Key Points
CD11c is highly expressed in the intracellular components of developing neutrophils.
CD11c regulates the survival and proliferation of developing neutrophils.
Abstract
Sepsis continues to be associated with high morbidity and mortality. Currently, sepsis is managed only conservatively. In sepsis, a substantial number of neutrophils is required, leading to accelerated neutrophil production. Immature neutrophils are released into the circulation to meet a demand, despite their less effective functioning in microbial eradication. Although an intervention to provide more mature neutrophils may serve as a potential sepsis treatment, the mechanism of neutrophil differentiation and maturation remains poorly understood. We discovered that CD11c, traditionally known as a dendritic cell marker, was expressed in neutrophils and regulated neutrophil maturation and effector functions. In the absence of CD11c, neutrophil maturation was impaired in the bone marrow, concomitant with a significant increase in the proliferation and apoptosis of preneutrophils, associated with less effector functions. Under lipopolysaccharide challenge, inducing an emergent neutrophil production in the bone marrow, CD11c deficiency exaggerated the release of immature neutrophils into the circulation, associated with a significant proliferation and apoptosis of preneutrophils. In contrast, constitutively active CD11c knock-in mice showed accelerated neutrophil maturation associated with enhanced effector functions, which further supports the notion that CD11c regulates neutrophil maturation. Furthermore, the constitutively active CD11c knock-in mice offered enhanced bacterial eradication. Taken together, we discovered that CD11c was critical for the regulation of neutrophil maturation, and CD11c activation could serve as a potential target for sepsis treatment.
Introduction
Sepsis continues to be associated with a high mortality of between 20% and 30% in both adults and children,1-5 with an annual cost of $20.3 billion, accounting for nearly a fifth of the total aggregate costs in all hospitalizations in the United States.6 Furthermore, sepsis incidence is expected to increase by 1.5% annually.7,8 Current sepsis management is supportive; numerous clinical trials to modulate inflammation have been unsuccesful.9 Although timely goal-directed initiation of supportive care has largely been advocated by the Surviving Sepsis campaign, the benefit of this approach has been questioned.10,11 Thus, developing therapeutic interventions for sepsis is an urgent task.
Neutrophils are innate immune cells that eradicate microbes via various effector functions as a first-line defense. Among leukocytes, neutrophils are remarkably short lived, with a circulating half-life of 6 to 8 hours.12 In a steady state, mature neutrophils are constantly produced in the bone marrow (BM) from hematopoietic stem cells through a series of intermediate progenitors and precursors and are then released into the circulation. In a sepsis setting, exaggerated neutrophil production, called emergency granulopoiesis, is induced,13 and not only mature neutrophils but also immature neutrophils are released into the circulation to meet the demand. However, immature neutrophils have been associated with reduced antimicrobial defense functions, leading to inferior host defense in patients with sepsis.14,15 Securing an adequate number of mature neutrophils for microbial eradication seems to be a promising intervention for sepsis; understanding the mechanism of how neutrophil maturation is controlled is necessary for the development of such an intervention. We, unexpectedly, identified that CD11c, a β2 integrin of the leukocyte adhesion molecule integrin family, intrinsically regulated neutrophil maturation. In a lipopolysaccharide (LPS) sepsis model, CD11c deficiency led to impaired neutrophil maturation and the release of significantly more immature neutrophils with less effector functions into the circulation. In contrast, constitutive activation of CD11c led to accelerated neutrophil maturation and the release of mature neutrophils both in a steady state and in sepsis. Furthermore, the constitutive activation of CD11c was associated with better bacterial eradication, suggesting that CD11c could be a potential druggable target for better host defense.
Material and methods
Mice
CD11a knockout (KO),16 CD11b KO, and CD11d KO mice were purchased from the Jackson Laboratory. C57BL/6J mice (wild type [WT]) were also purchased from the Jackson Laboratory. CD11c KO mice were kindly donated by Christie Ballantyne (Baylor University). CD11c I334G knock-in (KI) mice were generated by CRISPR-Cas9–based point mutagenesis by Biocytogen Co Ltd (Wakefield, MA). Animals were housed under specific pathogen-free conditions, with 12-hour light and dark cycles. All the animal protocols were approved by the Institutional Animal Care and Use Committee at Boston Children’s Hospital.
BM neutrophil isolation
BM cells were flushed from femurs. After red blood cell lyses, BM cells were fractionated by Percoll gradient (63% and 85%) centrifugation (400g, 30 minutes). The interface was collected and washed as total neutrophils with a purity of >90%. To compare protein expression, reactive oxygen species (ROS) generation, and phagocytosis between genotypes, immature and mature neutrophils from BM cells were also sorted by FACSAria system (BD Bioscience) and verified by Giemsa staining.
Cytospin and Giemsa staining of BM neutrophils
BM neutrophils were purified using a Ficoll-Paque gradient, spun onto glass slides using a Cytospin Cytocentrifuge kit (Thermo Fisher Scientific, Waltham, MA), and dried for 20 minutes. Slides were stained using a Hema 3 staining kit (Thermo Fisher Scientific).
Fluorescent microscopy of BM neutrophils
BM neutrophils were enriched by Percoll gradient (63% and 85%) centrifugation (400g, 30 minutes). Cytospins and slides of neutrophils were fixed with 2% paraformaldehyde/phosphate-buffered saline (PBS); permeabilized with 0.2% Triton-100 for 30 minutes; blocked with 5 μg/mL FcR blocker (2.93, Biolegend), 2% fetal calf serum (FCS), and 10% normal donkey serum in 0.2% Triton-100/PBS for 1 hour; stained with rabbit anti-human/mouse CD11c (1:40 diluted in 0.2% Triton-100/PBS containing 5% FCS; PA5-90208 Invitrogen) and goat anti-human/mouse myeloperoxidase (MPO) (1:40 diluted in 0.2% Triton-100/PBS containing 5% FCS; AF3667, R&D system) at 4°C, overnight; washed; and incubated for 1 hour with NL-493-donkey anti-rabbit antibody and NL-557-donkey anti-goat antibody, both diluted at 1:200 in buffer containing 2% FCS and 10% donkey serum in 0.2% Triton-100/PBS. The slides were washed and mounted with 4’,6-diamidino-2-phenylindole–labeled mounting medium (Vector Laboratories, Burlingame, CA) and evaluated using a Olympus IX81 Motorized Inverted research microscope system.
Electron microscopy of BM neutrophils
BM neutrophils were purified using Ficoll-Paque gradient and subjected to fixation using 2.5% glutaraldehyde, 1.25% paraformaldehyde, and 0.03% picric acid in 0.1 M sodium cacodylate buffer (pH 7.4). Cells were washed in 0.1 M cacodylate buffer and postfixed with 1% osmium tetroxide/1.5% potassium ferrocyanide. Cells were further washed with water and maleate buffer and then incubated in 1% uranyl acetate. Cells underwent dehydration in grades of alcohol and were then placed in propylene oxide. Samples were embedded in TAAB Epon and polymerized at 60°C for 48 hours. Ultrathin sections (60 nm slice) were cut on a Reichert Ultracut-S microtome, picked up onto copper grids stained with lead citrate and examined in a JEOL 1200EX transmission electron microscope or a TecnaiG2 Spirit BioTWIN, and images were recorded with an AMT 2k charged-couple device camera.
Flow cytometry
BM cells were isolated from mice, blocked with Fc receptor blocker (anti-mouse CD16/32 antibody, clone 93), and stained at 4°C with various antibodies. Lineage markers for mouse BM neutrophil maturation study include anti-mouse (m)CD3 (145-2C11), anti-mB220 (RA3-6B2), anti-mCD115 (AFS98), anti-mNK1.1 (PK136), and anti-mSiglecF (ES22-10D8, Miltenyi; Bergisch Gladbach, Germany); the other anti-mouse antibodies include anti-mCXCR2(SA045E1), anti-mCXCR4 (L276F12), anti-mc-Kit (2B8), anti-mGr1 (RB6-8C5), anti-mLy6G (1A8), anti-mCD11b (M1/70), anti-mCD11c (N418), and its isotype control hamster IgG1 (A19-3). BM neutrophils were stained with the aforementioned lineage markers, CD115, SiglecF, c-kit, CXCR2, and CXCR4. Preneutrophils were identified as Lin− Gr-1+CD11b+CXCR4+c-kitintCXCR2−, immature neutrophils were identified as Lin−Gr-1+CD11b+CXCR4−/lowc-kitlow/−CXCR2−, and mature neutrophils were identified as Lin−Gr-1+CD11b+CXCR4−/lowc-kitlow/−CXCR2+.17,18 Human peripheral blood neutrophils were identified by forward/side scatter and the anti-human (h) CD15 (W6D3). The other anti-human antibodies include anti-hCD11b (M1/70), anti-hCD11c (Bu15), and its isotype control (MG2b-57). If not specified, the antibodies were purchased from Biolegend (San Diego, CA). For intracellular staining, cells were permeabilized and stained intracellularly with fluorochrome-conjugated antibodies using fixation/permeabilization reagents and protocols from BD Bioscience (Franklin Lakes, NJ). For CD11c intracellular staining, we saturated cell surface CD11c (if any) with unlabeled anti-CD11c monoclonal antibody (N418), and then fixed, permeabilized cells, and stained the intracellular CD11c by fluorescence-labeled anti-CD11c antibody. For detection of phospho-Akt and phospho-Erk1/2, cells were fixed at 37°C for 10 minutes by Fix Buffer 1 (cat: 557870), and permeabilized by Perm Buffer III (cat: 558050), and then stained with isotype control (clone: MOPC-21) or anti-human/mouse Akt pS473 (clone: M89-61). Phosphoflow-related reagents and antibodies were purchased from BD Bioscience. In phosphoflow, owing to impaired signaling of CXCR2 and CXCR4 after robust fixation and permeabilization procedures, preneutrophils were identified as Lin− Gr-1+CD11b+c-kitint, immature neutrophils were identified as Lin−Gr-1lowCD11b+c-kitlow, and mature neutrophils were identified as Lin− Gr-1highCD11b+c-kit−. Data were acquired on a Canto II cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star, Ashland, OR).
Statistical analysis
Data were analyzed as indicated in the corresponding figure legends. Statistical significance was defined as P < .05. All the statistical calculations were performed using PRISM 9 software (GraphPad Software, La Jolla, CA).
Results and discussion
CD11c was expressed primarily in the intracellular location of neutrophils
CD11c/CD18 (CD11c is named αX; CD11c/CD18 is also called αXβ2, complement receptor 4 [CR4], p150,90; gene Itgax) is a member of the leukocyte adhesion molecule integrin family called β2 integrins and is primarily considered a marker and a major adhesion receptor of dendritic cells (DCs).19 However, CD11c messenger RNA expression (Itgax) has also been detected in both human and murine neutrophils (Immunological Genome Project20). Given this intriguing expression pattern, we sought to delineate the role of CD11c in neutrophil function. We initially examined CD11c protein expression on murine BM neutrophils using flow cytometry (Figure 1A and supplemental Figure 1A). CD11c cell surface expression was significantly limited on preneutrophils, immature, and mature neutrophils17 (Figure 1B). In contrast, CD11c was highly expressed intracellularly in preneutrophils, immature, and mature neutrophils (Figure 1B). As a comparison, cell surface and intracellular CD11c staining was performed in DCs (supplemental Figure 1B). CD11c was abundantly expressed on the cell surface of conventional and plasmacytoid DCs, as expected (Figure 1B). In addition, we observed intracellular expression of CD11c in the DCs but their CD11c expression was limited compared with their cell surface expression (Figure 1B). Intracellular CD11c expression was also confirmed by fluorescence microscopic examination of neutrophils (Figure 1C). We further separated neutrophils into nucleus and non-nucleus components. Nucleus fraction did not show CD11c expression (supplemental Figure 1C). CD11c expression was further examined in neutrophil-mimetic cells and human neutrophils. CD11c expression was greater intracellularly than on the cell surface in murine 32Dcl3 cells undergoing neutrophil differentiation (supplemental Figure 1D). Similarly, intracellular CD11c expression was more in human HL-60 cells undergoing neutrophil differentiation (supplemental Figure 2). Peripheral human neutrophils also demonstrated that intracellular CD11c expression was more than on cell surface expression (supplemental Figure 3). These intracellular expression data were also consistent with a previous report on CD11c that was detected in gelatinase granules by mass spectrometry.21
CD11c is expressed in neutrophils intracellularly. (A) Flow cytometry gating strategy identifying preneutrophils and immature and mature neutrophils in mouse BM. From the side scatter–A and forward scatter–A plot, live cells were selected (supplemental Figure 1A). Here, Lin includes CD3e, B220, and NK1.1. (B) Surface and intracellular CD11c expression detected by flow cytometry. Shown are representative data of at least 5 independent experiments with the same pattern. (C) Fluorescence microscopic examination (×60) of BM neutrophils. MPO, red; 4′,6-diamidino-2-phenylindole, blue; CD11c, green. Images show representative images of 3 independent experiments. c, conventional; p, plasmacytoid.
CD11c is expressed in neutrophils intracellularly. (A) Flow cytometry gating strategy identifying preneutrophils and immature and mature neutrophils in mouse BM. From the side scatter–A and forward scatter–A plot, live cells were selected (supplemental Figure 1A). Here, Lin includes CD3e, B220, and NK1.1. (B) Surface and intracellular CD11c expression detected by flow cytometry. Shown are representative data of at least 5 independent experiments with the same pattern. (C) Fluorescence microscopic examination (×60) of BM neutrophils. MPO, red; 4′,6-diamidino-2-phenylindole, blue; CD11c, green. Images show representative images of 3 independent experiments. c, conventional; p, plasmacytoid.
CD11c deficiency decreased neutrophil maturation in vivo
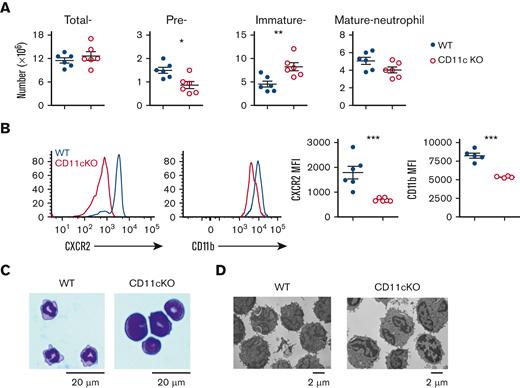
To examine the role of CD11c in neutrophil effector functions, BM neutrophils were purified from WT and CD11c KO16 mice using Ficoll-Paque gradient method.22 Classical neutrophils are heavier than Ficoll-Paque, called normal density neutrophils (NDNs), comprising preneutrophils, immature, and mature neutrophils.23,24 NDNs from CD11c KO mice BM demonstrated significant impairment of neutrophil effector functions including neutrophil chemotaxis, ROS25 production, phagocytosis, and neutrophil extracellular traps formation (supplemental Figure 4A-D). Because global neutrophil functions were impaired, we examined the composition of NDNs. Total neutrophil number (sum of preneutrophils, immature neutrophils, and mature neutrophils) was comparable between WT and CD11c KO mice (Figure 2A). The percentages and numbers of preneutrophils and mature neutrophils, however, were significantly higher in WT mice, whereas the percentage and number of immature neutrophils was significantly higher in CD11c KO mice (Figure 2A and supplemental Figure 5A). Similarly, splenic neutrophils in CD11c KO mice were less mature (Figure 2B). Giemsa staining and electron microscopy26 imaging also revealed higher abundance of less lobulated, immature neutrophils in CD11c KO mice BM (Figure 2C-D).
CD11c deficiency leads to impaired neutrophil maturation. (A) Number of total neutrophils, preneutrophils, and immature and mature neutrophils in the BM of WT and CD11c KO mice. Data are mean ± SEM of 3 experiments. (B) Maturation of splenic neutrophils. Left, representative histogram overlay analysis by flow cytometry; right, data are mean ± SEM of mean fluorescence intensity (MFI) of 3 experiments. (C-D) Giemsa staining and electron microscopy images of WT and CD11c KO BM neutrophils. Representative images of 3 independent experiments with the same pattern. SEM, standard error of the mean.
CD11c deficiency leads to impaired neutrophil maturation. (A) Number of total neutrophils, preneutrophils, and immature and mature neutrophils in the BM of WT and CD11c KO mice. Data are mean ± SEM of 3 experiments. (B) Maturation of splenic neutrophils. Left, representative histogram overlay analysis by flow cytometry; right, data are mean ± SEM of mean fluorescence intensity (MFI) of 3 experiments. (C-D) Giemsa staining and electron microscopy images of WT and CD11c KO BM neutrophils. Representative images of 3 independent experiments with the same pattern. SEM, standard error of the mean.
CD11c deficiency decreased neutrophil effector functions in vivo
In vivo bromodeoxyuridine incorporation showed that CD11c KO pre-neutrophils had higher proliferation (supplemental Figure 5B). They also had more apoptosis, probed by cleaved caspase-3 (supplemental Figure 5C), which explained the difference in the number of BM preneutrophils, immature, and mature neutrophils between WT and CD11c KO mice. Granulocyte colony-stimulating factor (G-CSF) is a major stimulant for neutrophil maturation. G-CSF stimulation leads to the activation of phosphoinositide 3-kinase (PI3K),27 janus kinase/signal transducers and activators of transcription, and mitogen-activated protein kinase pathways in neutrophils.28 Among them, PI3K-Akt pathway is reported responsible for preneutrophil survival.29 As expected, phospho-Akt level was lower in CD11c KO preneutrophils than in WT preneutrophils (Figure 3A), which corresponded with more apoptosis of CD11c KO preneutrophils (supplemental Figure 5C). The degree of Akt phosphorylation was not different between WT and CD11c KO immature/mature neutrophils. It is known that PI3K signal activates β2 integrin via “inside-out” signal.30 The “outside-in” signal, triggered as a result, can activate molecules such as spleen-associated tyrosine kinase (Syk),31 which also activates PI3K.32 The absence of CD11c, therefore, may modulate PI3K signal input by the loss of outside-in signal to the PI3K pathway.
CD11c deficiency leads to impaired neutrophil effector functions. (A) Akt phosphorylation in preneutrophils and immature and mature neutrophils in the BM of WT and CD11c KO mice. Data are mean ± SEM of 3 experiments. (B) ROS generation of sorted immature and mature neutrophils with or without PMA. BM cells from 3 mice were pooled as 1 biological sample. Upper panel: representative overlay analysis; bottom panel: mean ± SEM of mean fluorescence intensity (MFI) of 3 experiments. (C) Bulk RNA sequencing analysis comparing the transcriptome of sorted BM mature WT and CD11cKO neutrophils. Representative of 2 independent analyses with a similar pattern. (D-E) CD11b and ROS analysis of HL-60-differentiated neutrophils in vitro. Three independent experiments were collectively presented. CD11c knock out was conducted by CRISPR-Cas9 with 2 different guide RNAs (single guide [sg]1 and sg2). For ROS, we also presented the ratio of CD11c KO-sg1 and CD11cKO-sg2 transfected cells’ ROS compared with control-sg transfected cells. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. SEM, standard error of the mean.
CD11c deficiency leads to impaired neutrophil effector functions. (A) Akt phosphorylation in preneutrophils and immature and mature neutrophils in the BM of WT and CD11c KO mice. Data are mean ± SEM of 3 experiments. (B) ROS generation of sorted immature and mature neutrophils with or without PMA. BM cells from 3 mice were pooled as 1 biological sample. Upper panel: representative overlay analysis; bottom panel: mean ± SEM of mean fluorescence intensity (MFI) of 3 experiments. (C) Bulk RNA sequencing analysis comparing the transcriptome of sorted BM mature WT and CD11cKO neutrophils. Representative of 2 independent analyses with a similar pattern. (D-E) CD11b and ROS analysis of HL-60-differentiated neutrophils in vitro. Three independent experiments were collectively presented. CD11c knock out was conducted by CRISPR-Cas9 with 2 different guide RNAs (single guide [sg]1 and sg2). For ROS, we also presented the ratio of CD11c KO-sg1 and CD11cKO-sg2 transfected cells’ ROS compared with control-sg transfected cells. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. SEM, standard error of the mean.
As a member of β2 integrins, CD11c has 3 other related proteins, CD11a, CD11b, and CD11d, all of which couple with CD18. Among the 4 members, CD11a and CD11b have been studied extensively. CD11a is ubiquitously expressed on leukocytes and is involved in a number of important functions, including trafficking and immunological synapse formation.33,34 CD11b is mainly expressed on innate immune cells and plays a role in their recruitment, phagocytosis, and cell death.35-38 To date, the role of β2 integrins in neutrophil maturation is not known. To determine whether the maturation defect was specific to CD11c-deficient cells, we examined BM neutrophil maturation in CD11a KO, CD11b KO, and CD11d KO mice. The deficiency of CD11a, CD11b, or CD11d did not affect neutrophil maturation (supplemental Figure 6A-B), indicating that only CD11c affected neutrophil maturation among β2 integrins.
Next, we sought to compare the functions of immature and mature WT and CD11c KO neutrophils. They were sorted based on the surface markers as in Figure 1A. Giemsa staining showed that the maturity of neutrophils sorted based on the cell surface markers was comparable between WT and CD11c neutrophils (supplemental Figure 7A). In addition, MPO (primary granule protein), matrix metallopeptidase 9 (tertiary granule protein), and serpin B1 (intracellular protein) expression levels were comparable between immature WT and CD11c KO neutrophils, and between mature WT and CD11c KO neutrophils (supplemental Figure 7B). As expected, immature neutrophils showed very limited effector functions compared with mature neutrophils in general (Figure 3B and supplemental Figure 7C). Mature WT and CD11c KO neutrophils showed comparable level of phagocytosis (supplemental Figure 7C). Different from the phagocytosis result, WT mature neutrophils produced significantly higher ROS than mature CD11c KO mature neutrophils (Figure 3B). This indicates that the reduced BM neutrophil effector functions in CD11c KO mice were a result of both impaired maturation and an additional role of CD11c independent of maturation.
CD11c deficiency decreased neutrophil maturation and effector functions in vitro
To examine whether CD11c would regulate neutrophil maturation intrinsically, we stimulated sorted pre/immature WT and CD11c KO neutrophils with G-CSF in vitro. Compared with WT pre/immature neutrophils stimulated with G-CSF, CD11c KO neutrophils showed less CD11b expression (supplemental Figure 8A), indicating impaired maturation. RNA sequencing analysis showed that CD11c KO pre/immature neutrophils had significant upregulation of cell cycle and proapoptosis-related genes after G-CSF stimulation (supplemental Figure 8B-C), compatible with our in vivo findings of enhanced proliferation and apoptosis in the CD11c group (supplemental Figure 5B-C). RNA sequencing of mature neutrophils demonstrated 375 significantly downregulated genes in the CD11c KO group (Figure 3C). Ontology analysis indicated that cell proliferation (such as Cdk2ap1) and cell death (such as Bak1) were also enhanced in CD11c KO mature neutrophils (supplemental Figure 9A-B). The list of genes is included in supplemental Table 1.
To further confirm that CD11c would regulate neutrophil maturation intrinsically, the role of CD11c was examined in HL-60 cells using CRISPR-Cas9–based CD11c deletion (supplemental Figure 10A). Bulk cells with heterogeneous editing and isogenic cells derived from single clones (supplemental Figure 10B) were examined, and CD11c deletion was verified (supplemental Figure 10C). Then, we induced neutrophil differentiation using retinoic acid and dimethyl sulfoxide stimulation. CD11b expression has been used as a maturation marker for neutrophil differentiation in HL-60 cells.39 CD11c-deleted HL-60 cells showed impaired maturation demonstrated by a decrease in CD11b expression (Figure 3D and supplemental Figure 10D), in line with the murine neutrophil maturation experiments described earlier (supplemental Figure 8A). To corroborate the data of impaired maturation in CD11c-deleted HL-60 cells upon the differentiation stimulation, ROS production was significantly attenuated in CD11c-deleted HL-60 cells (Figure 3E and supplemental Figure 10E-F); similarly, phagocytosis was significantly attenuated in CD11c-deleted clones (supplemental Figure 10G). The regulatory role of conventional DCs (cDCs) in BM neutrophil homeostasis has been described.40 The number of cDCs regulates BM neutrophil homeostasis likely via affecting G-CSF levels. However, we previously demonstrated that G-CSF level was not different between WT and CD11c KO mice.41 In addition, there was no reported difference in the number of cDC1 and cDC2 cells between WT and CD11c KO mice in a previous study.42 We also confirmed that the number of cDC1 and cDC2 was comparable between the 2 strains (supplemental Figure 11A). In addition, our chimera analysis showed neutrophils from CD11c KO origin were less mature compared with those from WT origin (supplemental Figure 11B). Taken together, our data support that CD11c would intrinsically regulate neutrophil maturation.
CD11c deficiency led to an uncontrolled turnover of BM neutrophils into circulation upon LPS challenge
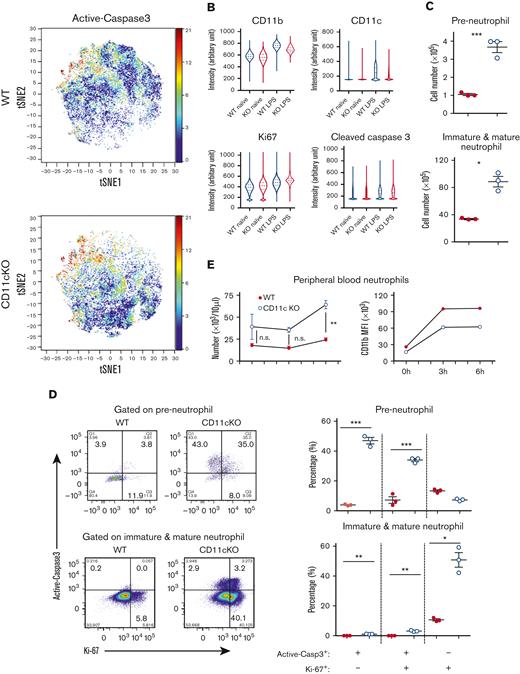
Neutrophils are massively generated and released as first-line defense cells in the setting of sepsis to provide an adequate number of mature neutrophils to infected tissues. In homeostatic conditions, BM total neutrophils did not show a significant difference between naive WT and CD11c KO mice, except slightly enhanced apoptosis and proliferation in CD11c KO neutrophils gated as Ly6G+CD11b+ cells (no lineage marker used) on CyTOF (supplemental Figure 12A). However, systemic LPS treatment led to significantly higher proliferation (Ki67) and apoptosis (cleaved caspase-3) with less maturation (CD11b) in CD11c KO total BM neutrophils gated as Ly6G+CD11b+ cells (no lineage marker used) on CyTOF (Figure 4A). In addition, we compared each expression level (Figure 4B). When gated using lineage markers with flow cytometry, significant apoptosis was similarly observed in CD11c KO neutrophils (supplemental Figure 12B). Further analysis using flow cytometry showed that there was a significant increase in the number of CD11c KO preneutrophils in the BM after LPS stimulation (Figure 4C) compared with the baseline number at the naive state (Figure 2A). The number of CD11c KO immature/mature neutrophils was also significantly higher than that of WT after LPS stimulation (Figure 4C). CD11c KO preneutrophils demonstrated significantly higher apoptosis and/or proliferation than WT counterparts, explaining, at least in part, the number of BM neutrophils for both strains (Figure 4D). This trend was also supported by Ly6G+CD11b+CXCR4hi population in the CyTOF (supplemental Figure 12C). Both WT and CD11c KO immature/mature neutrophils showed very limited apoptosis. Ki67 is expressed in all active phases of the cell cycles (G1, G2, S, and M), but not in G0. CD11c KO immature/mature neutrophils were significantly more in non-G0 phases (Figure 4D), suggesting the possibility that CD11c plays a role in neutrophil turnover. Of note, neutrophils in Figure 4B-C were gated with the use of lineage markers. In line with this BM phenotype, peripheral blood neutrophil counts were significantly higher in CD11c KO mice at 6 hours after LPS injection, and the mobilized neutrophils were significantly more immature (Figure 4E). Interestingly, average peripheral blood neutrophil counts showed a trend of being higher in CD11c KO mice at baseline, although statistically not significant (Figure 4E). This trend was also seen in complete blood count analysis (WT neutrophils 1.59 (±0.40) × 103/μL; CD11c KO neutrophils 2.83 (±0.19) × 103/μL; P = .4275); however, the difference was not statistically significant. The immaturity of neutrophils at the infection site was also evident after abdominal infection (supplemental Figure 12D). Collectively, these data revealed that CD11c was required for the generation of mature neutrophils in response to sepsis. LPS induces emergency granulopoiesis by engaging with TLR4 on the endothelial cells to produce G-CSF.25 Thus, G-CSF-mediated signaling events likely caused this exaggerated proliferation and apoptosis of CD11c KO preneutrophils in the LPS model, as in the case of naive mice. We also examined the profile of blood neutrophils after LPS stimulation. Interestingly, CD11c was transiently expressed on neutrophils at 4 hours after LPS stimulation (supplemental Figure 12E). This was associated with an increase in neutrophil phagocytosis (supplemental Figure 12F). The character of these CD11c cell surface–positive neutrophils will be examined in our future investigations.
CD11c deficiency leads to uncontrolled immature neutrophil mobilization upon LPS challenge. Both WT and CD11c KO mice were IV injected with LPS. (A) BM neutrophils were analyzed by CyTOF, 6 hours after LPS injection. Shown are viSNE plots of 1 of 3 biological samples with the same pattern. The color indicates expression level of labeled marker. (B) Violin plot of CD11b, CD11c, Ki67, and cleaved caspase-3 expressions. (C) The number of preneutrophils and immature/mature neutrophils in the BM. (D) Cleaved caspase-3 and Ki67 expression of preneutrophils and immature/mature neutrophils. Left panel: representative flow cytometry picture; right panel: statistical analysis. (E) Cell number and CD11b expression of blood neutrophils at indicated time points after LPS injection (3-5 mice in each time point). For panels C and D, Student t test was used; ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. For panel E, we used two-way analysis of variance. ∗∗P < .01, ∗∗∗P < .001. Experiments were repeated 3 times with a similar pattern. n.s., not significant.
CD11c deficiency leads to uncontrolled immature neutrophil mobilization upon LPS challenge. Both WT and CD11c KO mice were IV injected with LPS. (A) BM neutrophils were analyzed by CyTOF, 6 hours after LPS injection. Shown are viSNE plots of 1 of 3 biological samples with the same pattern. The color indicates expression level of labeled marker. (B) Violin plot of CD11b, CD11c, Ki67, and cleaved caspase-3 expressions. (C) The number of preneutrophils and immature/mature neutrophils in the BM. (D) Cleaved caspase-3 and Ki67 expression of preneutrophils and immature/mature neutrophils. Left panel: representative flow cytometry picture; right panel: statistical analysis. (E) Cell number and CD11b expression of blood neutrophils at indicated time points after LPS injection (3-5 mice in each time point). For panels C and D, Student t test was used; ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. For panel E, we used two-way analysis of variance. ∗∗P < .01, ∗∗∗P < .001. Experiments were repeated 3 times with a similar pattern. n.s., not significant.
Constitutive CD11c activation enhanced neutrophil maturation and effector functions
We observed that, compared with mature neutrophils, BM pre-neutrophils and immature neutrophils were more activated by phosphorylation in the BM of naive mice (the rank order of activation was preneutrophils >> immature neutrophils > mature neutrophils) (supplemental Figure 13A), compatible with the fact that most mature neutrophils are in G0 phase but preneutrophils and immature neutrophils are mostly in G1/S/G2/M phase. In addition, cleaved caspase-3 expression was significantly less in CD11c I334G preneutrophils (supplemental Figure 13A). It is known that integrins bind to their ligands upon activation for their functional contribution. We hypothesized that the activation of integrin CD11c would be important for neutrophil maturation in the BM, therefore constitutive activation of CD11c would further facilitate the production of mature neutrophils. To test this hypothesis, we created constitutive active CD11c mice. CD11c (α subunit) composes a heterodimeric molecule with CD18 (β subunit). Each subunit consists of a single transmembrane segment and a short cytoplasmic tail. The top domain of CD11c is called the αI domain and serves as a ligand binding domain, whereas other domains play regulatory roles. The top domain of CD18 is called the βI domain, which is structurally homologous to the αI domain. The ability of the αI domain to bind to its ligand is controlled by its conformational changes. The affinity of the αI domain to bind to its ligands is enhanced by a piston-like downward axial displacement of its C-terminal α7 helix, which is induced by the conformational signals relayed from the βI domain to the αI domain via the interdomain interaction (supplemental Figure 13B-C). In the inactive conformation, the region around the α7 helix forms the hydrophobic pocket to stabilize the α7 helix, hindering its ligand binding (supplemental Figure 13D-E). The αI domain is quite homologous between humans and mice. Both murine Ile-334 and human Ile-333 are located in the hydrophobic pocket (supplemental Figure 13F). This isoleucine was wedged into the hydrophobic pocket in the close conformation. However, Ile to Gly mutation displaced this residue from the hydrophobic pocket. Therefore, murine I344G mutant was expected to destabilize the hydrophobic pocket with the displacement of the α7 helix, leading to the activation as indicated by its constitutively activated phenotype (supplemental Figure 13G). Based on this structural and in vitro data, we created CD11c I334G KI mice (Figure 5A and supplemental Figure 13H). CD11c I334G KI–differentiated BM DCs showed significantly enhanced rosette formation under nonactivating conditions (magnesium/calcium ions) compared with WT (Figure 5B). Under activating conditions (manganese ions), both WT and CD11c I334G KI DCs showed significant rosette formation. In contrast, CD11c KO DCs showed limited rosette formation under both conditions. Furthermore, BM NDN neutrophils from CD11c I334G KI mice showed more lobulation on Giemsa staining (Figure 5C), indicating more maturation. BM total neutrophil counts (sum of preneutrophil, immature, and mature neutrophils) were comparable between WT and CD11c I334G KI mice (Figure 5D) but more mature neutrophils were seen in the KI mice (Figure 5E). Blood neutrophil counts were similar between the 2 strains but more maturation was seen in the KI mice as in the BM (Figure 5F). In the spleen, more neutrophils exist in the KI mice, along with more maturation (Figure 5G). The data from CD11c KO mice revealed that CD11c affected ROS irrespective of maturation stage (Figure 3B). In line with this finding, more ROS production was observed in mature KI neutrophils compared with mature WT neutrophils (Figure 5H). Under systemic LPS treatment, more neutrophils were seen in the blood, the spleen, and the BM of the KI mice (Figure 5I). As expected, neutrophils were more mature in the KI mice compared with WT controls (Figure 5J). Finally, in the setting of Escherichia coli intra-abdominal infection, bacterial burden was significantly less in the KI mice than in WT mice (Figure 5K), suggesting that the activation of CD11c helped to facilitate neutrophil maturation and effector functions for better microbial clearance in the setting of infection.
Constitutive activation of CD11c enhances neutrophil maturation and functions. (A) Scheme showing the strategy for creating the constitutively active CD11c KI mice (CD11cI334G). (B) Rosetting assay confirming the constitutive activation of CD11c molecule in CD11cI334G mice. Magnesium/calcium ions (1 mM) nonactivating condition, 1 mM manganese ions activating condition. (C) Giemsa staining of BM neutrophils. (D) Total BM neutrophil count. (E) Ratios of preneutrophils and immature and mature neutrophils. (F) Cell number and CXCR2 expression of blood neutrophils. (G) Cell number and CXCR2 expression of splenic neutrophils. (H) ROS detection in immature and mature neutrophils from naive WT and CD11c I334G mice. Left: representative flow cytometry overlay plot; right: data from 3 independent experiments. Cells from 3 to 4 mice were pooled together as 1 biological sample. (I) Neutrophil counts in the blood, the spleen, and the BM from WT and CD11cI334G mice, 6 hours after IV LPS stimulation. (J-K) WT and CD11c I334G mice were challenged intraperitoneally with E coli (1 × 108 colony-forming units) and sacrificed 6 hours later. (J) Giemsa staining of peritoneal neutrophils. (K) Bacterial loads of peritoneal cavity. ∗P < .05, ∗∗P < .01.
Constitutive activation of CD11c enhances neutrophil maturation and functions. (A) Scheme showing the strategy for creating the constitutively active CD11c KI mice (CD11cI334G). (B) Rosetting assay confirming the constitutive activation of CD11c molecule in CD11cI334G mice. Magnesium/calcium ions (1 mM) nonactivating condition, 1 mM manganese ions activating condition. (C) Giemsa staining of BM neutrophils. (D) Total BM neutrophil count. (E) Ratios of preneutrophils and immature and mature neutrophils. (F) Cell number and CXCR2 expression of blood neutrophils. (G) Cell number and CXCR2 expression of splenic neutrophils. (H) ROS detection in immature and mature neutrophils from naive WT and CD11c I334G mice. Left: representative flow cytometry overlay plot; right: data from 3 independent experiments. Cells from 3 to 4 mice were pooled together as 1 biological sample. (I) Neutrophil counts in the blood, the spleen, and the BM from WT and CD11cI334G mice, 6 hours after IV LPS stimulation. (J-K) WT and CD11c I334G mice were challenged intraperitoneally with E coli (1 × 108 colony-forming units) and sacrificed 6 hours later. (J) Giemsa staining of peritoneal neutrophils. (K) Bacterial loads of peritoneal cavity. ∗P < .05, ∗∗P < .01.
IQGAP1 as a novel protein interacting with CD11c
CD11c/CD18 functions by binding to its protein partners; therefore, we sought to identify a counterpart. Using the αI domain of CD11c in activated conformation (human I333G) tagged with glutathione S-transferase,43,44 we performed immunoprecipitation4 of HL-60 cell lysates. Mass spectrometry analysis demonstrated a range of top-hit proteins (supplemental Figure 14). Using previously published mass spectrometry analysis data of neutrophil granule proteins, we observed that half of the proteins exist in neutrophil granules.45 Using STRING software, we examined the interaction between proteins. CD11c was predicted to interact with IQGAP1 (Figure 6A). Immunoprecipitation of CD11c using HL-60 and 32Dcl3 cell lysates followed by IQGAP1 blotting confirmed their interaction (Figure 6B). The interaction between CD11c and IQGAP1 was also confirmed in primary human neutrophils (supplemental Figure 15B). Of note, IQGAP1, like CD11c, exists in gelatinase granules,21,45 supporting this interaction.
IQGAP1 interacts with CD11c. (A) Predicted interaction of top-hit proteins pulled down by immunoprecipitation (supplemental Figure 13) by STRING software. (B) Immunoprecipitation assay. Cell lysates of HL-60 cells were incubated with anti-human CD11c mAb (clone: CBR p150 2c1) for immunoprecipitation followed by blotting with anti-IQGAP-1 antibody or anti-CD11c polyclonal antibody. Cell lysates of 32D cl3 cells were incubated with anti-mouse CD11c mAb (clone: N417) for immunoprecipitation, followed by blotting with anti-IQGAP-1 antibody. What is shown is representative of 3 independent experiments. (C-E) CD11b (C), ROS (D), and phagocytosis (E) analyzed on HL-60 cells at 4 days under differentiation toward neutrophils. Three independent cell pools were analyzed in both WT and IQGAP1-KO groups. Shown are 1 of 3 independent experiments with the same pattern. (F) Apoptosis analysis by detecting cleaved caspase-3 at day 4 under differentiation toward neutrophils. HL-60 cells were treated with medium (control) or LPS (10 μg/mL) for 6 hours. Shown are 1 of 3 independent experiments with the same pattern. (G) Scheme of the role of CD11c-IQGAP1 interaction in neutrophil maturation. mAb, monoclonal antibody.
IQGAP1 interacts with CD11c. (A) Predicted interaction of top-hit proteins pulled down by immunoprecipitation (supplemental Figure 13) by STRING software. (B) Immunoprecipitation assay. Cell lysates of HL-60 cells were incubated with anti-human CD11c mAb (clone: CBR p150 2c1) for immunoprecipitation followed by blotting with anti-IQGAP-1 antibody or anti-CD11c polyclonal antibody. Cell lysates of 32D cl3 cells were incubated with anti-mouse CD11c mAb (clone: N417) for immunoprecipitation, followed by blotting with anti-IQGAP-1 antibody. What is shown is representative of 3 independent experiments. (C-E) CD11b (C), ROS (D), and phagocytosis (E) analyzed on HL-60 cells at 4 days under differentiation toward neutrophils. Three independent cell pools were analyzed in both WT and IQGAP1-KO groups. Shown are 1 of 3 independent experiments with the same pattern. (F) Apoptosis analysis by detecting cleaved caspase-3 at day 4 under differentiation toward neutrophils. HL-60 cells were treated with medium (control) or LPS (10 μg/mL) for 6 hours. Shown are 1 of 3 independent experiments with the same pattern. (G) Scheme of the role of CD11c-IQGAP1 interaction in neutrophil maturation. mAb, monoclonal antibody.
Next, IQGAP1 was successfully deleted in HL-60 cells using CRISPR-Cas9 technology (supplemental Figure 15A). CD11c expression did not differ after IQGAP1 knock out (supplemental Figure 15C). Under retinoic acid and dimethyl sulfoxide stimulation, IQGAP1-deleted HL-60 cells showed less CD11b expression, indicating that the cells were less mature (Figure 6C), corresponding with the finding in CD11c-deleted HL-60 cells (Figure 3D). As expected from the impaired maturation of IQGAP1-deleted cells, ROS and phagocytosis were significantly reduced in IQGAP1-deleted HL-60 cells (Figure 6D-E and supplemental Figure 15D). IQGAP1-deleted HL-60 cells also demonstrated an increased apoptosis during neutrophil differentiation stimulation (Figure 6F and supplemental Figure 15E), consistent with the phenotype of CD11c-deleted HL-60 cells. Collectively, our data reveal a novel function of CD11c in the regulation of neutrophil maturation by interacting with IQGAP1. CD11c interaction with IQGAP1 presumably provides antiapoptosis signal for more maturation of neutrophils, which leads to better neutrophil effector functions (Figure 6G). IQGAP1 is known as a scaffold protein that interacts with a number of partner proteins.46,47 We expect that PI3K-Akt pathway is responsible for the phenotype, but the interaction of the CD11c-IQGAP1 complex with PI3K pathway molecules needs to be determined in the future.
In summary, we found that CD11c regulates neutrophil maturation and effector functions in the BM and its constitutive activation helps to enhance neutrophil antimicrobial effector functions, which suggests that CD11c is a druggable target for sepsis. Full delineation of the CD11c signaling pathway to the regulate neutrophil maturation process will lead to a sophisticated intervention to modify the neutrophil maturation process during sepsis for the improvement of outcomes.
Acknowledgments
The authors acknowledge Mariel Bermudez and Mariel Manzor (both Boston Children’s Hospital) for technical support and Brice Gaudilliere (Stanford University) for discussion.
This study was funded by the National Institutes of Health, R21 HD099194 (K.Y. and S.K.) and the CHMC Anesthesia Foundation (K.Y. and L.H.).
Authorship
Contribution: L.H. and K.Y. conceptualized the study; L.H., R.A.V., S.K., Y.L, Y.C., M.S.-F., and K.Y. designed the study methodology; L.H., R.A.V., S.K., Y.L, M.S.-F., and K.Y. performed the investigations; S.K., L.H., and K.Y. acquired funding; K.Y. performed supervision; L.H., R.A.V., and K.Y. wrote the original manuscript draft; and L.H., R.A.V., H.L., and V.G.S. reviewed and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Koichi Yuki, Cardiac Anesthesia Division, Department of Anesthesiology, Critical Care and Pain Medicine, Boston Children’s Hospital, 300 Longwood Ave, Boston, MA 02115; e-mail: koichi.yuki@childrens.harvard.edu.
References
Author notes
Data are available on request from the corresponding author, Koichi Yuki (koichi.yuki@childrens.harvard.edu).
The full-text version of this article contains a data supplement.



![CD11c deficiency leads to impaired neutrophil effector functions. (A) Akt phosphorylation in preneutrophils and immature and mature neutrophils in the BM of WT and CD11c KO mice. Data are mean ± SEM of 3 experiments. (B) ROS generation of sorted immature and mature neutrophils with or without PMA. BM cells from 3 mice were pooled as 1 biological sample. Upper panel: representative overlay analysis; bottom panel: mean ± SEM of mean fluorescence intensity (MFI) of 3 experiments. (C) Bulk RNA sequencing analysis comparing the transcriptome of sorted BM mature WT and CD11cKO neutrophils. Representative of 2 independent analyses with a similar pattern. (D-E) CD11b and ROS analysis of HL-60-differentiated neutrophils in vitro. Three independent experiments were collectively presented. CD11c knock out was conducted by CRISPR-Cas9 with 2 different guide RNAs (single guide [sg]1 and sg2). For ROS, we also presented the ratio of CD11c KO-sg1 and CD11cKO-sg2 transfected cells’ ROS compared with control-sg transfected cells. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. SEM, standard error of the mean.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/7/10.1182_bloodadvances.2022007719/6/m_blooda_adv-2022-007719-gr3.jpeg?Expires=1763474084&Signature=RJiND0xLZ~ZZdxC4~EjDvrXk9puB9yONIwnxKLhXJecdtQ6HoCKTsyQyTcHdY3qus2Ukg-ccAEOm6QWdo9tgMaBt5qdDUr0Zb6hDX3cNM4A5Y3o5pLtlPInIIGqSLaJLDJO0qaO17Y8syHFmPjwWzaRlz~LKUui7J7FgxY9YQDuBqCnvKqeaX1zGaUWFaqWbTWKaIESON284DkuVP5qOAlVumG29jTkaAM1nHcuxzQ5XZuJjizApTynLM9E7g0DRjIWTpTo8MHJISZDhZcsU2v9u9-BpjtFMVQxLuW4Wko6Ytw515o2oHsAD1k2OEoY-nmkd0w6kXMIu3R12t7RUxQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)