Further validation of the modified RHtyper in the SJLIFE cohort
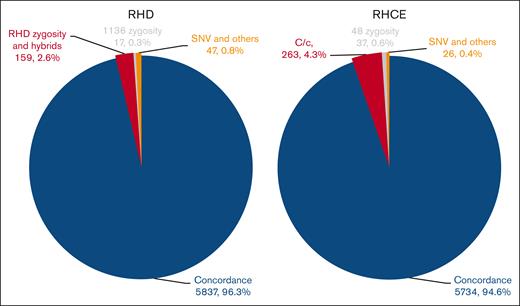
Next, we validated the modified RHtyper in a second available patient cohort, SJLIFE, consisting of 3030 cancer survivors. Among 2716 patients with racial information, 84.6% (2298) were White, 15.2% (413) Black or African American, 0.11% (3) Asian, 0.04% (1) American Indian or Alaska Native, and 0.04% (1) Native Hawaiian or Other Pacific Islander. The concordance rates between WES and WGS predictions were 96.3% (5837/6060 alleles) for RHD and 94.6% (5734/6060 alleles) for RHCE (Figure 4; supplemental Tables 2 and 4). Discrepancies included RHD zygosity and hybrid alleles (159 alleles, 2.6% of 6060 RHD alleles), RHCE∗C vs RHCE∗c differentiation (263 alleles, 4.3% of total 6060 RHCE alleles; 237 RHCE∗C misidentified as RHCE∗c, 3.9% of total RHCE alleles; 26 RHCE∗c as RHCE∗C, 0.4% of total RHCE alleles), RHCE c.48G>C zygosity (37 alleles, 0.6%), RHD c.1136C>T zygosity (17 alleles, 0.3%), and SNPs and other discrepancies (RHD, 47 alleles, 0.8%; and RHCE, 26 alleles, 0.4%). For 1036 patients with blood type information, the predicted RhD serological types using WES data were 99.8% (1034/1036 patients), consistent with the clinical serology results; notably, this comparison only assessed whether the modified RHtyper could correctly predict the presence or absence of RhD. The predicted frequency of C antigen was 65.23% (1499/2298 patients) per WGS and 58.96% (1355/2298 patients) per WES for White patients, and 23.24% (96/413 patients) per WGS and 24.21% (100/413 patients) per WES for Black or African American patients, consistent with the known racial distribution (68% of White people and 27% of Black people).21
Concordance between WES and WGS predictions for the SJLIFE study. The modified RHtyper achieved high concordance rates between the WES– and WGS–predicted RH genotypes of 96.3% for RHD and 94.6% for RHCE in the SJLIFE cohort consisting of 3030 cancer survivors. The number and percentage of concordant alleles and various types of discordant alleles are shown in parentheses.
Concordance between WES and WGS predictions for the SJLIFE study. The modified RHtyper achieved high concordance rates between the WES– and WGS–predicted RH genotypes of 96.3% for RHD and 94.6% for RHCE in the SJLIFE cohort consisting of 3030 cancer survivors. The number and percentage of concordant alleles and various types of discordant alleles are shown in parentheses.
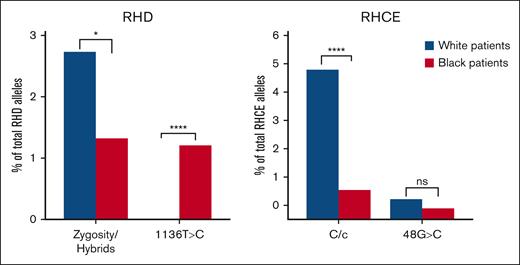
The modified WES-based RHtyper was trained primarily using data from Black or African American patients, whereas most patients in the SJLIFE cohort were White, for whom the frequency of RH variation is ∼1% to 2%.21 Therefore, we compared the concordance rates of White vs Black or African American patients in the SJLIFE cohort (Figure 5). Discrepancies were significantly higher among White patients for RHD zygosity and hybrid alleles (127 alleles or 2.8% of RHD alleles in White patients vs 11 alleles or 1.3% of RHD alleles in Black or African American patients; P = .0157), and RHCE∗C vs RHCE∗c differentiation (227 alleles or 5.0% of RHCE alleles in White patients vs 8 alleles or 1.0% of RHCE alleles in Black or African American patients; P < .0001). In contrast, the discrepancy in RHD c.1136C>T zygosity was significantly higher in Black or African American patients (10 alleles or 1.2% of RHD alleles in Black or African American patients vs 1 allele or 0.02% of RHD alleles in White patients; P < .0001), although the overall number of discrepant alleles was very low.
Discordance rates of trained SNPs/alleles among White and Black or African American patients in the SJLIFE study. ns, not significant; ∗P < .05; ∗∗∗∗P < .001.
Discordance rates of trained SNPs/alleles among White and Black or African American patients in the SJLIFE study. ns, not significant; ∗P < .05; ∗∗∗∗P < .001.
Discussion
The WGS-based RHtyper relies on sequencing coverage profiles to predict the zygosity of alleles and SNPs.6 This approach alone was less accurate for analyzing WES data because of uneven sequencing coverage and misalignment of sequencing reads. To improve the prediction accuracy using WES data, we optimized RHtyper by leveraging machine learning to target the 4 most affected SNPs and alleles: (1) RHD zygosity and hybrid alleles, (2) RHCE∗C vs RHCE∗c alleles, (3) RHD c.1136C>T zygosity, and (4) RHCE c.48G>C zygosity. Machine learning substantially increased the concordance of WES– with WGS–predicted RH genotypes when applied to 2 independent large patient cohorts, SCCRIP and SJLIFE, but a few limitations remained.
Manual or automated genotyping of RHD and RHCE from targeted exome sequencing and WES data has been performed by multiple groups.9,10,22-25 Prediction of RHD zygosity and hybrids and RHCE∗C vs RHCE∗c alleles by sequencing coverage have consistently been difficult with WES data. Schoeman et al reported that the sensitivity to detect a deletion in RHD and RHCE was 89.8%, and only 52.8% for duplications using sequencing coverage alone (n = 28).25 To overcome this limitation, Chou et al and Lane et al determined RHD zygosity using RHCE as a control, because nearly all individuals have 2 copies of RHCE, and RHCE∗C identification was based on decreased read coverage of RHCE exon 2 compared with RHCE exons 1 and 3.9,13 Chou et al reported that the approaches improved the concordance rate to 98% (n = 54).9 Lane et al developed the first automated algorithm for RBC antigen genotyping using WES data.13 Using copy number correction factors calculated from 20 individuals with known RHD zygosity and C/c antigen status to normalize the sequencing coverage of each exon, the authors were able to correctly genotype the remaining 55 individuals. The improvement strategies used by these studies involved creating predetermined rules based on data from a small cohort of individuals. However, this approach may not be comprehensive enough to capture all the necessary information for accurate prediction in a large number of individuals. WES data were also not reliable in predicting RHCE c.48G>C and RHD c.1136C>T owing to the misalignment of sequencing reads.9,25 The algorithm created by Lane et al was able to detect RHD c.1136C>T but could not distinguish homozygous from heterozygous ones.13
We optimized RHtyper for WES data using machine learning. The learning process allows the incorporation of diverse informative features and has been applied to complicated and high-dimensional data, including genomic sequencing data. It enables accurate predictions based on automated data learning rather than simple rule-based classification. In our study, informative features of per-base coverage and variant allele frequency from hundreds to thousands of exonic positions were used to identify the problematic SNPs and alleles. Training with almost 300 patients with SCD from the SCCRIP cohort allowed for the recognition of intricate patterns for accurate prediction. Machine learning markedly improved the concordance rates between WES and WGS predictions to 97.2% for RHD and 98.2% for RHCE in the SCCRIP cohort (n = 396) and 96.3% for RHD and 94.6% for RHCE in the SJLIFE cohort (n = 3030). Using similar machine learning approaches, RHtyper can be extended to analyze other blood group proteins encoded by highly homologous genes, for example, the MNS blood group.
Discordant predictions between WES and WGS remained despite machine learning. RHD zygosity and hybrid alleles, and RHCE∗C vs RHCE∗c alleles contributed to most discrepancies. Discordant RHD zygosity and hybrid allele calling occurred more often in patients with hemizygous RHD deletion or heterozygous RHD hybrid alleles, which require sufficient and even coverage for accurate identification and could remain challenging for certain patients even with machine learning. Because the sequencing coverage of RHD exon 2 is critical in differentiating RHCE∗C from RHCE∗c alleles, we initially suspected that the coverage might be erroneous owing to misalignment mediated by SNPs unique to certain patients. However, a comparison of RHD exon 2 and its surrounding intron sequences (50 bp into the surrounding introns) between patients with and without RHCE∗C vs RHCE∗c discrepancy revealed no SNPs that would have led to misalignment (data not shown). Furthermore, White patients in the SJLIFE cohort were more likely to have discordant predictions of RHD zygosity and hybrid alleles and RHCE∗C vs RHCE∗c alleles than Black or African American patients. The skewed discordance could be due to higher frequencies of D-negative and C-positive status in White (15% and 68%, respectively) than in Black or African American patients (8% and 27%, respectively).21 Racial differences between the training and validation cohorts could also provide an explanation, as all patients in the SCCRIP training cohort were Black or African American and 84.6% of the patients in the SJLIFE validation cohort were White. It is possible that individuals from different races may have slightly different sequencing coverage patterns, or that the informative features used to identify and/or differentiate those alleles vary slightly with race, for which future studies are warranted. Additional training using WES data from White individuals and individuals of other racial and ethical groups is needed to further improve the accuracy of RHtyper.
The clinical implementation of RHtyper may become increasingly relevant as more patients with chronic diseases are being interrogated by WES or WGS. It provides an analysis tool for data that may already exist or be obtained for other clinical care. RH genotyping can enhance transfusion safety by facilitating anti-Rh antibody identification and/or, in some cases, improve prophylactic RBC matching strategies. For example, for patients with the hybrid alleles of RHD∗01N.03 or RHD∗DIIIa-CEVS (4-7)-D, RHCE∗02.10.01 or RHCE∗CeRN, which encode partial C antigen, and no conventional RHCE∗Ce or RHCE∗CE allele, transfusion with C-negative RBCs is recommended to prevent anti-C formation.26 Genotyping blood donors, particularly frequent Black donors who support C/E/K-matched RBCs for patients with SCD, may facilitate RH genotype–matched blood transfusion and improve transfusion safety in the future. RHtyper achieved high concordance rates in 2 large validation cohorts after incorporating the machine learning models but was not 100% accurate. One limitation is that RHtyper may incorrectly predict RHCE∗C and RHCE∗c using WES data. Misidentification of RHCE∗C as RHCE∗c may result in C-positive patients receiving C-negative blood, which would not cause any harm to the patient, but from a resource perspective, it would be a poor allocation of C antigen-negative units. Conversely, the misidentification of RHCE∗C as RHCE∗c in blood donors may result in exposing C-negative recipients to C-positive blood and potential anti-C formation. Therefore, the use of RHtyper for RH genotyping of blood donors would need to be combined with other testing such as standard serologic typing. For clinical application, additional training and validation using samples from multiple racial groups with RH genotypes verified by RH SNP array, Sanger sequencing, and other molecular methods, as well as serological tests, are essential to further optimize RHtyper prediction.
There are limitations to our study. First, we used WGS-predicted genotypes by RHtyper as a reference. This seemed justified, as we previously demonstrated that the WGS-predicted genotypes were highly accurate compared with genotypes verified by multiple molecular methods and serological types for D and C/c antigens.6 Second, the SJLIFE cohort was only serologically typed for D antigen; thus, concordance with C antigen typing was not possible. However, the prevalence of C antigen in White and Black or African American patients derived from WGS and WES data were consistent with known frequencies, indicating that the genotyping results were likely accurate.21
In conclusion, we optimized RHtyper for WES data by adding machine learning to overcome the variable sequencing coverage and misalignment associated with WES data. The optimization improved RH genotyping accuracy and extended the application spectrum of RHtyper to include more widely available WES data.
Acknowledgments
The authors thank the Sickle Cell Clinical Research and Intervention Program and the St. Jude Lifetime Cohort Study teams, as well as the biorepository at St. Jude Children’s Research Hospital for sharing the whole-genome and whole-exome sequencing data and patient samples.
This study was supported by the National Institutes of Health/National Cancer Institute (CA021765), the American Lebanese Syrian Associated Charities (Y.Z.), National Blood Foundation Early-Stage Investigator’s Award (Y.Z.), and the National Institutes of Health/National Heart, Lung, and Blood Institute HL147879-01 (S.T.C. and C.M.W).
Authorship
Contribution: J.S.H., M.J.W., C.M.W., S.T.C., and Y.Z. designed the research; T-C.C. and G.W. developed and modified RHtyper; J.Y. performed confirmatory Sanger sequencing; J.S.H., M.J.W., and Z.W. provided patient samples and sequencing data; and T-C.C., C.M.W., S.T.C., and Y.Z. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yan Zheng, Department of Pathology, St. Jude Children's Research Hospital, 262 Danny Thomas Place, MS 342, Memphis, TN 38105; email: Yan.Zheng@stjude.org.
References
Author notes
Whole-genome and whole-exome sequencing data are available at St. Jude Cloud (https://platform.stjude.cloud/data/cohorts) for the 396 patients with sickle cell disease from the Sickle Cell Clinical Research and Intervention Program study (accession number: SJC-DS-1006), and the 3030 cancer survivors from St. Jude Lifetime Cohort Study (accession number: SJC-DS-1002).18 The RH genotypes of the patients are included in supplemental Tables 3 and 4. The source code and tutorial of RHtyper can be accessed via GitHub (https://github.com/disonchang/RHtyper).
The full-text version of this article contains a data supplement.