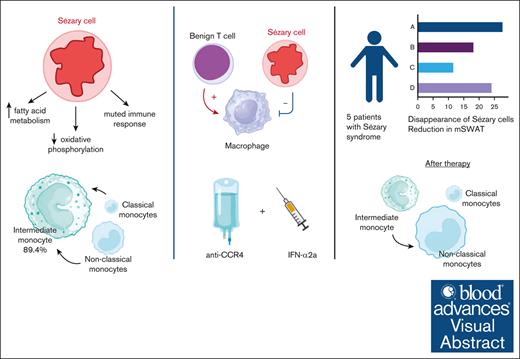
Dysfunctional monocytes with impaired phagocytosis and reduced cytokine response are key in Sézary syndrome.
Combination of anti-CCR4 with interferon-α2a augments phagocytosis of Sézary cells in vitro and in vivo leading to rapid clinical response.
Visual Abstract
Sézary syndrome (SS) is an aggressive leukemic expansion of skin-derived malignant CD4+ T cells. Drug monotherapy often results in disease relapse because of the heterogenous nature of malignant CD4+ T cells, but how therapies can be optimally combined remains unclear because of limitations in understanding the disease pathogenesis. We identified immunologic transitions that interlink mycosis fungoides with SS using single-cell transcriptome analysis in parallel with high-throughput T-cell receptor sequencing. Nascent peripheral CD4+ T cells acquired a distinct profile of transcription factors and trafficking receptors that gave rise to antigenically mature Sézary cells. The emergence of malignant CD4+ T cells coincided with the accumulation of dysfunctional monocytes with impaired fragment crystallizable γ-dependent phagocytosis, decreased responsiveness to cytokine stimulation, and limited repertoire of intercellular interactions with Sézary cells. Type I interferon supplementation when combined with a monoclonal antibody targeting the chemokine receptor type 4 (CCR4), unleashed monocyte induced phagocytosis and eradication of Sézary cells in vitro. In turn, coadministration of interferon-α with the US Food and Drug Administration–approved anti-CCR4 antibody, mogamulizumab, in patients with SS induced marked depletion of peripheral malignant CD4+ T cells. Importantly, residual CD4+ T cells after Sézary cell ablation lacked any immunologic shifts. These findings collectively unveil an auxiliary role for augmenting monocytic activity during mogamulizumab therapy in the treatment of SS and underscore the importance of targeted combination therapy in this disease.
Introduction
Sézary syndrome (SS) is an aggressive late-stage cutaneous T-cell lymphoma (CTCL) driven by circulating clonal malignant CD4+ T cells called Sézary cells. Mycosis fungoides (MF) is the most common form of CTCL, typically presenting with skin patches, plaques, and, in advanced stages, tumors. Unlike SS, which is defined by the presence of malignant T cells in the blood, MF is primarily characterized by skin manifestations and may progress to involve lymph nodes and internal organs. The 5-year overall survival of SS was ∼20% to 30% before 2003,1,2 which has since slightly improved to 30% to 43%.3,4 This raise incites discussions on whether advancements in therapeutic strategies or improved diagnostic precision, potentially introducing a lead-time survival bias, play the major role. Given the elusive nature of a definitive cure, relapse remains a significant challenge, with some cases reemerging within mere weeks.5 Current insights reveal that malignant Sézary cells possess considerable genomic, transcriptional, and functional heterogeneity, making single-drug treatments less effective.6-8 Coupled with the escalating incidence of SS,9 this necessitates an urgent exploration into combination therapies adept at addressing this heterogeneity.
Augmenting host immunity through blockade of checkpoint molecules has had unparalleled success in the treatment of solid tumor malignancies including melanoma,10 head and neck squamous cell carcinomas,11 non–small cell lung cancers,12 and colorectal cancers.13 For T cell–derived malignancies, the therapeutic potential of checkpoint inhibition is still unclear. For instance, in peripheral T-cell lymphoma, the programmed cell death protein 1 (PD-1) blockade with nivolumab induced a 40% overall response rate in a small subset of patients.14 A subsequent phase 2 study with the PD-1 antibody, pembrolizumab, reported a 27% overall response rate among 15 patients with SS.15 Approximately half of the patients with SS in this trial experienced a transient worsening of their erythroderma and pruritus that mostly resolved within 3 months with supportive management. This may be attributed to the paradoxical ability for PD-1 blockade to blunt the Th2 axis, simultaneously promoting the proliferation of Sézary cells.16 In turn, pembrolizumab has been implicated in inducing skin tumor formation in a patient with SS, which is characterized by the presence of large atypical T cells.17 Concerns regarding the tumorigenic potential of PD-1 immunotherapy have been circumvented with the development of mogamulizumab, a monoclonal antibody targeting the chemokine receptor 4 (CCR4). Mogamulizumab depletes T cells enriched for CCR4 in CTCL through antibody-dependent cellular cytotoxicity (ADCC), with a reported 37% objective response rate among patients with SS.18,19
Unraveling the origins of aberrant CD4+ T-cell clones in CTCL would provide a scientific premise for developing combination therapies that promote immune surveillance. MF and SS, despite earlier theories, might not originate from distinct T-cell subsets, as initially believed based on chemokine receptor expression patterns.20 Clinical findings challenge this concept, considering the short-lived responses in MF after electron beam radiation,21 and unique T-cell receptor (TCR) rearrangements can be found in the bone marrow years before being found in clinically evident skin disease.22 With the advent of highly efficient TCR sequencing technologies that allows comparison at each TCR-α/β/γ locus for individual cells, clonotypic overlap was found to be more prevalent between the peripheral blood and skin than between discrete skin lesions on the same patient with MF.23 Additionally, the inception of malignant transformation in T cells appears to be precocious in its timeline.24 This buttresses the proposition that cutaneous lymphoma progression is fueled by sequential infiltration of neoplastic T-cell clones with roots in primitive antecedents.
Here, we applied single-cell messenger RNA (mRNA) analysis in tandem with high-throughput TCR sequencing to identify the transcriptional programming that mediates the phenotypic transformation from smoldering cutaneous T-cell clones to aggressive Sézary cells that drive leukemic disease. Comprehensive immunologic profiling spotlighted an intriguing correlation: as MF and SS progress, there is an accrual of functionally impaired monocytes marked by compromised phagocytosis. This foundational understanding steered our exploration toward a combined therapeutic strategy, integrating type 1 interferon (IFN) to enhance monocytic functionality with mogamulizumab to thwart a key cutaneous homing receptor, presenting a novel treatment paradigm for SS.
Methods
Human participants
Four patients with clinically and histologically confirmed MF (supplemental Table 1) and 5 patients with relapsed/refractory SS (supplemental Table 2) were recruited. These 5 patients with SS were treated with mogamulizumab-kpkc 1 mg/kg IV and pegylated IFN-α2a (PEG–IFN-α2a) 90 μg subcutaneously weekly for 4 weeks, then biweekly. The median age of the patients was 67 years (range, 51-85 years). Body surface area involvement ranged from 45 to 90 (median, 75). Per the Declaration of Helsinki, all patients signed informed consent for blood collection before and 4 weeks after the initiation of the treatment (their samples were collected as a part of Tissue Banking Protocol) (Clinicaltrials.gov identifier: NCT00177268).
Sample processing for single-cell RNA sequencing
Patients with SS donated 15 mL of peripheral blood, and the peripheral blood mononuclear cells were prepared by gradient purification. For MF samples, 1-cm skin punch biopsies were collected, and single-cell suspensions were acquired using the Whole Skin Dissociation Kit (Miltenyi Biotec Inc, Auburn, CA) according to the manufacturer’s protocol.
Custom panel of genes relevant to SS
Conventional mRNA sequencing measures global gene abundance and splicing but is inconsistent for detecting and quantifying rare transcripts.25 To overcome these limitations for the detection of potentially scant genes relevant to the pathogenesis of SS, targeted libraries were used to allow for high-depth sequencing.26 A total of 61 genes were selected27-30 to augment the existing 399-gene BD Rhapsody Human Immune Response Panel. CBLB, FAS, and JUNB have already been present in BD Rhapsody Human Immune Response Panel, so they were not included in our custom panel to avoid duplications. Several transcripts were chosen for genes that had 1 poly-A site with variable coverage (ie, DNMT3A, EHD1, EPHA4, GLI3, ITPR2, PLCG1, RAG2, SGMS1, and TYK2) in order to maximize transcript capture. The custom panel of Sézary cell–specific genes is presented in supplemental Table 4.
Additional methods
Detailed methods of single-cell targeted transcriptome sequencing, targeted single-cell data processing and analysis,31-33 TCR monoclonality analysis,34-37 monocyte/macrophage profiling,38 macrophage isolation and culture, treatment with SS sera, cytokine and chemokine profiling, flow cytometry analysis, and monocyte phagocytosis assay are presented in supplemental Methods.
Statistical analysis
Statistical tests were performed using GraphPad Prism 10. Differences between the 2 groups were compared by the unpaired 2-sample Wilcoxon test, unless otherwise indicated. When >2 groups are simultaneously compared with one another, the 1-way analysis of variance was used with Šidák post-hoc test for multiple pairwise comparisons. The log-rank (Mantel-Cox) test was used for the survival comparisons. Statistical significance was determined as P < .05.
Results
An atlas of peripheral immune cells in MF and SS
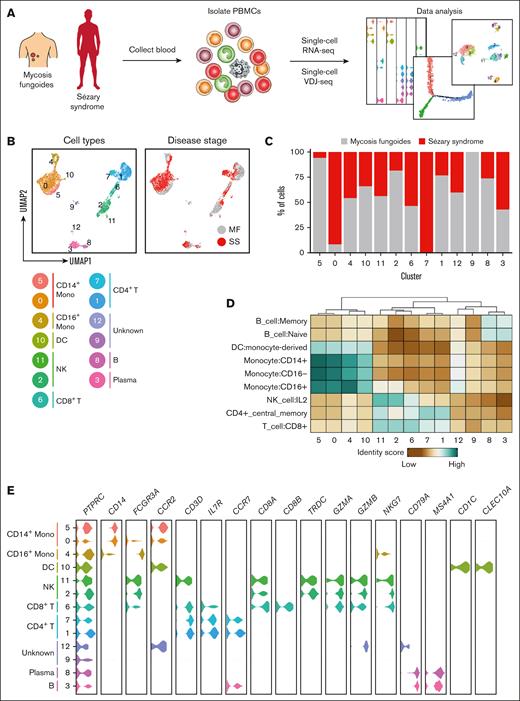
We obtained peripheral blood samples from 4 patients with stage IA or IB MF (clinical characteristics of those patients presented in supplemental Table 1). Peripheral blood mononuclear cells isolated from the blood were subjected to targeted single-cell mRNA sequencing in tandem with variable diversity joining sequencing to calculate clonality and compare clonotype overlap (Figure 1A). Targeted single-cell mRNA sequencing was enriched by genes specific for SS27-30 (supplemental Table 4) allowing for high-depth sequencing to detect and quantify potentially rare transcripts that would be inconsistent with conventional mRNA sequencing.26 To confirm the presence of neoplastic clones in the blood of patients with MF, we also obtained punch biopsy specimens from their affected skin lesions to perform single-cell mRNA and TCR sequencing. Clonal heterogeneity based on TCR-β rearrangement was observed in each cutaneous sample, with at least 1 clonotype shared between the skin and blood (supplemental Figure 1A-B). These shared clonotypes appeared phenotypically similar because no significant differences in enrichment for type 1 T helper (Th1), Th2, or central memory markers were observed among CD4+ T cells with overlapping TCR-β clonotypes between the skin and blood (supplemental Figure 1C). However, within the peripheral blood, TCR-β clonotypes that overlapped with the skin were enriched for T-cell lymphoma genes including CD79A and CD24 when compared with the remaining top 25 most frequent clonotypes (supplemental Figure 1D).39-41 Gene Ontology analysis of genes enriched among peripheral TCR-β clonotypes that overlapped with the skin demonstrated involvement in regulation of leukocyte activation, lymphocyte activation, and regulating cell surface receptor signaling (supplemental Figure 1E). Although it cannot be deduced from these studies whether T cell clones from the blood seeded the skin or vice versa, these shared TCR-β clonotypes nevertheless are transcriptionally distinct from the remaining peripheral CD4+ T cells.
An atlas of peripheral immune cells in MF and SS. (A) Experimental schematic depicting the overall study design. (B) UMAP representations of integrated single-cell transcriptomes derived from the peripheral blood of patients with MF and SS. (C) Bar plots of the proportion of each cluster identified in panel B for patients with MF and SS. (D) Heat map of the identity score for the indicated immune cell type for each cluster identified in panel B. (E) Violin plots of selected genes for multiple immune cell types.
An atlas of peripheral immune cells in MF and SS. (A) Experimental schematic depicting the overall study design. (B) UMAP representations of integrated single-cell transcriptomes derived from the peripheral blood of patients with MF and SS. (C) Bar plots of the proportion of each cluster identified in panel B for patients with MF and SS. (D) Heat map of the identity score for the indicated immune cell type for each cluster identified in panel B. (E) Violin plots of selected genes for multiple immune cell types.
To investigate shifts in the immune cell distribution from MF to SS, peripheral blood was obtained from 5 patients with SS (supplemental Table 2) and processed for tandem single-cell mRNA and TCR variable diversity joining. MF and SS single-cell transcriptomes were integrated with Seurat.42 After gene expression normalization, 13 unique cell subsets and their frequency relative to disease stage were identified and visualized via uniform manifold approximation and projection (UMAP; Figure 1B-C; individual patients’ UMAPs presented in supplemental Figure 2). Cell lineages including monocytes, dendritic cells, natural killer cells, CD8+ and CD4+ T cells, plasma cells, and B cells were identified based on reference transcriptomic data sets using SingleR43 in combination with the expression of known marker genes (Figure 1D-E; supplemental Figure 3). Collectively, this analysis represents the landscape of circulating immune cells in MF and SS.
Transcriptional profile of malignant Sézary cells
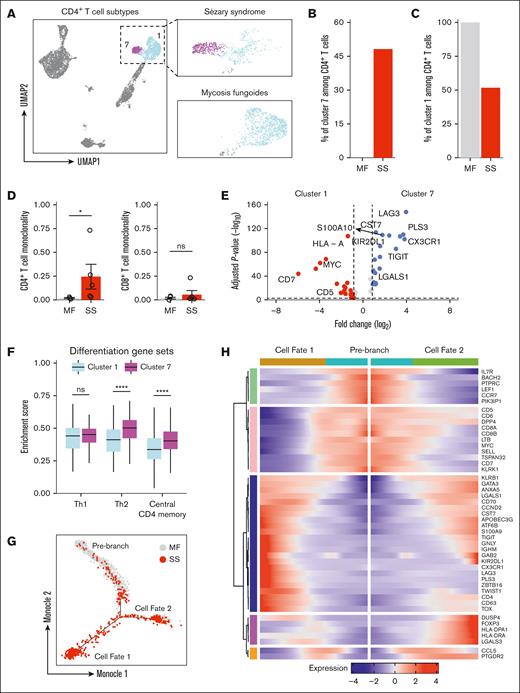
UMAP visualization displayed that all peripheral CD4+ T cells were segregated into 2 populations (Figure 2A). One subpopulation (cluster 7) was only present for patients with SS and accounted for 48.1% of all CD4+ T cells (Figure 2B). Reciprocally, the other CD4+ T-cell subpopulation (cluster 1) represented 100% of all CD4+ T cells among patients with MF (Figure 2C). To quantify T-cell monoclonality in the peripheral blood for patients with MF and SS, a log-normalized Shannon’s entropy of TCR distribution was used.44,45 A monoclonality score of 0 represents a maximally polyclonal population with each TCR represented equally, whereas values close to 1 indicate a repertoire driven by a single dominant clone. CD4+ and CD8+ T cells in the peripheral blood of patients with early-stage MF were nearly uniformly polyclonal, with a monoclonality score nearing 0 (Figure 2D). CD4+ T cells in SS, by contrast, were selectively more clonal, with CD8+ T cells largely unaffected. Thus, the clonal expansion of CD4+ T cells can be detected with TCR sequencing and supports the notion that SS-specific cluster 7 represents this malignant repertoire.
A unique CD4 T-cell subpopulation emerges during SS. (A) UMAP plot demonstrating 2 clusters of CD4 T cells. Cells are color coded by clusters. (B) Bar plots of the proportion of CD4 T cells in cluster 7 from patients with MF and SS. (C) Bar plots of the proportion of CD4 T cells in cluster 1 from patients with MF and SS. (D) Bar plots enumerating monoclonality for CD4 (left) and CD8 (right) in MF and SS. (E) Box plots comparing enrichment scores for the indicated phenotype among cluster 1 and cluster 7 cells. Center line, median; box limits, upper and lower quartiles. (F) Violin plots of selected genes for multiple immune cell types. (G) Single-cell trajectory plot showing lineage relationships among cluster 1 and 7 cells from patients with MF and SS using the Monocle algorithm. (H) Heat map depicting temporal marker expression intensity for significantly branch-dependent genes (Branch Expression Analysis Modeling test; false discovery rate of <1 × 10−10) for the transition identified in panel G. ∗P < .05 by 2-tailed t test. ns, nonsignificant.
A unique CD4 T-cell subpopulation emerges during SS. (A) UMAP plot demonstrating 2 clusters of CD4 T cells. Cells are color coded by clusters. (B) Bar plots of the proportion of CD4 T cells in cluster 7 from patients with MF and SS. (C) Bar plots of the proportion of CD4 T cells in cluster 1 from patients with MF and SS. (D) Bar plots enumerating monoclonality for CD4 (left) and CD8 (right) in MF and SS. (E) Box plots comparing enrichment scores for the indicated phenotype among cluster 1 and cluster 7 cells. Center line, median; box limits, upper and lower quartiles. (F) Violin plots of selected genes for multiple immune cell types. (G) Single-cell trajectory plot showing lineage relationships among cluster 1 and 7 cells from patients with MF and SS using the Monocle algorithm. (H) Heat map depicting temporal marker expression intensity for significantly branch-dependent genes (Branch Expression Analysis Modeling test; false discovery rate of <1 × 10−10) for the transition identified in panel G. ∗P < .05 by 2-tailed t test. ns, nonsignificant.
To investigate the transcriptional programming that identifies Sézary cells, differential gene expression was performed comparing the putative malignant cells (cluster 7) with remaining CD4+ T cells (cluster 1). The complete differential expression results are available in supplemental Table 5. This analysis confirmed that previously identified positive selectors PLS3, CX3CR1, KIR2DL1, TIGIT, LGALS1, CCND2, S100A10, and CD527,46-51 and negative markers including CD2, CD5, CD7, CD44, PTPRC, and SELL46,47,52 could distinguish between normal and malignant cells (Figure 2E; supplemental Figure 4). Additionally, Sézary cells were enriched for BTG1 and TRIB2 that drive drug resistance and oncogenesis in acute lymphoblastic or myelogenous leukemias,53,54 suggesting there may be overlapping pathways that drive disease between leukemias of differing origins.
Metascape analysis55 identified enrichment of genes with annotations related to regulation of leukocyte activation, regulation of cytokine production, adaptive immune response, wingless-related integration site signaling, and NF-κβ signaling in malignant compared with benign CD4+ T cells (supplemental Figure 5). In turn, the Sézary cell cluster 7 was enriched for Th2 markers and central CD4+ memory phenotype and metabolic alterations that promoted fatty acid metabolism with reciprocally decreased oxidative phosphorylation when compared with the remaining CD4+ T-cell cluster 1 (Figure 2F; supplemental Figure 6A). These metabolic shifts coincided with an overall dampening in the inflammatory response and in particular the IFN-α, tumor necrosis factor α (TNF-α), and interleukin-2 (IL-2)/signal transducer and activator of transcription 5 signaling pathways among malignant CD4+ T cells (supplemental Figure 6B).
Upregulation of the coinhibitory molecule, LAG3, along with increased expression of central memory markers and decreased cytokine responsiveness suggested Sézary cells may represent an exhausted or chronic antigen experienced population. To evaluate the transcriptional evolution from nonleukemic to malignant CD4 T cells from MF to SS, we applied Monocle56 that provides robust trajectory inference without any knowledge a priori. Unsupervised trajectory analysis with this approach correctly ordered cells from patients with MF and SS without our input and predicted 1 branch point that gave rise to 2 cell fates (Figure 2G; supplemental Figure 7A). The temporal expression of state-defining genes as cells transition from the prebranch to cell fate 1 or 2 are compiled in Figure 2H. Progressive upregulation of a cluster of genes including FOXP3, DUSP4, and LGALS3 that are each involved in acquisition of regulatory T-cell function57-60 characterized the trajectory from prebranch to cell fate 2. The branch to cell fate 1 was defined by sequential downregulation of CD5, CD7, DPP4, CD8A, CD8B, MYC, and SELL and reciprocal upregulation of TOX, TWIST1, PLS3, LAG3, ZBTB16, KIR2DL1, GATA3, TIGIT, and CX3CR1 that have each been implicated in SS pathogenesis (supplemental Figure 7B). Thus, based on the transcriptional landscape and trajectory analysis, nascent CD4+ T cells in MF and SS exhibit patterns suggesting they could potentially differentiate into either regulatory T cells or adopt a malignant transcriptome. These observations are based on the present data and do not claim to capture the full temporal evolution of these cells.
Accumulation of dysfunctional monocytes in SS
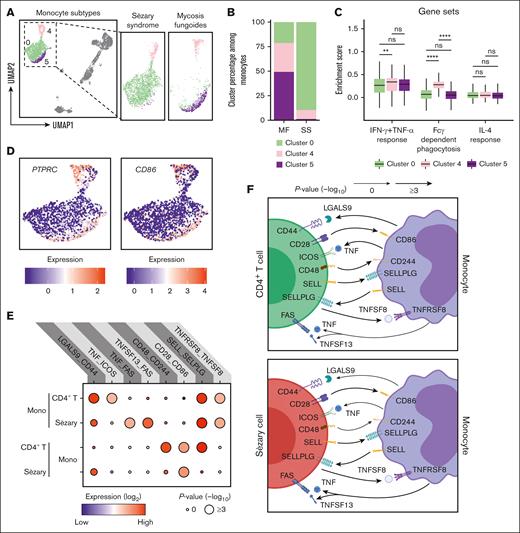
Enhancing monocyte activation represents an attractive innate immune cell target because stimulating phagocytosis can promote eradication of malignant leukemic cells.61,62 The features and functions of monocyte subpopulations were explored. UMAP visualization demonstrated 3 distinct clusters comprising CD14+ (clusters 0 and 5) and CD16+ (cluster 4) monocytes (Figures 1E and 3A). The upregulation of CD14, CD163, CLEC4E, S100A9, and S100A12 suggests that cluster 0 represents both proinflammatory and anti-inflammatory cells, which are commonly referred to as intermediate monocytes. The upregulation of CD36, CD86, RNASE2, CCR2, and ALDH1A1 in cluster 5 suggests that these monocytes belong to the classical monocyte subset. The upregulation of CXCL16, FCGR3A (which encodes for CD16), ADGRE1 (which encodes for EMR1), TNFRSF8, and LYN in cluster 4 suggests that these monocytes belong to the nonclassical monocyte subset. Cluster 0 of intermediate monocytes expanded from 21.2% of all monocytes in MF to 89.4% in SS, whereas cluster 4 of nonclassical monocytes decreased from 29.5% to 9.5%, and cluster 5 of the classical monocytes dramatically reduced from 49.2% to 1.1% (Figure 3B). Cluster 0 of intermediate monocytes that accumulates in SS was relatively deficient in genes related to IFN-γ and TNF-α responsiveness as well fragment crystallizable γ (Fcγ)-dependent phagocytosis (Figure 3C). Interestingly, each subset was comparably responsive to IL-4. In support of a dysfunctional state, cluster 0 of intermediate monocytes harbored low expression of PTPRC, CD86, and IFITM3, with reciprocal upregulation of the IL-1 receptor antagonist, IL1RN (supplemental Figure 8). Using publicly available CITE-seq data (GSE171811),63 we performed a comprehensive analysis of peripheral blood monocytes from healthy controls (HC) and patients with SS. The UMAP projection distinctly highlighted the gene expression divergence between SS and HC monocytes, with SS-derived monocytes showcasing a unique clustering pattern. This variation in clustering is further emphasized by the differential gene expression, with genes like CXCL8, CLEC7A, and CD83 prominently upregulated in SS, contrasting with the heightened expression of LYZ, HLA-DBQ1, and S100A9 in HC monocytes. A subsequent gene ontology enrichment analysis cemented these findings, revealing that although HC monocytes predominantly partake in major histocompatibility complex class 2 processing, NF-κB signaling, and antimicrobial humoral response, SS monocytes demonstrate significant involvement in pathways associated with apoptotic signaling, chemical stress response, and negative regulation of signal transduction. Collectively, in SS, there's a pronounced shift in monocyte populations, with a significant expansion of the intermediate monocytes that show reduced responsiveness to specific immune pathways. These monocytes are distinctively different from their healthy counterparts, highlighting a unique immune landscape inherent to SS.
Emergence of exhausted monocytes with unique intercellular interactions in SS. (A) UMAP plot demonstrating 3 clusters of monocytes. Cells are color coded by clusters. (B) Bar plots enumerating the proportion of monocytes in the indicated cluster for patients with MF and SS. (C) Box plots comparing enrichment scores for the indicated phenotype among cells in the indicated cluster. Center line, median; box limits, upper and lower quartiles. ∗∗P < .01; ∗∗∗∗P < .0001 by 2-tailed t test with Bonferroni multiple comparisons correction. (D) UMAP plots showing the expression of selected surface markers in all monocyte clusters. (E) Dot plot of the predicted interactions between monocytes and the indicated immune cell types. P values are indicated by the circle sizes, as shown in the scale on the bottom right (permutation test). The means of the average expression level of each interacting molecules are indicated by the color. (F) Cartoon depicting the potential receptor/ligand interactions between monocytes and other types of peripheral immune cells. ns, nonsignificant.
Emergence of exhausted monocytes with unique intercellular interactions in SS. (A) UMAP plot demonstrating 3 clusters of monocytes. Cells are color coded by clusters. (B) Bar plots enumerating the proportion of monocytes in the indicated cluster for patients with MF and SS. (C) Box plots comparing enrichment scores for the indicated phenotype among cells in the indicated cluster. Center line, median; box limits, upper and lower quartiles. ∗∗P < .01; ∗∗∗∗P < .0001 by 2-tailed t test with Bonferroni multiple comparisons correction. (D) UMAP plots showing the expression of selected surface markers in all monocyte clusters. (E) Dot plot of the predicted interactions between monocytes and the indicated immune cell types. P values are indicated by the circle sizes, as shown in the scale on the bottom right (permutation test). The means of the average expression level of each interacting molecules are indicated by the color. (F) Cartoon depicting the potential receptor/ligand interactions between monocytes and other types of peripheral immune cells. ns, nonsignificant.
To evaluate how monocytes regulate CD4+ T and Sézary cells, intercellular molecular interactions were analyzed using the accumulated ligand/receptor interaction database CellPhoneDB64 (Figure 3E). Interactions unique to each subset were identified (Figure 3F). TNF and TNFSF13 each binding to FAS was selective for monocytes that interacted with Sézary cells. Benign CD4+ T cells by contrast exhibited unique costimulatory interactions with monocytes involving CD44:LGALS9, CD48:CD244, and ICOS:TNF. Thus, the emergence of malignant CD4+ T cells evidently leads to a shift in functionality of monocyte from costimulatory activated to proapoptotic immunosuppressed.
Type I IFN promotes phagocytosis of Sézary cells via modulation of macrophage activation state in vitro
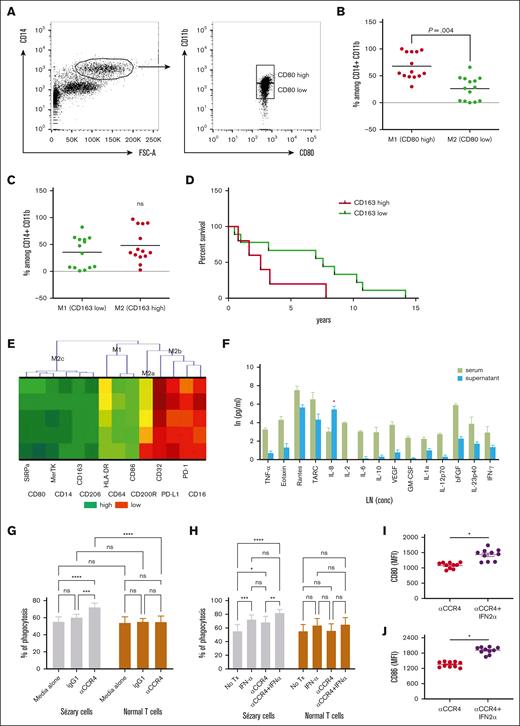
Mogamulizumab, an anti-CCR4 antibody, has recently gained US Food and Drug Administration approval for the treatment of patients with relapsed/refractory MF and SS65 because of its ability to deplete target cells through ADCC.66 However, the observed dysfunctional phenotype of macrophages in SS could impede the efficiency of ADCC. It is challenging to accurately determine macrophage polarization in the blood of patients with SS solely based on CD80 (M1) or CD163 (M2) markers because of their variable expression (Figure 4A-B). Although the expression of CD163, commonly associated with M2 polarization, was variable among patients (Figure 4C), a higher percentage of CD163low macrophages correlated with improved patient survival (Figure 4D). To overcome this, we coincubated healthy monocytes with serum from patients with SS, aiming to investigate the impact of the SS microenvironment. Our results demonstrated a significant shift toward an M2c phenotype in these monocytes, indicating a potential adaptation to the immunosuppressive milieu of SS (Figure 4E). This shift was characterized by the high expression of SIRPα, an inhibitory receptor that interacts with the CD47 “don't eat me” signal on tumor cells, potentially reducing phagocytosis. The elevated production of IL-8 after exposure to patients’ sera, as shown in Figure 4E, suggests that IL-8 might primarily serve as a chemoattractant in the context of SS rather than promoting classical macrophage activation. Next, we sought to determine whether type I IFNs could overcome this functional impairment and enhance mogamulizumab-mediated Sézary cell depletion. In our in vitro assays, we demonstrated that CD14+ macrophages cocultured with Sézary cells engage in phagocytosis of malignant cells upon treatment with anti-CCR4 (Figure 4G). This phagocytic activity was significantly amplified when anti-CCR4 treatment was augmented with IFN-α2a, leading to a marked diminution of Sézary cells as shown in Figure 4H. The additive effect of IFN-α2a with anti-CCR4 not only bolstered the phagocytic capacity of macrophages against Sézary cells but also did not nonspecifically target normal lymphocytes. The upregulation of macrophage activation markers, including CD80 and CD86 (Figure 4I-J), in conjunction with IFN-α2a treatment suggests a shift toward a more activated macrophage state, despite the inherent functional inactivation noted in the patient-derived macrophages. In summary, our data elucidate that type I IFN, particularly IFN-α2a, synergizes with anti-CCR4 to not only enhance the phagocytic capability of macrophages but also to modulate their activation status. This heightened state of activation may potentially counteract the dysfunctional phenotype observed in macrophages derived from patients with SS. Thus, type I IFN could be a crucial adjunct to improve the efficacy of ADCC mediated by mogamulizumab, providing a more robust therapeutic strategy in the treatment of SS.
IFN-α2a enhances macrophage-medicated depletion of Sézary cells by anti-CCR4 antibodies. (A) Gating strategy to identify CD80high and CD80low CD14+CD11b+ cells in the peripheral blood of 14 patients with SS. (B) Percentage of CD80high and CD80low among CD14+CD11b+ cells in the peripheral blood of 14 patients with SS. (C) Percentage of CD163high and CD163low among CD14+CD11b+ cells in the peripheral blood of 14 patients with SS. (D) Overall survival of 14 patients with SS based on the level of CD163 expression on CD14+CD11b+ cells in the peripheral blood. (E) Hierarchical clustering analysis by the 2-way joining of mean fluorescent intensity (MFI) of various differentiation markers on the Mф after exposure to sera of patients with SS in comparison with healthy sera. (F) Cytokine profile of sera of patients with SS and supernatant obtained 7 days after coincubation of healthy Mф with sera from patients with SS. ∗P < .05. (G) Phagocytosis of Sézary cells vs normal T cells in the presence or absence of anti-CCR4 antibody. ∗∗∗P < .001; ∗∗∗∗P < .0001 by analysis of variance (ANOVA) with Šidák multiple pairwise comparisons. (H) Phagocytosis of Sézary cells vs normal T cells after coculture in anti-CCR4 antibody with or without IFN-α2a. ns, nonsignificant; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001 by analysis of ANOVA with Šidák multiple pairwise comparisons. (I-J) Flow cytometry MFI of (I) CD80 and (J) CD86 on macrophages after coculture with Sézary cells in anti-CCR4 antibody with or without IFN-α2a supplementation. ns, nonsignificant; ∗P < .05 by ratio paired t test.
IFN-α2a enhances macrophage-medicated depletion of Sézary cells by anti-CCR4 antibodies. (A) Gating strategy to identify CD80high and CD80low CD14+CD11b+ cells in the peripheral blood of 14 patients with SS. (B) Percentage of CD80high and CD80low among CD14+CD11b+ cells in the peripheral blood of 14 patients with SS. (C) Percentage of CD163high and CD163low among CD14+CD11b+ cells in the peripheral blood of 14 patients with SS. (D) Overall survival of 14 patients with SS based on the level of CD163 expression on CD14+CD11b+ cells in the peripheral blood. (E) Hierarchical clustering analysis by the 2-way joining of mean fluorescent intensity (MFI) of various differentiation markers on the Mф after exposure to sera of patients with SS in comparison with healthy sera. (F) Cytokine profile of sera of patients with SS and supernatant obtained 7 days after coincubation of healthy Mф with sera from patients with SS. ∗P < .05. (G) Phagocytosis of Sézary cells vs normal T cells in the presence or absence of anti-CCR4 antibody. ∗∗∗P < .001; ∗∗∗∗P < .0001 by analysis of variance (ANOVA) with Šidák multiple pairwise comparisons. (H) Phagocytosis of Sézary cells vs normal T cells after coculture in anti-CCR4 antibody with or without IFN-α2a. ns, nonsignificant; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001 by analysis of ANOVA with Šidák multiple pairwise comparisons. (I-J) Flow cytometry MFI of (I) CD80 and (J) CD86 on macrophages after coculture with Sézary cells in anti-CCR4 antibody with or without IFN-α2a supplementation. ns, nonsignificant; ∗P < .05 by ratio paired t test.
Monocyte engagement with coadministration of IFN-α2a and anti-CCR4 in a SS cohort
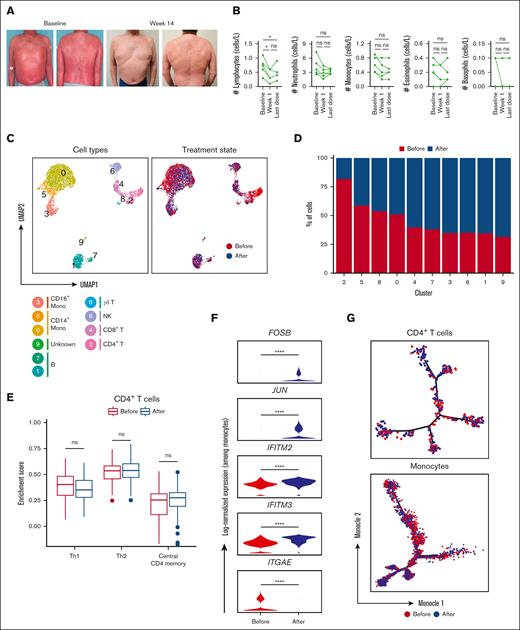
In our preliminary in vitro studies, we established the capacity of IFN-α2a to enhance the mogamulizumab-induced depletion of Sézary cells. Moving from bench to bedside, we assessed the clinical implications of this potentiation. In our cohort, 5 patients with SS, who had previously failed 1.8 ± 2.4 (mean ± standard deviation) therapeutic approaches and had been diagnosed for an average duration of 1.5 ± 0.8 (mean ± standard deviation) years, received weekly treatments combining mogamulizumab and PEG–IFN-α2a. Remarkably, as early as 3 weeks into the combination therapy, complete resolution was evident in the peripheral blood of all patients, corroborated by a negative flow and the absence of a dominant clone per Biomed2 polymerase chain reaction. Concurrently, there was a significant partial response in the skin, with a reduction of ≥50% in mSWAT (modified severity-weighted assessment tool) scores from baseline as soon as 2.9 months of treatment on average (Figure 5A).
Single-cell analysis of immune cells among patients with SS before and after the combination treatment with anti-CCR4 and IFN-α2a. (A) Representative clinical images at baseline and 14 weeks after initiating dual therapy with IFN- α2a and mogamulizumab. (B) Kinetics of peripheral immune cells for the indicated time relative after initiating dual therapy. (C) UMAP representations of integrated single-cell transcriptomes derived from the peripheral blood before and after treatment with anti-CCR4 and IFN-α2a. (D) Bar plots of the proportion of each cluster identified in panel B before and after treatment with anti-CCR4 and IFN-α2a. (E) Box plots comparing enrichment scores for the indicated phenotype among CD4 T cells before and after treatment with anti-CCR4 and IFN-α2a. Center line, median; box limits, upper and lower quartiles. (F) Log-normalized expression of the indicated genes among monocytes before and after treatment with anti-CCR4 and IFN-α2a. (G) Single-cell trajectory plot showing lineage relationships among monocytes before and after treatment with anti-CCR4 and IFN-α2a using the Monocle algorithm. ns, nonsignificant.
Single-cell analysis of immune cells among patients with SS before and after the combination treatment with anti-CCR4 and IFN-α2a. (A) Representative clinical images at baseline and 14 weeks after initiating dual therapy with IFN- α2a and mogamulizumab. (B) Kinetics of peripheral immune cells for the indicated time relative after initiating dual therapy. (C) UMAP representations of integrated single-cell transcriptomes derived from the peripheral blood before and after treatment with anti-CCR4 and IFN-α2a. (D) Bar plots of the proportion of each cluster identified in panel B before and after treatment with anti-CCR4 and IFN-α2a. (E) Box plots comparing enrichment scores for the indicated phenotype among CD4 T cells before and after treatment with anti-CCR4 and IFN-α2a. Center line, median; box limits, upper and lower quartiles. (F) Log-normalized expression of the indicated genes among monocytes before and after treatment with anti-CCR4 and IFN-α2a. (G) Single-cell trajectory plot showing lineage relationships among monocytes before and after treatment with anti-CCR4 and IFN-α2a using the Monocle algorithm. ns, nonsignificant.
This cutaneous clearance was harmoniously accompanied by a marked reduction in circulating lymphocytes, as elucidated in Figure 5B. An interesting facet of this therapy was its impact on the monocyte population. Single-cell transcriptomic analysis was executed 1 month after combination treatment and benchmarked against baseline (supplemental Figure 10). Upon integrating and normalizing gene expression using Seurat, 11 distinct cell subsets were spotlighted, along with their frequency in relation to the treatment timeline, as illustrated via UMAP (Figure 5C-D). Using reference transcriptomic data sets via SingleR43 and assessing known marker genes, we mapped out the cell lineages (supplemental Figure 11).
Notably, the residual CD4+ T cells, after dual therapy, presented no remarkable transcriptional shifts from the pretreatment phase (supplemental Figure 12). The expression spectrum of CD4 T cells, before and after therapy, remained unchanged with respect to Th1, Th2, and central CD4+ memory markers (Figure 5E). Our attention, however, gravitated toward the monocytes. Specific monocyte subpopulations displayed differential proportions after treatment, as detailed in Figure 5D and supplemental Figure 10C. The classic monocytes (cluster 0) maintained a relatively stable proportion after treatment. Cluster 5 overexpresses FCGR3A (which encodes for CD16), CD1C, and CLEC10A. The expression of CD16 (FCGR3A) alongside CD14 suggests that these are intermediate monocytes. The intermediated monocytes showed a decreased proportion after therapy. The cells in cluster 3 (nonclassical monocytes) exhibited an increased proportion after treatment. These findings suggest that the treatment affects specific monocyte subpopulations differently. In patients with SS, there is a notable shift in monocyte subtypes, with an increased proportion of intermediate monocytes and a reduced number of classic monocytes. After treatment with anti-CCR and IFN-α2b, a significant rise in nonclassical monocytes was observed, coupled with a decline in the intermediate monocyte population. A selective group of 5 genes, FOSB, JUN, IFITM2, IFITM3, and ITGAE, associated with adhesion and type I IFN induction, exhibited differential expression patterns before and after therapy (Figure 5F). But intriguingly, there was no detectable alteration in the branch representation for lineage trajectories either before or after the treatment phase (Figure 5G).
In summary, our observations emphasize that the synergy of mogamulizumab and IFN-α2a not only holds promise in eliminating peripheral Sézary cells but also plays a pivotal role in engaging CD16+ monocytes, without causing discernible transcriptional alterations in the residual CD4+ T cells.
Discussion
The intricacies of SS and MF highlight the overarching importance of individualized therapeutic strategies. Through our investigation, we identified distinct metabolic and inflammatory alterations in Sézary cells. These cells display a preference for fatty acid metabolism, reduced oxidative phosphorylation, and a muted inflammatory response, notably within the IFN-α, TNF-α, and IL-2/signal transducer and activator of transcription 5 signaling pathways. The phenotypic and metabolic variations observed in Sézary cells, paired with their trajectory analysis, suggest that a 1-size-fits-all treatment approach might be inadequate. The muted inflammatory response, particularly within the IFN-α pathway, points to the potential of individualized therapies targeting specific immune pathways. Additionally, the potential exhaustion or chronic antigen exposure of Sézary cells necessitates therapies addressing these immune suppressive tendencies.
The pathogenesis of SS has historically been centered around the aberrant behavior of T cells. However, recent investigations have spotlighted the integral role of monocytes, especially in modulating the dynamics between T cells and the microenvironment. Central to our findings is the profound shift in monocyte populations in SS. Leveraging UMAP visualizations, we identified a stark accumulation of intermediate monocytes, which constituted a staggering 89.4% of all monocytes in SS. This expansion appears to be at the expense of classical and nonclassical monocytes, which saw dramatic declines. Delving into their genetic architecture, the intermediate monocytes in SS exhibited a notable deficiency in genes tied to IFN-γ, TNF-α responsiveness, and Fcγ-dependent phagocytosis. This was further underscored by their downregulated expression of PTPRC, CD86, and IFITM3 and the elevation of IL-1 receptor antagonist, IL1RN. A comparison using the CITE-seq data set painted an even more distinct picture: although monocytes from HC were prominently engaged in major histocompatibility complex class 2 processing, NF-κB signaling, and antimicrobial responses, SS-derived monocytes leaned heavily into pathways related to apoptosis and stress response.
This alteration in monocyte behavior has profound implications on their interactions with CD4+ T cells and Sézary cells. Our exploration using the CellPhoneDB revealed that in the presence of benign CD4+ T cells, monocytes engaged in costimulatory interactions. However, when interacting with Sézary cells, they adopted proapoptotic, immunosuppressive functionalities, particularly evident with TNF and TNFSF13 each binding to FAS. Such data suggest that the rise of malignant CD4+ T cells instigates a shift in monocyte behavior from a traditionally activated state to a state that favors immunosuppression.
The integration of mogamulizumab, an anti-CCR4 antibody, into the therapeutic arsenal for SS has presented a promising approach since the US Food and Drug Administration approval for relapsed/refractory MF and SS cases.18,19 Its principal mechanism revolves around depleting target cells via ADCC. However, given our observations of a potentially altered phenotype of monocytes in SS, questions arise about their optimal role in this process. Specifically, PEG–IFN-α2a, which has demonstrated effectiveness in clinical practice,67 particularly when combined with skin-directed therapies, may bolster the phagocytic capabilities of CD14+ macrophages in anti-CCR4 treatment settings. Furthermore, IFN-α2a invoked an activation state in macrophages, evidenced by the upregulated presence of CD80, CD86, and CD163. This combined effect of increased phagocytosis and macrophage activation suggests that the combination of mogamulizumab and IFN-α2a might offer a refined therapeutic strategy for SS.
Building on our in vitro insights regarding the synergy between mogamulizumab and IFN-α2a, the focus was shifted to a clinical setting to ascertain the implications of these bench-side observations. Within the cohort, consisting of 5 patients with SS, a therapeutic regimen combining mogamulizumab and PEG–IFN-α2a was administered. Clinical data revealed a complete disappearance of Sézary cells in peripheral blood by the third week, paralleled by a discernible partial response in cutaneous manifestations, evidenced quantitatively by a reduction in mSWAT scores in the initial months of treatment.
Furthermore, the translational investigation delineated alterations in the monocyte landscape after therapy. Using single-cell transcriptomic analysis, differential proportions in monocyte subpopulations were discerned. Classic monocytes demonstrated a relatively invariant posttreatment proportion. In contrast, a decline in intermediate monocytes was registered, juxtaposed with an increase in nonclassical monocytes. Moreover, differential gene expression patterns were evident, particularly concerning genes such as FOSB, JUN, IFITM2, IFITM3, and ITGAE, which are implicated in adhesion and type I IFN induction. Intriguingly, lineage trajectory analyses before and after therapy did not indicate significant deviations, suggesting that although the combined regimen modulates monocyte subtypes and their genetic transcription profiles, their intrinsic developmental paths remain largely unaffected.
Although the primary emphasis of this investigation centered on monocyte dynamics, it is noteworthy to highlight the observed stability in the transcriptional landscape of residual CD4+ T cells subsequent to therapeutic intervention. The data indicated a static transcriptional profile of these cells, with no remarkable shifts before and after the dual therapy. Specifically, the expression spectrum concerning Th1, Th2, and central CD4+ memory markers remained consistent after therapy. This preservation of the CD4+ T cell transcriptional status, even in the face of significant monocyte modulation, suggests a potential selective specificity of the combined treatment. The fact that the phenotype of other circulating lymphocytes remained largely unaltered under dual modality underscores the therapeutic precision, potentially mitigating off-target effects and paving the way for targeted therapeutic strategies.
In summary, our investigation contributes to the foundational understanding of the interactions between monocytes and CD4+ T cells in SS and offers a preliminary insight into the potential of combining mogamulizumab with PEG–IFN-α2a for therapeutic intervention. By dissecting the metabolic, phenotypic, and transcriptional alterations intrinsic to Sézary cells, we have unveiled potential therapeutic avenues and highlighted the significance of individualized treatment strategies. Although our in vitro results suggest a synergistic effect between mogamulizumab and IFN-α2a, we must exercise caution in translating these findings to clinical practice. The encouraging outcomes observed in our small patient cohort provide a rationale for further investigation but cannot be considered conclusive evidence of clinical efficacy. Recognizing this, we stress the imperative for a comprehensive clinical trial, with proper controls and a larger patient population, to rigorously evaluate the therapeutic benefits, safety, and long-term effectiveness of this combination therapy. Only through such controlled studies can we truly ascertain the value and applicability of this treatment strategy in the broader context of SS management.
Acknowledgments
The authors thank the patients who donated blood samples that made this work possible. The authors also thank the University of Pittsburgh Health Science Sequencing Core at the UPMC Children’s Hospital of Pittsburgh (William A. MacDonald) for sequencing. The authors are grateful to Mac Hopkins (BD Bioscience), Sreejith Ramakrishnan (BD Bioscience), and Ian Taylor (BD Bioscience) for excellent technical assistance in developing a custom Sézary gene panel.
This work was financially supported by the Martin and Dorothy Spatz Foundation and Kyowa Kirin.
Authorship
Contribution: O.K. performed the experiments; T.T.J. performed the data analysis; O.E.A. acquired samples and treated patients with Sézary syndrome; T.T.J. and O.E.A. wrote the manuscript with editorial input from all the authors; and O.E.A. acquired funding for this study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Oleg E. Akilov, Department of Dermatology, University of Pittsburgh, 3708 5th Ave, 5th Floor, Suite 500.68, Pittsburgh, PA 15213; email: akilovoe@upmc.edu.
References
Author notes
The single-cell RNA-sequencing data set was deposited in Gene Expression Omnibus (accession number GSE192836).
The full-text version of this article contains a data supplement.