TO THE EDITOR:
Bruton’s tyrosine kinase (BTK) plays an important role in platelet function downstream of immunoreceptor tyrosine-based activation motif receptors such as collagen receptor glycoprotein VI (GPVI) and C-type lectin-like receptor 2 (CLEC2) and has therefore been proposed as a novel target to prevent thrombosis in patients that are at increased risk.1,2 However, BTK inhibitors that are presently approved for clinical use are associated with bleeding, likely to be owing to off-target effects. In this study, we therefore explored, whether platelet BTK can be selectively targeted using chemical degraders named Proteolysis Targeting Chimeras (PROTACs) that are heterobifunctional molecules that contain an E3 ligand, a linker molecule, and a ligand for the protein of interest. These degraders mediate ubiquitination and proteasomal degradation of target proteins and multiple groups have confirmed the presence of a functional proteasomal system in human platelets.3 We hereby demonstrate that, BTK and TEC can be targeted for proteasomal degradation in human platelets, thereby impairing GPVI-mediated platelet function and primary aggregate formation under flow. Our findings confirm that chemical degraders can be successfully used in anucleate human platelets, cells that are unable to resynthesize proteins or are genetically manipulated, making this is an exciting and promising new research and therapeutic approach to modulate human platelet function and thrombosis.
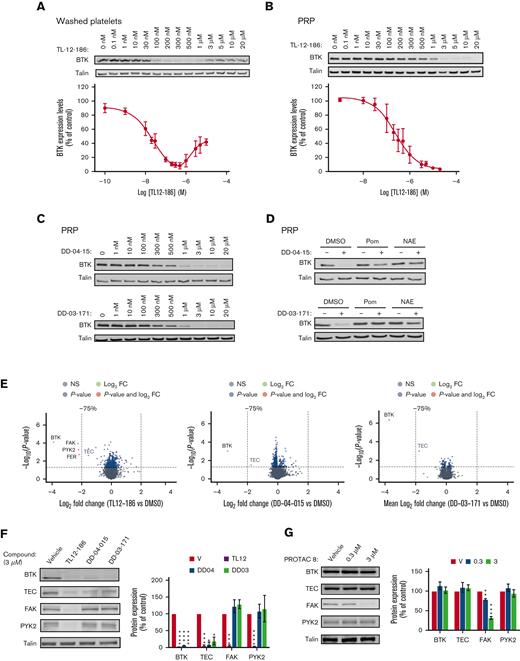
Studies on mouse models with megakaryocyte/platelet specific deletion of BTK, pharmacological inhibitors, and platelets from patients with a functional mutation in BTK (X-linked Bruton agammaglobulinemia), demonstrated an essential role of BTK downstream of immunoreceptor tyrosine-based activation motif receptors such as GPVI and CLEC2, whereas having little effect on GPCR-mediated platelet function.2,4 In addition, BTK contributes to FcRIIa-mediated spontaneous platelet aggregation5 and responses to bacteria.6 Oral BTK inhibitors furthermore impair thrombus formation triggered by plaques and collagen, proposing BTK is a promising target as an antiplatelet drug.7,8 To date, it has not been possible to remove proteins from anuclear human platelets, but the recent development of PROTAC molecules have now made this an exciting feasibility.9 Therefore, in this study, we explored the potential of PROTAC molecules to degrade BTK in human platelets. E3 ligase Cereblon (CRBN), but not von Hippel-Lindau, is expressed in human platelets,10 and CRBN ligands such as thalidomide and pomalidomide were therefore, our initial choice of E3 ligase ligands. The Gray lab developed the multikinase degrader TL12-186, which contains pomalidomide attached via a linker molecule to the versatile ATP-site–directed pharmacophore scaffold TAE684.11,12 This degrader has previously been shown to potently target BTK for degradation in addition to 27 other proteins depending on the cell line used.11 The effect of TL12-186 on BTK expression in human platelets was evaluated by western blotting, demonstrating targeted protein degradation in human platelets (Figure 1A). Incubation of washed platelets with TL12-186 for 4 hours resulted in potent and effective degradation of BTK with maximal degradation reached at 200 to 500 nM (Figure 1A), confirming the following: (1) chemical degraders are effective in human platelets, and (2) BTK is susceptible to protein degradation. Interestingly, higher concentrations of TL12-186 resulted in less effective protein degradation, a “hook effect” phenomenon caused by the three-part system being over-saturated with degrader molecules at higher concentrations.13 Platelet BTK degradation required over 10 times higher concentrations when incubated in platelet-rich plasma for 20 hours (PRP, Figure 1B), suggesting compound binding to plasma components.
PROTAC-mediated, concentration-dependent degradation of target proteins of interest in human platelets. Washed platelets at 4 × 108/ml (A) or platelets in PRP (B-G) were incubated with the indicated concentrations of PROTACs for 4 h/30°C or 20 h/30°C, respectively. Platelets in PRP were pretreated with DMSO, 50 μM Pom, or 10 μM NAE inhibitor for 4 h/30°C before incubation with 10 μM of DD-04-015 or 3 μM of DD-03-171 for 20 h/30°C (D) Platelets in PRP were subsequently washed in CGS and resuspended in Hepes Tyrodes at 4 × 108/ml (B-G). Washed platelets were lysed in either 4× NuPage sample buffer containing 0.5M DTT and subjected to SDS-PAGE/immunoblotting with the indicated antibodies (A-D, F-G) or lysed in RIPA buffer for TMT proteomics analysis (E). Volcano plots of the TMT protein data show the log2 fold change in protein expression of PROTAC-treated platelets compared to vehicle (DMSO) treated controls against the -Log10 (P-value) from a paired t-test (E). Each dot is representative of 1 protein. The grey dots represent proteins with a P-value > .05; blue indicates proteins with a P-value < .05 that are <75% degraded; red indicates proteins with a P-value < .05 that are >75% degraded. Volcano plots were made using the “EnhancedVolcano” R package. TEC is indicated in grey as it did not pass the analytical software internal threshold for TMT quantification (E). Western blot analysis confirmed the TMT proteomics data (F). The graphs represent quantification of the results using Odyssey Licor software, expressed as percentage of vehicle control ± SEM (A,B,F-G). Curves (A,B) were fitted by non-linear regression using GraphPad Prism 9. The statistical analysis of (F,G) was performed using a repeated measures two-way ANOVA or mixed-effects analysis, ∗P ≤ .05, ∗∗P ≤ .01, ∗∗∗ P ≤ .005, and ∗∗∗∗ P ≤ .001. Shown are representative blots and graphs (n = 3-4; A:3, B:3, C:4, D:3, E:3, F:3, G:4). DMSO, dimethyl sulfoxide; PRP, platelet-rich plasma; Pom, pomalidomide; NAE, NEDD8- activating enzyme.
PROTAC-mediated, concentration-dependent degradation of target proteins of interest in human platelets. Washed platelets at 4 × 108/ml (A) or platelets in PRP (B-G) were incubated with the indicated concentrations of PROTACs for 4 h/30°C or 20 h/30°C, respectively. Platelets in PRP were pretreated with DMSO, 50 μM Pom, or 10 μM NAE inhibitor for 4 h/30°C before incubation with 10 μM of DD-04-015 or 3 μM of DD-03-171 for 20 h/30°C (D) Platelets in PRP were subsequently washed in CGS and resuspended in Hepes Tyrodes at 4 × 108/ml (B-G). Washed platelets were lysed in either 4× NuPage sample buffer containing 0.5M DTT and subjected to SDS-PAGE/immunoblotting with the indicated antibodies (A-D, F-G) or lysed in RIPA buffer for TMT proteomics analysis (E). Volcano plots of the TMT protein data show the log2 fold change in protein expression of PROTAC-treated platelets compared to vehicle (DMSO) treated controls against the -Log10 (P-value) from a paired t-test (E). Each dot is representative of 1 protein. The grey dots represent proteins with a P-value > .05; blue indicates proteins with a P-value < .05 that are <75% degraded; red indicates proteins with a P-value < .05 that are >75% degraded. Volcano plots were made using the “EnhancedVolcano” R package. TEC is indicated in grey as it did not pass the analytical software internal threshold for TMT quantification (E). Western blot analysis confirmed the TMT proteomics data (F). The graphs represent quantification of the results using Odyssey Licor software, expressed as percentage of vehicle control ± SEM (A,B,F-G). Curves (A,B) were fitted by non-linear regression using GraphPad Prism 9. The statistical analysis of (F,G) was performed using a repeated measures two-way ANOVA or mixed-effects analysis, ∗P ≤ .05, ∗∗P ≤ .01, ∗∗∗ P ≤ .005, and ∗∗∗∗ P ≤ .001. Shown are representative blots and graphs (n = 3-4; A:3, B:3, C:4, D:3, E:3, F:3, G:4). DMSO, dimethyl sulfoxide; PRP, platelet-rich plasma; Pom, pomalidomide; NAE, NEDD8- activating enzyme.
Because our data demonstrated targeted BTK degradation, we next incubated PRP with the selective BTK degraders; DD-04-1511 and DD-03-171.14 These degraders contain the reversible BTK inhibitors RN486 and CGI-1746 as warheads and are linked to pomalidomide and thalidomide, respectively. Platelets were washed, lysed, and BTK expression levels were analyzed by western blotting. BTK was degraded with similar degradation potency profile to TL12-186 (Figure 1C and supplemental Figures 1A-B for details). Pomalidomide and NEDD8-activating enzyme (NAE) inhibitor largely abolished PROTAC-mediated BTK proteolysis by DD-04-015 and DD-03-171 (Figure 1D), confirming that BTK is targeted for degradation through the CRBN-mediated ubiquitin proteasomal pathway in human platelets. Blood was obtained from healthy, drug-free volunteers according to local NHS research ethics (20/SC/0222) and the University of Bristol approval. The study was conducted in accordance with the Declaration of Helsinki.
To evaluate the specificity of BTK degradation, we implemented a multiplexed quantitative proteomic approach employing TMT. This method allows an unbiased assessment and comparison of the range of targets, and the extent of degradation of the proteins of interest by the BTK degraders. TL12-186 selectively degraded BTK/TEC, focal adhesion kinase (FAK)/protein tyrosine kinase 2-beta (PYK2), and FER, demonstrating that these tyrosine kinases can be targeted for PROTAC-mediated degradation in human platelets (Figure 1E, supplemental Figure 2, and supplemental Table1). Interestingly 4 of these kinases were also degraded by TL-12-186 in the leukemic cell line MOLM-14, but not MOLM-4.11 The BTK degraders DD-04-015 and DD-03-171 had remarkable selectivity, with strong degradation of its primary target BTK and TEC, but limited effect on other platelet proteins detected (Figure 1E, supplemental Table 1). Western blotting further confirmed our proteomics results with BTK and TEC being degraded by all compounds, whereas FAK and PYK2 were targeted by TL12-186 only (Figure 1F). The high specificity of the CRBN targeted compounds may be explained by the anucleate nature of human platelets, avoiding changes in protein levels associated with degradation of neosubstrate transcription factors.14 The PROTAC approach can also be applied to human platelet proteins other than BTK, because the FAK PROTAC-8,15 based on the FAK inhibitor defactinib linked to E3-ligase ligand thalidomide, selectively degraded FAK, but not BTK and TEC, in human platelets (Figure 1G).
Having demonstrated successful degradation of BTK and TEC in human platelets by DD-04 to 015, we next evaluated collagen-related peptide (CRP)-mediated integrin αIIbβ3 activation, P-selectin expression (a marker of α-granule secretion), and phosphatidyl serine (PS) exposure and compared this to the extent of BTK proteolysis. Figure 2A demonstrates that degradation of BTK by DD-04 to 015 led to a concentration-dependent inhibition of CRP-mediated integrin αIIbβ3 activation, α-granule secretion, and PS exposure. Interestingly, PAR-1–mediated platelet function was unaffected by the TL12 to 186 compound and the BTK degraders (Figure 2B,C), confirming that BTK degradation targets the GPVI pathway and not the PAR1 pathway, in agreement with functional data of platelets from patients containing a functional mutation in BTK.4 In contrast, FAK degradation had no effect on CRP and PAR-mediated integrin αIIbβ3 activation and P-selectin expression (Figure 2B,C). The BTK inhibitor CGI-1746 (warhead of DD-03-171) and ibrutinib reduced CRP-mediated platelet function, but not PAR-1–mediated platelet function, with CGI-1746 being the least potent in inhibiting CRP-mediated platelet function (Figure 2D). Lastly, BTK degradation leads to impaired primary platelet aggregate formation under flow on a collagen-coated surface to a similar extent as the BTK inhibitors CGI-1746 and ibrutinib (Figure 2E). Control studies demonstrated that overnight incubation did not affect platelet functionality (supplemental Figure 3). Together, our data shows highly selective degradation of platelet BTK and TEC by protein degraders. Future studies will focus on optimization of the following: (1) BTK PROTAC design to prevent degradation of TEC, potentially avoiding bleeding; (2) experimental conditions to enhance potency and efficacy of target degradation; and (3) platelet selectivity. Because the PROTAC field is developing rapidly, the latter may ultimately be achieved by targeting a platelet specific E3 ligase, and/or use of a platelet specific delivery vehicle. In summary, to the best of our knowledge, we demonstrate for the first time the enormous potential of chemical degraders in modulating platelet function. With the PROTAC field growing exponentially and the inability of platelets to resynthesize proteins, PROTAC-mediated protein degradation is an exciting and promising therapeutic approach to modulate platelet function and thrombosis.
The effect of PROTACs upon platelet functionality and thrombi formation. Platelets in PRP were incubated with vehicle (DMSO) or the indicated concentration of PROTAC TL12-186, DD-04-015, and DD-03-171 at 3 μM for 20 h/30°C (A-C,E). Alternatively, PRP was incubated with the indicated inhibitors for 10 min (D,E). Platelets were washed and resuspended at 2 × 107/ml for FACS analysis (A-D). Washed platelets were stimulated with 2 μg/ml CRP-XL (A-D) or 10 μM TRAP-6 (B-D) for 10 minutes in the presence of PAC1-FITC/CD62P-PE to assess integrin αIIbβ3 activation and P-selectin expression, respectively. Alternatively, platelets were stimulated by a combination of 2 μg/ml CRP and 1 U/ml thrombin in the presence of Annexin-V-488 fluorescent dye to measure levels of PS exposure (A). Results are expressed as mean % of maximal normalized signal intensity ± SEM, n = 4 (A-D). For in vitro flow thrombosis experiments PRP was treated with vehicle (DMSO), DD-03-171, CGI-1764, and ibrutinib and recombined with the red cell layer of fresh blood from the same donor. Whole blood was then flown over a collagen Vena8 GCS Cellix biochip using a laminar flow rate of 3.07 ml/h for a sheer rate of 1000/s. Statistical analysis was performed using a repeated measures two-way ANOVA, ∗P ≤ .05, ∗∗P ≤ .01 (B-D); or two-way ANOVA, ∗∗∗∗P < .0001, ∗∗∗P ≤ .001 (E). Shown are representative blots and graphs (n = 3-4; A:4, B:4, C:4, D:4, E: 3). Max, maximum; PAR-1, protease activated receptor-1.
The effect of PROTACs upon platelet functionality and thrombi formation. Platelets in PRP were incubated with vehicle (DMSO) or the indicated concentration of PROTAC TL12-186, DD-04-015, and DD-03-171 at 3 μM for 20 h/30°C (A-C,E). Alternatively, PRP was incubated with the indicated inhibitors for 10 min (D,E). Platelets were washed and resuspended at 2 × 107/ml for FACS analysis (A-D). Washed platelets were stimulated with 2 μg/ml CRP-XL (A-D) or 10 μM TRAP-6 (B-D) for 10 minutes in the presence of PAC1-FITC/CD62P-PE to assess integrin αIIbβ3 activation and P-selectin expression, respectively. Alternatively, platelets were stimulated by a combination of 2 μg/ml CRP and 1 U/ml thrombin in the presence of Annexin-V-488 fluorescent dye to measure levels of PS exposure (A). Results are expressed as mean % of maximal normalized signal intensity ± SEM, n = 4 (A-D). For in vitro flow thrombosis experiments PRP was treated with vehicle (DMSO), DD-03-171, CGI-1764, and ibrutinib and recombined with the red cell layer of fresh blood from the same donor. Whole blood was then flown over a collagen Vena8 GCS Cellix biochip using a laminar flow rate of 3.07 ml/h for a sheer rate of 1000/s. Statistical analysis was performed using a repeated measures two-way ANOVA, ∗P ≤ .05, ∗∗P ≤ .01 (B-D); or two-way ANOVA, ∗∗∗∗P < .0001, ∗∗∗P ≤ .001 (E). Shown are representative blots and graphs (n = 3-4; A:4, B:4, C:4, D:4, E: 3). Max, maximum; PAR-1, protease activated receptor-1.
Acknowledgments: The authors thank the healthy blood donors within the Faculty of Life Sciences, University of Bristol, for their generous blood donations and Nathanael S. Gray (Chemical and Systems Biology, Chem-H, Stanford Cancer Institute, Stanford Medicine, Stanford University, Stanford, CA) and Graig Crews and John Hines (Department of Molecular, Cellular and Developmental Biology, Yale Science Building, Yale University, New Haven, CT) for generously providing DD-04-1511 and FAK PROTAC-815, respectively.
This work was supported by the National Centre for the Replacement, Refinement and Reduction of Animals in Research, Bristol Alumni and the British Heart Foundation (grant RG/15/16/31758, FS/16/27/32213, PG/16/3/31833, PG/16/21/32083, FS/17/60/33474, SP/F/21/150023, AA/18/1/34219). The graphical abstract was generated using resources provided by BioRender.com.
Contribution: I.H., S.F.M., and V.K.A. conceptualized the study; I.H. and K.J.H. curated the data; J.S.T., A.M., K.M.S., J.V, and L.J.G. worked on the formal analysis; I.H. and V.K.A. acquired funding for the study; J.S.T., A.M., K.M.S., J.V., M.L.J., and I.H. performed the investigation for the study; J.S.T., A.M., K.M.S., S.F.M., and B.N. contributed to the study methodology; I.H. led and supervised the study; I.H. wrote the original draft; and J.S.T., K.M.S, L.J.G., A.W.P., B.N., V.K.A, and I.H. wrote, reviewed, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ingeborg Hers, School of Physiology, Pharmacology and Neuroscience, Biomedical Sciences Building, University of Bristol, Bristol BS8 1TD, United Kingdom; e-mail: i.hers@bristol.ac.uk.
References
Author notes
∗J.S.T., A.M., and K.M.S. contributed equally to this study.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (accession number PXD034440).
Data are available on request from the corresponding author, Ingeborg Hers (i.hers@bristol.ac.uk).
The full-text version of this article contains a data supplement.