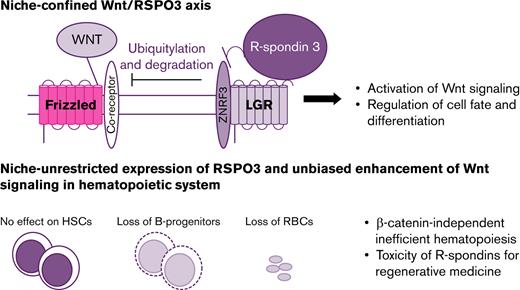
Key Points
Disruption of niche-confined expression of RSPO3 leads to refractory anemia and depletion of early B-cell progenitors.
Inefficient hematopoiesis associated with excessive RSPO3 stimulation uncovers potential challenges with the therapeutic use of RSPOs.
Abstract
Self-renewal and differentiation of stem and progenitor cells are tightly regulated to ensure tissue homeostasis. This regulation is enabled both remotely by systemic circulating cues, such as cytokines and hormones, and locally by various niche-confined factors. R-spondin 3 (RSPO3) is one of the most potent enhancers of Wnt signaling, and its expression is usually restricted to the stem cell niche where it provides localized enhancement of Wnt signaling to regulate stem cell expansion and differentiation. Disruption of this niche-confined expression can disturb proper tissue organization and lead to cancers. Here, we investigate the consequences of disrupting the niche-restricted expression of RSPO3 in various tissues, including the hematopoietic system. We show that normal Rspo3 expression is confined to the perivascular niche in the bone marrow. Induction of increased systemic levels of circulating RSPO3 outside of the niche results in prominent loss of early B-cell progenitors and anemia but surprisingly has no effect on hematopoietic stem cells. Using molecular, pharmacologic, and genetic approaches, we show that these RSPO3-induced hematopoietic phenotypes are Wnt and RSPO3 dependent and mediated through noncanonical Wnt signaling. Our study highlights a distinct role for a Wnt/RSPO3 signaling axis in the regulation of hematopoiesis, as well as possible challenges related to therapeutic use of RSPOs for regenerative medicine.
Introduction
Stem cells reside in defined niches within tissues where their self-renewal and differentiation are tightly regulated through the coordinated activity of a number of extrinsic factors.1-4 These include potent secreted molecules such as Hedgehogs, Notch ligands, Wnts, R-spondins (RSPOs), and others.1-6 In addition to expression limited to the niche, diffusion of these factors is restrained through various mechanisms such as attachment of lipid moieties or their ability to bind to extracellular matrix components such as heparan sulfate proteoglycans.7 Mutations in stem cells leading to constitutive activation of the corresponding pathways can lead to stem cell independence from the niche and the development of cancer.8,9
RSPOs are recently discovered enhancers of both canonical and noncanonical Wnt signaling.10-14 They inactivate the E3 ubiquitin ligases ZNRF3 and RNF43,15,16 which control the turnover of the Wnt receptor Frizzled, either indirectly by binding to leucine-rich repeat-containing, G protein–coupled receptors 4, 5, and 6 (LGR4, LGR5, and LGR6),11,17 or via direct binding in the presence of heparan sulfate.18 Four RSPOs capable of amplifying Wnt signaling have been identified.19 However, it remains to be determined whether these RSPOs have additional distinct and/or redundant activities in various tissues.
Under normal conditions, RSPO expression is primarily localized to stromal components of the niche, such as platelet-derived growth factor receptor–positive stromal cells located within intestinal crypts,20 myofibroblasts positioned proximal to the stem cell compartment in the stomach,21 central vein endothelial cells in the liver,22,23 and stromal components of the adrenal gland capsule.24 This niche-confined mode of expression provides localized enhancement of Wnt signaling, allowing for tightly controlled regulation of self-renewal and differentiation processes in stem and progenitor cells. In the gut, R-spondin 3 (RSPO3), one of the most potent enhancers of Wnt signaling, locally potentiates canonical Wnt signaling and actively drives Lgr5+ intestinal stem cell expansion.11,25,26 Although they act cooperatively, Wnt and RSPO3 do not act interchangeably in the gut: Wnt proteins act as priming factors by maintaining RSPO3 receptor expression, whereas RSPO3 drives and specifies the extent of stem cell expansion.26 Dualism of priming and self-renewal factors and spatial restriction of RSPO3 expression allows for remarkably precise control of the gut homeostasis. In the liver, RSPO3 is expressed specifically in central vein endothelial cells and is implicated in Wnt/β-catenin–dependent metabolic zonation.22,23 In adrenal glands, capsular RSPO3 signals enable both proper replenishment of damaged cells and maintenance of endocrine zonation.24
Altered expression of RSPOs can disrupt the precise signaling associated with Wnt gradients and leads to uncontrolled activation of canonical and noncanonical Wnt signaling. Recurrent RSPO gene fusions leading to overexpression of RSPOs have been observed in a subset of colorectal, stomach, and liver tumors.27-29 On the contrary, exogenous treatments with RSPO1, RSPO2, and RSPO3 support ex vivo growth of organoids derived from multiple tissues,30-33 making it an attractive approach for regenerative medicine to address damage to tissues such as lung, liver, or intestine. Although ex vivo data and limited in vivo data support the therapeutic potential of these factors, the consequences of increased circulating levels of RSPO on an organism remain unknown.
To explore the consequences of disrupting the niche-restricted expression of RSPO3, we used a transgenic approach to induce increased systemic levels of circulating RSPO3 protein. RSPO3 overexpressing mice exhibit Wnt-ligand–dependent phenotypes such as hyperproliferation in the gut and impaired liver metabolic zonation. In addition, we detected previously unreported development of ineffective hematopoiesis. Although RSPO3 overexpression has no effect on hematopoietic stem cell (HSC) proliferation and differentiation, it induces the loss of specific progenitors, leading to anemia and lymphopenia. Those clinical features are often observed in low-risk myelodysplastic syndrome (MDS); a subset of patients with low-risk MDS display increased RSPO3 expression. Our data suggest that niche restriction of RSPO3 is required to protect from hematologic toxicities. These findings will need to be taken into account in the development of therapeutics targeting tissue regeneration.
Methods
Generation of Rosa26.CAGGs.LSL.hRSPO3-IRES-Luc2.cki mice
We used recombination mediated cassette exchange to modify embryonic stem cells for the generation of Rosa26.CAGGs.LSL.hRSPO3-IRES-Luc2.cki mice. Properly modified embryonic stem cells were injected into blastocysts, and germline transmission was obtained after crossing the resulting male chimeras with C57BL/6N female mice.
In situ hybridization
RNA in situ hybridization (ISH) was performed with the RNAscope method according to manufacturer’s protocol (Advanced Cell Diagnostics) using the RNAscope 2.5 HD Reagent Kit-RED (322350) or RNAscope Multiplex Fluorescent Detection Reagent version 2 (323110). Probes used were MmRspo3 (402011), MmLepr-C2 (402731), and HsRspo3-O3 (491468). For human samples, quantification of ISH had been performed as follows: ISH-stained slides were scanned on a NanoZoomer XR whole slide imager (Hamamatsu) at 200× magnification. Tissue regions were identified by using standard thresholding and morphologic operations in MATLAB R2020b (MathWorks). ISH-positive staining was identified by using HSV color thresholding, with standard morphologic filtering with the requirement that ISH-positive staining was colocated with hematoxylin-positive areas, which were also identified by HSV thresholding.
RSPO3 immuno–polymerase chain reaction assay
The RSPO3 immuno–polymerase chain reaction (PCR) assay34 was developed by using biotinylated anti-RSPO3 2F8 antibody (Genentech) for capture and DNA-labeled anti-RSPO3 4A6 antibody (Genentech) for detection. The capture and detection antibodies were incubated with either serially diluted recombinant RSPO3 (R&D Systems) standards or diluted mouse serum or human plasma in streptavidin-coated 384-well PCR plates; captured RSPO3 was detected by using real-time PCR.
Tissue harvesting and processing for single-cell RNA-sequencing
Tissue harvesting and processing for single-cell RNA-sequencing (scRNA-seq) have been described elsewhere.35
Mouse strains and in vivo treatments
We generated a conditional RSPO3 overexpressing mouse model by crossing Rosa26.CAGGs.LSL.hRSPO3-IRES-Luc2.cki.B6N mice to Rosa26-CreER.T2 or Mx1.Cre animals. RSPO3 overexpression was induced in 5- to 8-week-old animals, both male and female, by intraperitoneal (IP) injections of 2 mg tamoxifen (MilliporeSigma) or PolyI:C (400 μg, injections every other day for 5 days; MilliporeSigma). No tamoxifen was given to control animals. B6.SJL-PtprcaPepcb/BoyJ (C57BL6/Ly5.1; The Jackson Laboratory) mice or bone marrow–derived cells from Mx1Cre–β-cateninfl/fl or Mx1Cre+-β-catenin fl/fl (mouse model development and functional testing were previously described36-38) were used for bone marrow transplantation experiments, and PolyI:C induction was performed to induce β-catenin deletion (400 μg via IP injections every other day for 5 times). The porcupine inhibitor LGK974 (Calbiochem) was resuspended in MCT (0.5% methyl cellulose, 0.2% Tween-80) vehicle and administered at 5 mg/kg twice a day via oral gavage for 14 consecutive days. The tankyrase inhibitor G007-LK (MilliporeSigma) was resuspended in dimethyl sulfoxide and administered at 30 mg/kg via IP injections once a day for 7 consecutive days. All mouse experiments were performed according to animal use guidelines of the Genentech Institutional Animal Care and Use Committee.
Flow cytometry and cell sorting
Analytical flow cytometry and sorting were performed on LSRFortessa and FACS Aria instruments (all, BD Biosciences). The following antibodies were used for analysis and sorting of hematopoietic cell populations: CD71-FITC, Ter119-PE, CD45RA-FITC, IgM-BV421, CD19-PE, CD43-APC, CD11b-PerCP-Cy7, Gr-1-APC-Cy7, CD41-FITC, CD45-APC-Cy7, APC-Lin– antibody cocktail, sca1-APC-Cy7, c-kit-PE, CD34-FITC, CD16/32-BV605, IL7R-BV421, CD150-BV421, CD48-FITC, CD45-FITC, CD3-BV421, CD19-PE, NK1.1-APC, CD3-BV421, CD45.1-FITC, CD45.2-PE, VCAM1-APC, and F4/80-FITC. Viable cells were isolated based on propidium iodide (MilliporeSigma) exclusion, and apoptotic cells were discriminated based on APC Annexin V (Pharmingen) and propidium iodide staining.
5-Bromodeoxyuridine incorporation and assessment of proliferation
The animals received a single IP injection of 1 mg 5-bromodeoxyuridine 1 hour before they were killed. Bone marrow cells were stained with antibody cocktails for erythroid progenitors (CD71-FITC/Ter119-PE) and LSK cells (APC-Lin– antibody cocktail, sca1-APC-Cy7, c-kit-PE) as described above; they were then sorted and subsequently stained with the APC BrdU Flow kit (BD Pharmingen) to determine their proliferation status.
Enzyme-linked immunosorbent assay
The Quantikine Mouse Epo Immunoassay (R&D Systems) was used to detect erythropoietin levels in serum from control and RSPO3-induced mice according to manufacturer’s instructions.
Colony formation assays
Bone marrow cells (2.5 × 103) were seeded into methylcellulose-containing media to quantify the absolute number of myeloid progenitors (MethoCult GF3434) and pre–B cells (MethoCult M3630; both, Stem Cell Technologies).
Detailed descriptions of RNA-sequencing, single-cell RNA-sequencing, and data analysis are presented in the supplemental Methods.
In vitro stimulation with Wnt ligands
Ter119+ erythroid progenitors were isolated and seeded in vitro in Iscove modified Dulbecco medium with 20% fetal bovine serum (both, Gibco). CD45RA+IgM+/− early B progenitors were isolated and seeded in vitro in RPMI 1640 with 20% fetal bovine serum (both, Gibco) and 5 ng/mL of recombinant IL-7 (PeproTech) and were maintained in the presence of OP9 stromal cells (ATCC). Progenitor cells were stimulated with RSPO3 (50 ng/mL; Genentech), Wnt5a (500 ng/mL), Wnt5b (500 ng/mL) and Wnt3a (150 ng/mL) (all, R&D Systems) or combinations thereof.
MDS patient samples
This report includes bone marrow samples collected from 29 hypomethylating agent-naive patients or patients with relapse/refractory MDS enrolled in a clinical study sponsored by Hoffmann–La Roche (#NCT02508870). The protocol was approved by institutional review boards where applicable, and patients gave written informed consent. Bone marrow core biopsy specimens were collected before initiation of treatment. Bone marrow mononuclear cell fractions were submitted for RNA-sequencing (TruSeq, Illumina). Normal donor bone marrow samples and plasma samples from control donors and patients with MDS with a verified diagnosis were procured from a vendor. Bone marrow core biopsy samples for RSPO3 ISH assessment were collected from normal donors (n = 2) and patients with MDS (n = 12).
Statistical analysis
Statistical analyses were performed by using GraphPad Prism 8.0 (GraphPad Software). Experiments were repeated at least twice. No statistical method was used to predetermine sample size. No animals or samples were excluded from data analysis. Animals were age and sex matched and randomly assigned in control and treatment groups. Group allocation and outcome assessment were not performed in a blinded manner. Data are represented as mean ± standard error of the mean of independent biological replicates. P < .05 was considered statistically significant.
Results
RSPO3 overexpression causes phenotypic changes in the gut, liver, and hematopoietic system
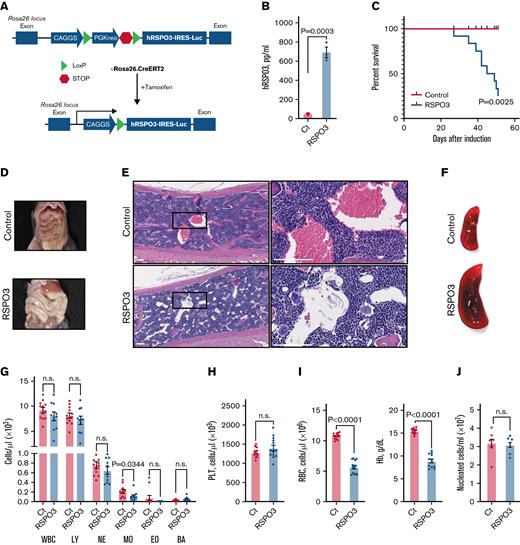
To model the disruption of niche-restricted RSPO3 expression and elucidate the effect of elevated RSPO3 levels on overall organism homeostasis, we generated a conditional overexpression mouse model that allows for inducible systemic overexpression of RSPO3 upon tamoxifen treatment (Figure 1A). One month after induction of RSPO3, overexpression was confirmed by immuno-PCR, and increased levels of circulating RSPO3 were detected in the serum (Figure 1B). Furthermore, animals became moribund and showed a rapid decline in survival (Figure 1C).
Widespread RSPO3 expression impairs survival. (A) Schematic depicting the generation of our conditional RSPO3 overexpression mouse model. (B) Circulating human RSPO3 (hRSPO3) levels in serum from control and RSPO3 animals measured by using immune-PCR; n = 3 per group. (C) Kaplan-Meyer survival curves for control and RSPO3 animals; n = 12 per group. Representative images of guts (D), hematoxylin, and eosin-stained sternum slides (E), and spleens (F) from control and RSPO3 animals. Rectangles mark regions shown at higher magnification; n = 10 per group. Quantification of white blood cells (WBCs) and major subpopulations (G), platelets (PLT) (H), and RBC and hemoglobin levels (Hb) (I) in the peripheral blood from control and RSPO3 mice. n = 12 for control and n = 10 for RSPO3. (J) Bone marrow cellularity in control and RSPO3 mice (n = 6 per group). Tissues were analyzed 1 month after tamoxifen induction, and data are represented as mean ± standard error of the mean. Two-tailed unpaired t test (panels B, G, H, and J), Mann-Whitney test (panel I), and log-rank test (panel C). Scale bars, 90 μm. BA, basophils; Ct, control; EO, eosinophils; LY, lymphocytes; MO, monocytes; NE, neutrophils; n.s., not significant (P > .05); RSPO3, tamoxifen-induced animals.
Widespread RSPO3 expression impairs survival. (A) Schematic depicting the generation of our conditional RSPO3 overexpression mouse model. (B) Circulating human RSPO3 (hRSPO3) levels in serum from control and RSPO3 animals measured by using immune-PCR; n = 3 per group. (C) Kaplan-Meyer survival curves for control and RSPO3 animals; n = 12 per group. Representative images of guts (D), hematoxylin, and eosin-stained sternum slides (E), and spleens (F) from control and RSPO3 animals. Rectangles mark regions shown at higher magnification; n = 10 per group. Quantification of white blood cells (WBCs) and major subpopulations (G), platelets (PLT) (H), and RBC and hemoglobin levels (Hb) (I) in the peripheral blood from control and RSPO3 mice. n = 12 for control and n = 10 for RSPO3. (J) Bone marrow cellularity in control and RSPO3 mice (n = 6 per group). Tissues were analyzed 1 month after tamoxifen induction, and data are represented as mean ± standard error of the mean. Two-tailed unpaired t test (panels B, G, H, and J), Mann-Whitney test (panel I), and log-rank test (panel C). Scale bars, 90 μm. BA, basophils; Ct, control; EO, eosinophils; LY, lymphocytes; MO, monocytes; NE, neutrophils; n.s., not significant (P > .05); RSPO3, tamoxifen-induced animals.
We therefore performed a detailed survey of various tissues and organs for overt phenotypic abnormalities after ubiquitous RSPO3 overexpression. Livers showed no discernible overt abnormalities upon RSPO3 overexpression (supplemental Figure 1A), despite high expression of Lgr4 and Lgr5 in subsets of hepatocytes.22 Subsequent staining of liver sections for glutamine synthetase and Cyp2e1 revealed impaired metabolic zonation (supplemental Figure 1B), consistent with a previous report on stimulation with RSPO1.22 Key liver enzymes were not significantly altered, but additional biochemical analyses revealed modest protein loss potentially related to the zonation defect (supplemental Figure 1C). Unexpectedly, we found no alterations in adrenal gland size or morphology (supplemental Figure 1D) but detected atrophy of the ovaries (supplemental Figure 1E). Although endothelial cells are sensitive to reduced RSPO3 levels, we found no difference in the composition and marker expression of tight junctions in the gut and bone marrow (supplemental Figure 1F). Vascular integrity was also intact (supplemental Figure 1G), further suggesting that elevated RSPO3 levels do not affect endothelial cells. Later time points could not be evaluated due to the observed mortality.
As previously reported for similar transgenic animals,25,33 RSPO3 mice developed an enlarged abdomen caused by a hyperproliferative response in the intestine driven by canonical Wnt pathway activation (Figure 1D; supplemental Figure 1H-I). Although this is potentially life-threatening, mice with Lgr5-driven RSPO3 overexpression display a similar intestinal phenotype but do not show such a rapid decline in survival,25 suggesting that additional mechanisms are involved.
Although a detailed survey of various tissues following RSPO3 overexpression failed to show an obvious cause of lethality, histologic analysis revealed previously unreported reduction in red blood cells (RBCs) in the bone marrow (Figure 1E) and pronounced splenomegaly (Figure 1F). Subsequent complete blood counts showed no major difference within leukocyte subpopulations (Figure 1G) and platelets (Figure 1H) but uncovered severe anemia (Figure 1I), with no change in overall bone marrow cellularity (Figure 1J). These findings suggest that RSPO3 overexpression leads to impaired hematopoietic function, which consequently may reduce viability in mice. Development of specific hematopoietic phenotype upon induction of RSPO3 overexpression was further confirmed in the Mx1.Cre model (supplemental Figure 2).
RSPO3 is expressed by leptin receptor–positive stromal cells in the bone marrow
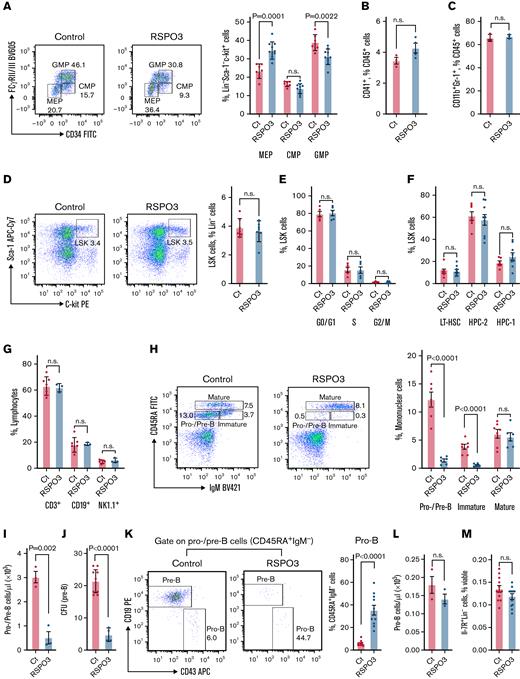
The role of Wnt/RSPO signaling has not been previously characterized for homeostatic hematopoiesis. To first elucidate whether RSPOs are niche-confined factors in the hematopoietic system, we examined their expression in murine bone marrow cells34 using scRNA-seq (Figure 2A). We found that Rspo2 and Rspo3 are expressed by a subpopulation of mesenchymal cells that express high levels of the leptin receptor Lepr (Figure 2B-D). Leptin receptor–positive (LepR+) cells form the main component of the perisinusoidal niche that is important for maintenance of HSCs and early B-progenitors in the bone marrow.1,39 These stromal cells are also the predominant source of RSPO3 in a publicly available bone marrow scRNA-seq data set40 that includes LepR+ stromal cells, collagen Col2.3+ osteoblasts, and vascular endothelial cadherin VE-cad+ endothelial cells (Figure 2E). Using ISH, we confirmed that Rspo3 is expressed by a small number of LepR+ stromal cells located primarily in perivascular areas (Figure 2F-G).
RSPO3 is expressed by LepR+stromal cells in the bone marrow. (A) A t-distributed stochastic neighbor embedding (t-SNE) plot with color-coded clusters of various bone marrow–derived cells is shown. t-SNE plots of various bone marrow–derived cell populations with color-coded expression levels of Rspo2 (B) and Rspo3 (C). (D) Markov Affinity-based Graph Imputation of Cells (MAGIC) plot showing co-expression of Rspo2 and Rspo3 with Lepr in a subset of bone marrow cells. (E) t-SNE plot of bone marrow–derived osteoblasts (Col2.3+), perivascular cells (Lepr+), and vascular endothelial cells (VE-cad+) from Tikhonova et al40 with color-coded expression levels of Rspo3. (F) Representative image of an ISH for Rspo3 transcript distribution in the bone marrow of n = 3 animals. Rectangles mark regions shown at higher magnification; arrows point out Rspo3 transcripts near blood vessels (BV). (G) Representative image of a double ISH for Rspo3 (blue) and Lepr (red) transcript distribution in the bone marrow of n = 3 animals. (H) Hierarchical clustering of human hematopoietic progenitor populations from the Immunological Genome Project data set41 based on the expression profile of common Wnt receptors. Expression data are scaled column-wise for each gene across cell types. Scale bars, 100 μm (panel F, large panel) and 50 μm (panel F, smaller panels; panel G). DC, dendritic cells; Ery/Meg prog, erythroid-megakaryocytic progenitors; Fbl, fibroblasts; l. pre-B, large pre–B cells; LMPPs, lymphoid-primed multipotent progenitors; max, maximum; min, minimum; MSC, mesenchymal stromal cells; NK, natural killer cells; prog, progenitor; s. pre-B, small pre–B cells; T, T cells.
RSPO3 is expressed by LepR+stromal cells in the bone marrow. (A) A t-distributed stochastic neighbor embedding (t-SNE) plot with color-coded clusters of various bone marrow–derived cells is shown. t-SNE plots of various bone marrow–derived cell populations with color-coded expression levels of Rspo2 (B) and Rspo3 (C). (D) Markov Affinity-based Graph Imputation of Cells (MAGIC) plot showing co-expression of Rspo2 and Rspo3 with Lepr in a subset of bone marrow cells. (E) t-SNE plot of bone marrow–derived osteoblasts (Col2.3+), perivascular cells (Lepr+), and vascular endothelial cells (VE-cad+) from Tikhonova et al40 with color-coded expression levels of Rspo3. (F) Representative image of an ISH for Rspo3 transcript distribution in the bone marrow of n = 3 animals. Rectangles mark regions shown at higher magnification; arrows point out Rspo3 transcripts near blood vessels (BV). (G) Representative image of a double ISH for Rspo3 (blue) and Lepr (red) transcript distribution in the bone marrow of n = 3 animals. (H) Hierarchical clustering of human hematopoietic progenitor populations from the Immunological Genome Project data set41 based on the expression profile of common Wnt receptors. Expression data are scaled column-wise for each gene across cell types. Scale bars, 100 μm (panel F, large panel) and 50 μm (panel F, smaller panels; panel G). DC, dendritic cells; Ery/Meg prog, erythroid-megakaryocytic progenitors; Fbl, fibroblasts; l. pre-B, large pre–B cells; LMPPs, lymphoid-primed multipotent progenitors; max, maximum; min, minimum; MSC, mesenchymal stromal cells; NK, natural killer cells; prog, progenitor; s. pre-B, small pre–B cells; T, T cells.
RSPOs are thought to mediate their activity through binding to LGR4-6 and/or the E3 ubiquitin ligases RNF43 and ZNRF3.11 Analysis of the public human hematopoietic data set from the Immunological Genome Project41 revealed expression of Lgr5 and Rnf43 in a subset of early B-progenitors, whereas Lgr4 expression was mostly restricted to erythroid cells (Figure 2H).
Elevated RSPO3 levels do not affect HSCs
Given the ability of RSPO3 to drive proliferation of stem cells in the gut and the newly discovered HSC niche-restricted expression pattern of RSPO3, we evaluated the consequence of systemic RSPO3 overexpression in the bone marrow. Analyzing the relative abundance and functionality of HSCs and early progenitors, we observed equal proportions of common myeloid progenitors in control and RSPO3 overexpressing mice; however, the proportions of granulocyte/megakaryocyte progenitors and megakaryocyte/erythrocyte progenitors were significantly altered (Figure 3A). Consistent with our complete blood count data showing no difference in platelets or granulocytes, we detected comparable proportions of CD41+CD45+ megakaryocytes (Figure 3B) and CD11b+Gr-1+ myeloid progenitors (Figure 3C). Unexpectedly, no significant alteration in the HSC compartment was detected, as the proportion of HSCs (Figure 3D), proliferation status (Figure 3E), and proportion of long-term HSCs (Figure 3F) were unchanged among LSK (Lin–Sca-1+c-kit+) cells. Together, these results indicate that, in contrast to findings with intestinal stem cells, RSPO3 overexpression does not affect proliferation and numbers of HSCs. Observed skewing in downstream megakaryocyte/erythrocyte progenitor differentiation with no other lineages being affected is likely compensatory to severe anemia.
Effects of elevated RSPO3 levels on HSCs and early B-progenitors. (A) Representative fluorescence-activated cell sorting (FACS) plot and corresponding bar graphs quantifying the relative abundance of megakaryocyte-erythrocyte progenitors (MEP), granulocyte-macrophage progenitors (GMP), and common myeloid progenitors (CMP) among Lin–Sca-1–Il-7R–c-kit+ bone marrow cells from n = 7 control and n = 10 RSPO3 animals. Bar graphs depicting relative abundance of megakaryocytes (CD41+CD45+FSChigh) (B) and myeloid progenitors (CD11b+Gr-1+) (C) among CD45+ bone marrow cells from n = 3 control and n = 3 to 4 RSPO3 animals. (D) Representative FACS plots and corresponding bar graphs quantifying the relative abundance of LSK (Lin–c-kit+Sca-1+) cells in the bone marrow of control and RSPO3 mice; n = 7 per group. (E) Bar graphs depicting the proportion of LSK cells from control and RSPO3 animals at various stages of cell cycle determined by using 5-bromodeoxyuridine incorporation; n = 5 per group. (F) Bar graphs showing the proportion of long-term HSCs (LT-HSC, CD150+CD48–) and restricted hematopoietic progenitor cell fractions (HPC-1, CD150–CD48+; HPC-2, CD150+CD48+) among LSK cells from n = 6 control and n = 9 RSPO3 animals. (G) Bar graphs depicting the relative abundance of 3 major lymphocyte subpopulations in peripheral blood of n = 7 control and n = 3 RSPO3 mice. (H) Representative FACS plots and corresponding bar graph showing the relative abundance of pro–/pre–B cells (CD45RA+IgM–), immature B cells (CD45RA+IgM+), and mature B cells (CD45RA++IgM+) among mononuclear cells from n = 7 control and n = 6 RSPO3 animals. (I) Bar graph depicting the absolute number of pro–/pre–B cells (CD45RA+IgM–) per microliter of peripheral blood from control and RSPO3 mice; n = 3 per group. (J) Bar graph showing the number of colony-forming units (CFU) (pre–B cell) after culturing equal numbers of total bone marrow cells from n = 10 control and n = 4 RSPO3 mice for 10 days in methylcellulose. (K) Representative FACS plots and corresponding bar graph quantifying the combined relative number of pro–B cells (CD43+CD19–) and pre–B cells (CD43–CD19+) among CD45RA+IgM– B-progenitors from control and RSPO3 mice; n = 10 per group. (L) Bar graph depicting the absolute number of pro–B cells (CD45RA+IgM–CD43+CD19–) per microliter of peripheral blood from control and RSPO3 mice; n = 3 per group. (M) Bar graph depicting the percent common lymphoid progenitors (CLP; Lin–Il-7R+) among viable bone marrow cells from control and RSPO3 animals; n = 12 per group. Bone marrow was analyzed 1 month after tamoxifen induction, and data are represented as mean ± standard error of the mean. Two-tailed unpaired t test. APC, allophycocyanin; BV, brilliant violet; Ct, control; FcγRII/III, Fc gamma receptor II/III; FITC, fluorescein isothiocyanate; IgM, immunoglobulin M; n.s., not significant (P > .05); PE, phycoerythrin; RSPO3, tamoxifen-induced animals.
Effects of elevated RSPO3 levels on HSCs and early B-progenitors. (A) Representative fluorescence-activated cell sorting (FACS) plot and corresponding bar graphs quantifying the relative abundance of megakaryocyte-erythrocyte progenitors (MEP), granulocyte-macrophage progenitors (GMP), and common myeloid progenitors (CMP) among Lin–Sca-1–Il-7R–c-kit+ bone marrow cells from n = 7 control and n = 10 RSPO3 animals. Bar graphs depicting relative abundance of megakaryocytes (CD41+CD45+FSChigh) (B) and myeloid progenitors (CD11b+Gr-1+) (C) among CD45+ bone marrow cells from n = 3 control and n = 3 to 4 RSPO3 animals. (D) Representative FACS plots and corresponding bar graphs quantifying the relative abundance of LSK (Lin–c-kit+Sca-1+) cells in the bone marrow of control and RSPO3 mice; n = 7 per group. (E) Bar graphs depicting the proportion of LSK cells from control and RSPO3 animals at various stages of cell cycle determined by using 5-bromodeoxyuridine incorporation; n = 5 per group. (F) Bar graphs showing the proportion of long-term HSCs (LT-HSC, CD150+CD48–) and restricted hematopoietic progenitor cell fractions (HPC-1, CD150–CD48+; HPC-2, CD150+CD48+) among LSK cells from n = 6 control and n = 9 RSPO3 animals. (G) Bar graphs depicting the relative abundance of 3 major lymphocyte subpopulations in peripheral blood of n = 7 control and n = 3 RSPO3 mice. (H) Representative FACS plots and corresponding bar graph showing the relative abundance of pro–/pre–B cells (CD45RA+IgM–), immature B cells (CD45RA+IgM+), and mature B cells (CD45RA++IgM+) among mononuclear cells from n = 7 control and n = 6 RSPO3 animals. (I) Bar graph depicting the absolute number of pro–/pre–B cells (CD45RA+IgM–) per microliter of peripheral blood from control and RSPO3 mice; n = 3 per group. (J) Bar graph showing the number of colony-forming units (CFU) (pre–B cell) after culturing equal numbers of total bone marrow cells from n = 10 control and n = 4 RSPO3 mice for 10 days in methylcellulose. (K) Representative FACS plots and corresponding bar graph quantifying the combined relative number of pro–B cells (CD43+CD19–) and pre–B cells (CD43–CD19+) among CD45RA+IgM– B-progenitors from control and RSPO3 mice; n = 10 per group. (L) Bar graph depicting the absolute number of pro–B cells (CD45RA+IgM–CD43+CD19–) per microliter of peripheral blood from control and RSPO3 mice; n = 3 per group. (M) Bar graph depicting the percent common lymphoid progenitors (CLP; Lin–Il-7R+) among viable bone marrow cells from control and RSPO3 animals; n = 12 per group. Bone marrow was analyzed 1 month after tamoxifen induction, and data are represented as mean ± standard error of the mean. Two-tailed unpaired t test. APC, allophycocyanin; BV, brilliant violet; Ct, control; FcγRII/III, Fc gamma receptor II/III; FITC, fluorescein isothiocyanate; IgM, immunoglobulin M; n.s., not significant (P > .05); PE, phycoerythrin; RSPO3, tamoxifen-induced animals.
Elevated RSPO3 levels disrupt early lymphoid development and lead to anemia
Although overall lymphocyte development showed no maturation defect (Figure 3G), a systemic loss of early B-progenitors in the bone marrow of RSPO3 overexpressing mice was observed (Figure 3H-J). This loss occurred primarily at the pre–B-cell stage and not at the stage of pro–B cells or Il-7R+Lin– common lymphoid progenitors, as shown by assessment of phenotype and colony-forming activity (Figure 3I-M). These results suggest that the RSPO3-induced B-lymphoid phenotype is distinct from the differentiation arrest at the Il-7R+Lin– common lymphoid progenitor stage observed upon constitutive hematopoietic specific β-catenin stabilization.42,43
Assessment of the erythroid lineage in RSPO3 overexpressing mice revealed a rapid and progressive reduction in CD71–Ter119+ RBCs (Figure 4A-C). RSPO3 overexpressing mice present with significantly elevated erythropoietin levels (Figure 4D) yet display only a modest increase in the formation of burst-forming units during in vitro culturing of bone marrow (Figure 4E) and intact differentiation into pro-erythroblasts (Figure 4F), indicating that early erythropoiesis proceeds normally.
RSPO3 overexpression impairs erythropoiesis. Representative FACS plots analyzing erythroid progenitors in bone marrow (A) and spleens (B) from control and RSPO3 mice; n = 20 per group. (C) Timeline of anemia development in RSPO3 animals measured as the percentage of CD71– Ter119+ erythroid cells among mononuclear cells; n = 3 to 5 per time point. (D) Erythropoietin concentration in serum from control and RSPO3 animals; n = 6 per group. (E) Bar graph depicting the number of burst-forming unit–erythroid (BFU-E), colony-forming unit–granulocyte macrophages (CFU-GM), and colony-forming unit–granulocyte erythrocyte monocyte/macrophage megakaryocyte (CFU-GEMM) after culturing equal numbers of total bone marrow cells from control and RSPO3 mice for 10 days in methylcellulose; n = 6 per group. (F) Quantification of CD71+ Ter119– pro-erythroblasts in control and RSPO3 mice; n = 9 per group. (G) Representative FACS plots and corresponding bar graph analyzing various stages of terminal erythropoiesis in control and RSPO3 mice; n = 9 per group. Gates from right to lower left represent basophilic (baso-), polychromatic (poly-), and orthochromatic (ortho-) erythroblasts, followed by reticulocytes (Ret) and RBCs. (H) Absolute number of reticulocytes in peripheral blood from control and RSPO3 mice; n = 14 per group. (I) Bar graph depicting the relative number of erythroid-macrophage islands in the bone marrow of control and RSPO3 animals measured as the percent CD11b+FSChigh among Ter119+ cells by flow cytometry; n = 3 to 4 per group. Tissues were analyzed 1 month after tamoxifen induction, and data are represented as mean ± standard error of the mean. One-way ordinary analysis of variance for panel C, Mann-Whitney test for panel D, and two-tailed unpaired t test for all other analyses. Ct, control; FITC, fluorescein isothiocyanate; FSC, forward scatter; n.s., not significant (P > .05); PE, phycoerythrin; RSPO3, tamoxifen-induced animals.
RSPO3 overexpression impairs erythropoiesis. Representative FACS plots analyzing erythroid progenitors in bone marrow (A) and spleens (B) from control and RSPO3 mice; n = 20 per group. (C) Timeline of anemia development in RSPO3 animals measured as the percentage of CD71– Ter119+ erythroid cells among mononuclear cells; n = 3 to 5 per time point. (D) Erythropoietin concentration in serum from control and RSPO3 animals; n = 6 per group. (E) Bar graph depicting the number of burst-forming unit–erythroid (BFU-E), colony-forming unit–granulocyte macrophages (CFU-GM), and colony-forming unit–granulocyte erythrocyte monocyte/macrophage megakaryocyte (CFU-GEMM) after culturing equal numbers of total bone marrow cells from control and RSPO3 mice for 10 days in methylcellulose; n = 6 per group. (F) Quantification of CD71+ Ter119– pro-erythroblasts in control and RSPO3 mice; n = 9 per group. (G) Representative FACS plots and corresponding bar graph analyzing various stages of terminal erythropoiesis in control and RSPO3 mice; n = 9 per group. Gates from right to lower left represent basophilic (baso-), polychromatic (poly-), and orthochromatic (ortho-) erythroblasts, followed by reticulocytes (Ret) and RBCs. (H) Absolute number of reticulocytes in peripheral blood from control and RSPO3 mice; n = 14 per group. (I) Bar graph depicting the relative number of erythroid-macrophage islands in the bone marrow of control and RSPO3 animals measured as the percent CD11b+FSChigh among Ter119+ cells by flow cytometry; n = 3 to 4 per group. Tissues were analyzed 1 month after tamoxifen induction, and data are represented as mean ± standard error of the mean. One-way ordinary analysis of variance for panel C, Mann-Whitney test for panel D, and two-tailed unpaired t test for all other analyses. Ct, control; FITC, fluorescein isothiocyanate; FSC, forward scatter; n.s., not significant (P > .05); PE, phycoerythrin; RSPO3, tamoxifen-induced animals.
Detailed analysis of terminal erythropoiesis revealed a relative increase in pro-erythroblasts and early erythroid progenitors in RSPO3 overexpressing mice (Figure 4G). The normal 1:2:4 ratio for basophilic, polychromatic, and orthochromatic erythroblasts was fully preserved, indicating that these nucleated erythroid progenitors mature properly. A near doubling of circulating reticulocytes (Figure 4H) and their preserved interaction with macrophages important for enucleation (Figure 4I) suggest that the RSPO3-induced anemia is unlikely to be due to a differentiation arrest or an enucleation defect but is likely to be linked to loss of RBCs.
To determine whether the aforementioned hematopoietic phenotypes require sustained levels of RSPO3, bone marrow from RSPO3 donors was transplanted into control recipient mice (supplemental Figure 3A). The transplants yielded multilineage engraftment, and both anemia and lymphopenia were fully reversed to baseline (supplemental Figure 3B-E), illustrating their dependency on continuous high RSPO3 levels from a non-hematopoietic source. The reverse transplantation of normal bone marrow into RSPO3 animals only slightly prolonged the survival of recipients and resulted in the development of the same erythroid and lymphoid phenotypes (supplemental Figure 3F-I).
Wnt ligands are essential for the RSPO3-induced hematopoietic phenotypes through noncanonical signaling
To assess the Wnt dependency of RSPO3-induced hematopoietic phenotypes, we used porcupine inhibitor LGK974 (Figure 5A), which blocks posttranslational acylation of Wnt ligands and inhibits their secretion. The hyperproliferative response in the gut of RSPO3 overexpressing animals was completely abolished after 14 days of continuous LGK974 treatment (Figure 5B). Similarly, we observed a complete reversal of the erythroid phenotype (Figure 5C), including normalization of absolute RBC counts (Figure 5D) and bone marrow morphology (Figure 5E). Analysis of the lymphoid lineage confirmed recovery of B-progenitors to normal levels after LGK974 treatment (Figure 5F).
The RSPO3-induced hematopoietic phenotype requires Wnt ligands. (A) Schematic depicting the LGK974 dosing regimen. (B) Image of representative pieces of small intestine from control and RSPO3 animals of each treatment group; n = 3 to 10 per group. (C) Representative FACS plots and corresponding bar graph quantifying erythroid progenitors in control and RSPO3 animals of each treatment group; n = 3 to 10 per group. (D) Absolute number of RBCs in peripheral blood before and after RSPO3 induction and LGK974 treatment; n = 3 to 9 per group. (E) Representative images of hematoxylin and eosin–stained sternum sections showing a reversal of the RSPO3-induced bone marrow phenotype after LGK974 treatment; n = 3 to 10 per group. (F) Representative FACS plots and corresponding bar graph quantifying early B-progenitors in bone marrow from control and RSPO3 animals of each treatment group; n = 3 to 10 per group. Data shown are mean ± standard error of the mean and are representative of 2 independent experiments. One-way analysis of variance followed by Dunnett’s test for multiple comparisons. Scale bar, 500 μm. FITC, fluorescein isothiocyanate; IgM, immunoglobulin M; n.s., not significant (P > .05); PE, phycoerythrin; p.o., by mouth.
The RSPO3-induced hematopoietic phenotype requires Wnt ligands. (A) Schematic depicting the LGK974 dosing regimen. (B) Image of representative pieces of small intestine from control and RSPO3 animals of each treatment group; n = 3 to 10 per group. (C) Representative FACS plots and corresponding bar graph quantifying erythroid progenitors in control and RSPO3 animals of each treatment group; n = 3 to 10 per group. (D) Absolute number of RBCs in peripheral blood before and after RSPO3 induction and LGK974 treatment; n = 3 to 9 per group. (E) Representative images of hematoxylin and eosin–stained sternum sections showing a reversal of the RSPO3-induced bone marrow phenotype after LGK974 treatment; n = 3 to 10 per group. (F) Representative FACS plots and corresponding bar graph quantifying early B-progenitors in bone marrow from control and RSPO3 animals of each treatment group; n = 3 to 10 per group. Data shown are mean ± standard error of the mean and are representative of 2 independent experiments. One-way analysis of variance followed by Dunnett’s test for multiple comparisons. Scale bar, 500 μm. FITC, fluorescein isothiocyanate; IgM, immunoglobulin M; n.s., not significant (P > .05); PE, phycoerythrin; p.o., by mouth.
Porcupine inhibition effectively blocks secretion of all Wnt ligands and therefore interferes with both canonical and noncanonical Wnt signaling. Canonical Wnt signaling acts via β-catenin and can also be blocked by tankyrase 1/2 inhibitors,44 which stabilize Axin, thereby promoting β-catenin degradation. We treated RSPO3 overexpressing mice with the tankyrase 1/2–specific inhibitor G007-LK for 7 days (supplemental Figure 4A-B). Treatment led to a complete reversal of the hyperproliferative gut phenotype, known to be fully dependent on canonical Wnt signaling. However, G007-LK treatment had only minor or no effect on the anemia or loss of B-progenitors in RSPO3 overexpressing mice (supplemental Figure 4C-F), although it induced a remarkable shift toward the myeloid lineage (supplemental Figure 4G-H).
To evaluate the contribution of canonical Wnt signaling to the RSPO3-induced hematopoietic phenotype using a genetic approach, we took advantage of the previously developed Mx1Creβ-cateninfl/fl mouse model36-38 that upon induction with PolyI:C allows for predominantly hematopoietic deletion of β-catenin. We transplanted RSPO3 transgenic mice either with autologous bone marrow or with bone marrow derived from Mx1Creβ-cateninfl/fl mice (Figure 6A). Upon engraftment and subsequent induction with PolyI:C, efficient deletion of β-catenin was confirmed in Mx1Cre+β-cateninfl/fl animals (Figure 6B). Assessment of key hematopoietic phenotypes after tamoxifen induction showed that deletion of β-catenin did not prevent anemia (Figure 6C-E) or loss of early B-progenitors (Figure 6F) in these animals. This provides strong evidence that the canonical Wnt signaling is not involved in mediating the observed RSPO3-induced hematopoietic phenotypes.
β-catenin is not required for RSPO3-induced hematopoietic phenotypes. (A) Schematic depicting the experimental approach to test the effect of RSPO3 overexpression on inducible inactivation of β-catenin in hematopoietic cells. (B) Genomic PCR-based quantification of β-catenin deletion efficiency in various experimental groups. Genomic DNA was collected from sorted CD45+ cells 28 days after completion of PolyI:C induction. (C) Representative FACS plots and corresponding bar graph quantifying erythroid progenitors in recipients of autologous bone marrow (BM), Mx1Cre–β-cateninfl/fl–derived BM (Mx1Cre–), or Mx1Cre+β-cateninfl/fl–derived BM (Mx1Cre+) with or without induction of RSPO3 overexpression; n = 5 to 10 per group. (D) Representative images of hematoxylin and eosin–stained sternum sections showing no reversal of the RSPO3-induced BM phenotype upon loss of β-catenin; n = 5 to 10 per group. (E) Absolute number of RBCs in recipients of autologous BM, Mx1Cre–β-cateninfl/fl–derived BM, or Mx1Cre+β-cateninfl/fl–derived BM with or without induction of RSPO3 overexpression; n = 5 to 10 per group. (F) Representative FACS plots and corresponding bar graph quantifying early B-progenitors in BM from control and RSPO3 animals of each treatment group; n = 5 to 10 per group. Data shown are mean ± standard error of the mean. One-way analysis of variance followed by Dunnett’s test for multiple comparisons. Scale bar, 500 μm. AutoBMT, autologous bone marrow transplant; BMT, bone marrow transplant; FITC, fluorescein isothiocyanate; IgM, immunoglobulin M; n.s., not significant (P > .05); Tam, tamoxifen.
β-catenin is not required for RSPO3-induced hematopoietic phenotypes. (A) Schematic depicting the experimental approach to test the effect of RSPO3 overexpression on inducible inactivation of β-catenin in hematopoietic cells. (B) Genomic PCR-based quantification of β-catenin deletion efficiency in various experimental groups. Genomic DNA was collected from sorted CD45+ cells 28 days after completion of PolyI:C induction. (C) Representative FACS plots and corresponding bar graph quantifying erythroid progenitors in recipients of autologous bone marrow (BM), Mx1Cre–β-cateninfl/fl–derived BM (Mx1Cre–), or Mx1Cre+β-cateninfl/fl–derived BM (Mx1Cre+) with or without induction of RSPO3 overexpression; n = 5 to 10 per group. (D) Representative images of hematoxylin and eosin–stained sternum sections showing no reversal of the RSPO3-induced BM phenotype upon loss of β-catenin; n = 5 to 10 per group. (E) Absolute number of RBCs in recipients of autologous BM, Mx1Cre–β-cateninfl/fl–derived BM, or Mx1Cre+β-cateninfl/fl–derived BM with or without induction of RSPO3 overexpression; n = 5 to 10 per group. (F) Representative FACS plots and corresponding bar graph quantifying early B-progenitors in BM from control and RSPO3 animals of each treatment group; n = 5 to 10 per group. Data shown are mean ± standard error of the mean. One-way analysis of variance followed by Dunnett’s test for multiple comparisons. Scale bar, 500 μm. AutoBMT, autologous bone marrow transplant; BMT, bone marrow transplant; FITC, fluorescein isothiocyanate; IgM, immunoglobulin M; n.s., not significant (P > .05); Tam, tamoxifen.
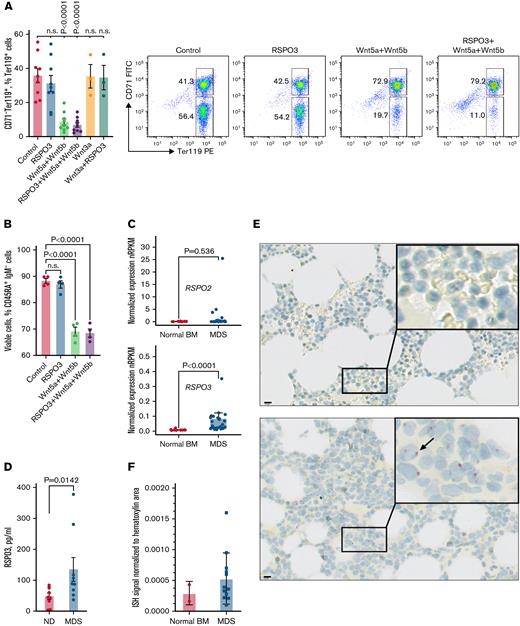
To further verify that the observed hematopoietic phenotypes are mediated by noncanonical Wnt/RSPO3 signaling, we sorted the total Ter119+ population from normal bone marrow and determined the effect of noncanonical Wnt ligands Wnt5a and Wnt5b on erythroid progenitors in vitro. Although RSPO3 alone did not affect the number of cells, treatment with noncanonical Wnt ligands led to a profound loss of CD71–Ter119+ RBCs, implicating these cells as a key responsive population (Figure 7A). This loss did not occur after treatment with Wnt3a, a ligand that activates the canonical Wnt pathway. Sorted early B-progenitors are also sensitive to stimulation with Wnt5a and Wnt5b in vitro, albeit to a lesser extent than the CD71–Ter119+ RBCs (Figure 7B). Collectively, these data indicate that in the hematopoietic system, excessive exposure to RSPO3 can lead to loss of progenitors through the potentiation of noncanonical Wnt signaling.
A subset of patients with MDS express high levels of RSPO3. (A) Representative FACS plots and corresponding bar graph quantifying the proportion of CD71–Ter119+ RBCs after in vitro culturing in the absence or presence of indicated growth factors; n = 3 to 9 per condition. (B) Bar graph quantifying the proportion of viable CD45RA+IgM– B-progenitors from control animals after culturing for 48 hours in the absence or presence of indicated growth factors; n = 4 per condition. (C) Expression of RSPO2 and RSPO3 in bone marrow (BM) of n = 13 control donors and n = 29 patients with MDS. Gene expression was obtained in the form of normalized reads per kilobase gene model per million total reads (nRPKM). (D) Circulating RSPO3 levels in plasma from control donors and patients with MDS; n = 9 per group. (E) Representative images of ISH for RSPO3 transcript distribution in BM core biopsy samples from n = 2 control donors and n = 12 patients with MDS. Rectangles mark regions shown at higher magnification; arrow points at a hematopoietic cell with an RSPO3 transcript. (F) Bar graph quantifying the expression of RSPO3 in normal BM and MDS BM. ISH-positive signal is normalized to hematoxylin areas. Data are represented as mean ± standard error of the mean. One-way analysis of variance followed by Dunnett’s test for multiple comparison (panels A and B), Mann-Whitney test for (panels C and D). Scale bar, 10 μm. FITC, fluorescein isothiocyanate; ND, normal donor; n.s., not significant (P > .05); PE, phycoerythrin.
A subset of patients with MDS express high levels of RSPO3. (A) Representative FACS plots and corresponding bar graph quantifying the proportion of CD71–Ter119+ RBCs after in vitro culturing in the absence or presence of indicated growth factors; n = 3 to 9 per condition. (B) Bar graph quantifying the proportion of viable CD45RA+IgM– B-progenitors from control animals after culturing for 48 hours in the absence or presence of indicated growth factors; n = 4 per condition. (C) Expression of RSPO2 and RSPO3 in bone marrow (BM) of n = 13 control donors and n = 29 patients with MDS. Gene expression was obtained in the form of normalized reads per kilobase gene model per million total reads (nRPKM). (D) Circulating RSPO3 levels in plasma from control donors and patients with MDS; n = 9 per group. (E) Representative images of ISH for RSPO3 transcript distribution in BM core biopsy samples from n = 2 control donors and n = 12 patients with MDS. Rectangles mark regions shown at higher magnification; arrow points at a hematopoietic cell with an RSPO3 transcript. (F) Bar graph quantifying the expression of RSPO3 in normal BM and MDS BM. ISH-positive signal is normalized to hematoxylin areas. Data are represented as mean ± standard error of the mean. One-way analysis of variance followed by Dunnett’s test for multiple comparison (panels A and B), Mann-Whitney test for (panels C and D). Scale bar, 10 μm. FITC, fluorescein isothiocyanate; ND, normal donor; n.s., not significant (P > .05); PE, phycoerythrin.
RSPO3 levels are elevated in a subset of patients with MDS
Refractory anemia and lymphopenia are clinical features that are often observed in patients with low-risk MDS.45 Aberrant activation of Wnt signaling in the bone marrow stroma has been closely linked to MDS.46-51 We therefore investigated the expression of RSPO2 and RSPO3, two of the most potent Wnt enhancers of the RSPO family, in bone marrow samples from 13 healthy donors and 29 patients with MDS enrolled in a study sponsored by Hoffmann–La Roche (supplemental Tables 1 and 2). Several patients with MDS displayed significant upregulation of RSPO3, but not RSPO2, compared with healthy donors (Figure 7C). We also procured plasma samples and detected elevated levels of circulating RSPO3 levels in several patients with MDS with verified diagnosis (Figure 7D). Importantly, for some of the patients with MDS, the levels of circulating RSPO3 were only twofold lower than the levels observed in our RSPO3 overexpressing mouse model.
To further validate our findings, we performed ISH on bone marrow core biopsy samples from normal donors and patients with MDS that were collected before treatment initiation and noted increased expression of RSPO3 in some of the patients with MDS (Figure 7F). RSPO3 appears to be expressed by hematopoietic cells, suggesting autocrine RSPO3 production to achieve niche independence. Because MDS is a highly heterogeneous group of myeloid disorders, further studies are required to dissect the contribution of stroma-derived excessive RSPO3 secretion vs niche-independent autocrine production of RSPO3 by abnormal myeloid cells. Although anemia in MDS has been primarily linked to a maturation arrest of erythroid progenitors,52 our data suggest that enhanced loss of RBCs due to an excessive Wnt/RSPO3 exposure can also contribute to anemia development. This may partially explain resistance to erythropoietin and luspatercept therapies observed in many patients with MDS.
Discussion
The role of niche-derived factors in the regulation of stem cell numbers and their role in driving tissue recovery has been studied for many factors, but only a few of them translated into medical treatments. Although this may be due in part to their challenging pharmacologic properties, the pleiotropic nature of their activity may also lead to toxicity when used systemically and exposing cells outside the niche to their activity.
The current study focused on RSPOs, a family of recently discovered Wnt signaling enhancers with promising use in regenerative medicine. Because of the potential for compensation, as all four RSPO members have the ability to enhance Wnt signaling, we used an inducible mouse model to evaluate the effect of niche-unrestricted overexpression of RSPO3 on multiple tissues. Although previous studies have focused on known sites of RSPO3 activity, an unbiased assessment of RSPO3-induced phenotypes revealed previously unrecognized defects in hematopoiesis in parallel to the known hyperproliferative response in the gut25,26 and impaired liver zonation.22,23 Hematopoiesis is tightly regulated and depends on the interaction of stem and progenitor cells with components of the niche, including stromal cells, secreted signaling factors, and extracellular matrix components. Contribution of Wnt signaling in maintaining hematopoiesis has been extensively studied,36-38,42,43 although the role for RSPO3 in the hematopoietic system remains uncharacterized.
Here, we show that, similar to the intestine and liver, bone marrow RSPO3 expression is confined to a niche, where hematopoietic stem and progenitor cells reside. Strikingly, disruption of niche-confined RSPO3 expression had no profound effect on HSC proliferation or differentiation, suggesting that in the bone marrow, Wnt/RSPO signaling is not limiting for HSC regulation. Overexpression of secreted factors, as shown for erythropoietin, growth hormone, vascular endothelial growth factor, and others, accurately predicts their physiological role, primarily because the phenotype is driven by relevant expression of the cognate receptor and engagement of physiological downstream targets.53,54 This is in contrast to transcription factors, which may bind non-physiological motifs or reflect pathway activity in a cell where it cannot be physiologically activated because of lack of an upstream component.55,56 Here we indeed show that the effect of RSPO3 overexpression in the hematopoietic system is limited to the populations expressing receptors for RSPO3, similar to the pattern observed in the gut and liver.
Although the current study revealed previously unidentified consequences of niche-unrestricted RSPO3 expression, it also highlights novel biologic features driven by enhanced Wnt signaling in hematopoiesis. Current approaches to understanding the role of augmented Wnt signaling in hematopoiesis in vivo rely on conditional expression of stabilized β-catenin42,43 or loss-of-function Apc mutations.57 This results in prominent loss of HSC repopulation and a block in multilineage differentiation.42,43,57 In the model described here, unrestricted RSPO3 expression led to profound hematopoietic phenotypes distinct from those induced by constitutive β-catenin activation. Although RSPO3 effectively promoted canonical Wnt signaling in these mice as evidenced by the prominent intestinal hyperproliferation, RSPO3 potentiated Wnt ligand/receptor complexes in the bone marrow that do not signal via β-catenin. These contrasting results further emphasize the limitations of previous gain-of-function models for understanding the role of Wnt signaling in hematopoiesis. In particular, they extrapolate the effects of enhanced Wnt signaling to all hematopoietic lineages without considering the specificity of receptor/ligand interactions. In addition, they are limited to activation of the canonical pathway, while bone marrow stromal cells predominantly express noncanonical Wnt ligands, and they do not account for complexity of Wnt signaling regulation that may occur in the niche.58-60 Currently, few tools exist to dissect the exact mechanism of noncanonical Wnt signaling, which has been shown to affect the NFAT and Cdc42 signaling pathways via modulation of intracellular Ca2+ levels. Because RSPO3 overexpression leads to loss of both nucleated B-progenitors and enucleated erythroid cells, we speculate that the potential underlying mechanism might be related to Ca2+-induced cell death.61,62 Although the physiological role of RSPOs during hematopoiesis remains unclear, our data highlight the importance of niche-restricted RSPO3 expression, as aberrant upregulation of RSPO3 can sensitize RBC and B-progenitors to noncanonical Wnt signaling and lead to loss of these cells.
Various conditions might be associated with increased RSPO3 levels, including general infection and inflammation,63 local de-regulation of Wnt signaling, hematologic malignancies, or systemic therapy with RSPO3 or RSPO3 mimetics. In the bone marrow, where noncanonical Wnt ligands are highly expressed,58-60 increased local or circulating levels of RSPO3 may sensitize cells to their activity and lead to anemia and loss of early B-progenitors. Although its contribution to MDS pathogenesis requires further evaluation, our study provides a clear example of a pathology in which RSPO3 is expressed outside of the context of the stem cell niche. Similarly, elevated levels of RSPO and RSPO mimetics may therefore be challenging to use for therapeutic applications because of the potential for hematologic toxicity.
Acknowledgments
The authors thank Alice Tan, C.K. Poon, Terrence Ho, George Tweet, and Jovencio Borneo for FACS support; Angela Martzal and Lorie Leong for analysis of clinical blood and serum samples; Kerstin Seidel for liver immunostaining; Jian Jiang for tissue processing; Linda Rangell for coordinating ISH of bone marrow core biopsy samples; Cecile Chalouni for assistance with imaging; Michael Townsend and Nandhini Ramamoorthi for procuring plasma samples; Jeff Eastham for ISH data quantification; Keith Anderson for engineering embryonic stem cells; Laboratory Animal Resources for animal care; and members of the de Sauvage laboratory for comments on the manuscript.
Authorship
Contribution: A.V.K. and F.J.d.S. conceptualized the project; A.V.K. designed, conducted, and analyzed the majority of experiments; M.H. and N.M.K. assisted with in vivo experiments; S.H. and L.V. conceived, performed, and analyzed scRNA-seq data; G.J.P.D. and C.M. developed the conditional RSPO3 overexpression mouse model; E.E.S. performed ISH; S.B. and A.M. assisted with ex vivo experiments; P.H. and A.H. performed endothelial phenotype analysis; H.K. provided pathology support; M.D. and Q.Y. analyzed clinical and RNA-seq data from patients with MDS; J.Z. performed immuno-PCR; U.K. and F.R. provided bone marrow samples from Mx1Cre bcatfl/fl mice; Z.M. contributed next-generation sequencing data; R.P. performed RNA-seq analysis; F.J.d.S. supervised the study; and A.V.K. and F.J.d.S. wrote the manuscript.
Conflict-of-interest disclosure: A.V.K., M.H., S.H., L.V., G.J.P.D., E.E.S., N.M.K., S.B., P.H., A.H., A.M., H.K., M.D., Q.Y., J.Z., Z.M., C.M., R.P., F.J.d.S are employees of Genentech or Roche and hold Roche shares. The remaining authors declare no competing financial interests.
Correspondence: Frederic J. de Sauvage, Department of Molecular Oncology, Genentech, MS37, 1 DNA Way, South San Francisco, CA 94080; e-mail: desauvage.fred@gene.com.
References
Author notes
Single-cell RNA-sequencing data are available for browsing at https://nicheview.shiny.embl.de/. Raw sequencing data are available through the Gene Expression Omnibus database (accession number GSE122467). RNA-sequencing data from the myelodysplastic syndrome patient samples are not yet publicly available due to restrictions associated with the ongoing clinical trial but are available on request from the corresponding author, Frederic J. de Sauvage (desauvage.fred@gene.com).
The full-text version of this article contains a data supplement.