Key Points
NPM1 haploinsufficiency in collaboration with MEIS1 overexpression is sufficient to induce complete AML in mice.
The MEIS1-SMC4 axis is a potential therapeutic target in NPM1c AML.
Abstract
NPM1 is among the most frequently mutated genes in acute myeloid leukemia (AML). Mutations in the NPM1 gene result in the increased export of NPM1 to the cytoplasm (NPM1c) and are associated with multiple transforming events including the aberrant upregulation of MEIS1 that maintains stem cell and cell cycle–associated pathways in NPM1c AML. However, another consequence of the NPM1c mutation is the inadequate levels of NPM1 wild-type in the nucleus and nucleolus, caused by the loss of one wild-type allele in addition to enforced NPM1 nuclear export. The contribution of NPM1 haploinsufficiency independently of the NPM1 mutation to AML development and its relationship with MEIS1 function is poorly understood. Using mouse models, our study shows that NPM1 haploinsufficiency paired with MEIS1 overexpression is sufficient to induce a fully penetrant AML in mice that transcriptionally resembles human NPM1c AML. NPM1 haploinsufficiency alters MEIS1-binding occupancies such that it binds the promoter of the oncogene structural maintenance of chromosome protein 4 (SMC4) in NPM1 haploinsufficient AML cells but not in NPM1 wild-type–harboring Hoxa9/Meis1-transformed cells. SMC4 is higher expressed in haploinsufficient and NPM1c+ AML cells, which are more vulnerable to the disruption of the MEIS1-SMC4 axis compared with AML cells with nonmutated NPM1. Taken together, our study underlines that NPM1 haploinsufficiency on its own is a key factor of myeloid leukemogenesis and characterizes the MEIS1-SMC4 axis as a potential therapeutic target in this AML subtype.
Introduction
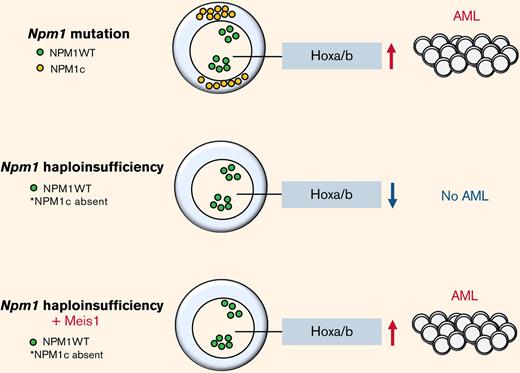
Nucleophosmin 1 (NPM1) is a nucleolar protein required for genome stability and ribosome biogenesis.1 In acute myeloid leukemia (AML), NPM1 is mutated in ∼60% of cytogenetically normal AML patients.2 Although NPM1 mutations are absent in clonal hematopoiesis and rare in myelodysplastic syndrome (MDS) (2%), they are considered AML-initiating lesions that recur at relapse in the majority of NPM1 mutation harbouring AML patients.2-5 Mutations in the NPM1 gene largely result in insertions in the terminal exon leading to a functionally stronger nuclear export and aberrant cytoplasmic localization (NPM1c) and reduction in nuclear protein levels.1,2,5 Previous work has shown that NPM1c contributes to leukemia development and maintenance by multiple mechanisms such as upregulation of HOX/MEIS1 expression, dislocation of myeloid differentiation transcription factors PU.1 and CTCF, and recently, disruption of promyelocytic leukemia protein nuclear body formation.6-9 However, the relevance of insufficient levels of wild-type NPM1 in the nucleolus/nucleus caused by the enforced export of NPM1c in AML biology is not precisely known so far. It is notable that not just 30% of heterozygous Npm1c mutation–harboring mice develop delayed-onset AML with maturation,10 but also 16% of mice lacking one allele of Npm1 (Npm1 haploinsufficient) develop a delayed-onset myeloproliferative disease,11 suggesting that Npm1 haploinsufficiency, in concert with secondary events, contributes to disease.
A hallmark of NPM1c AML is the overexpression of homeobox genes, including the three-amino acid extension loop homeobox cofactor MEIS1, triggered by cytoplasmic localization of the mutant NPM1c protein.12-17 Studies have proven that Meis1 overexpression itself is not leukemogenic in mouse models, but it is required for AML growth.18-20 Moreover, recent studies indicate that MEIS1 expression can be predictive for treatment response to Menin (MEN1) and MLL (MLL1) interaction inhibitors.20-23 Preclinical in vivo studies strongly suggest that the efficacy of Menin-MLL interaction inhibitors on NPM1c AML cells relies on its suppression of MEIS1-driven leukemic stem cell program, reaffirming that MEIS1 is a critical factor for the sustenance of NPM1c AML.
In this study, we demonstrate that Npm1 haploinsufficiency and Meis1 overexpression can collaboratively act as oncogenic events in the absence of Npm1c mutation to generate rapid myeloid leukemia that is transcriptionally akin to human NPM1c AML. We describe that in the context of Npm1 haploinsufficiency, MEIS1 regulates a cell cycle gene expression program required for leukemic growth that is also found enriched in NPM1c AML patients. In particular, Npm1 haploinsufficiency alters Meis1 binding, inducing upregulation of SMC4 and resistance to Menin-MLL inhibition.
Methods
Cell culture
All cells were grown in a humidified incubator at 37°C and 5% CO2. GP+E86 and 293T LentiX cells were grown in Dulbecco’s modified Eagle medium media (41966-029, Gibco) supplemented with 10% fetal calf serum (FCS) and 1% pen/strep. Primary 5FU-enriched murine bone marrow (BM) was cultured in Dulbecco’s modified Eagle medium supplemented with 15% FCS and 1% pen per strep and 10 ng/mL interleukin-3 (IL-3), 10 ng/mL IL-6, and 50 ng/mL stem cell factor. Murine cell lines established from leukemic BM of NPM1+/− mice overexpressing Meis1 or of mice expressing H9M1 were established in 20% RPMI supplemented with 10 ng/mL Il-3.
OCI-AML2 and OCI-AML3 cells were cultured in RPMI 1640 media (31870074, Life Technologies) supplemented with 20% FCS and 1% pen per strep. Primary AML cells were cultured in Iscove modified Dulbecco medium supplemented with 20% BIT 9500 serum substitute, 100 ng/mL stem cell factor, 50 ng/mL FLT3L, 20 ng/mL IL-3, 20 ng/mL granulocyte colony-stimulating factor, 10-4 M β-mercaptoethanol, and ciprofloxacin (10 μg/mL).
Murine models
All mouse strains were bred and maintained in the University of Ulm animal facility except for the Npm1flox-cA/+; Mx1-Cre+ mice (hereafter referred to as Npm1cA/+), which were bred and housed at the Wellcome Trust Sanger Institute, United Kingdom. The humanized Npm1+/c+ mice and Npm1+/− mice were previously described.10,11,24 Murine BM was transplanted IV into lethally irradiated (1025 cGy) Npm1+/+ mice as previously described.25
The AML cell line OCI-AML3 was injected into 8- to 12-week-old NOD scid gamma (NSG) mice (NOD.Cg-PrkdcScidIl2rgtm1Wjl/SzJ) which were sublethally irradiated (325 cGy) and injected intraperitoneally with 20 mg anti-intravenous immune globulin antibody (Privigen, CSL Behring GmbH) a day prior. NSG mice were checked each day for signs of disease. Mice exhibiting signs of disease were euthanized, and their blood, spleen, and BM were analyzed for human leukemic engraftment.
Retroviral and lentiviral transductions
5-FU stimulated BM was harvested and cultured for 48 hours before culturing on top of irradiated GP+E86 cells genetically engineered to produce virus from an MSCV-based retroviral vector for Meis1 or Hoxa9, after which the cells were harvested and FACS sorted for GFP, yellow fluorescent protein (YFP,) or double positivity and injected directly into lethally irradiated recipient mice along with steady-state BM as helper cells.
Short hairpin RNA (shRNA) against SMC4 (murine: TRCN00001008940; human: TRCN0000154901) and MEIS1 (murine: TRCN0000321054 and TRCN0000321055) cloned in p.LKO.1 lentiviral vector were transfected with helper plasmids pMD2.G and pSPAX2 using TransLT1 (Mirus) in LentiX cells. Virus particles were harvested 48 hours and 72 hours posttransfection.
Please see supplemental Methods for complete materials and methods.
Results
MEIS1 is overexpressed in NPM1c AML patients and collaborates with NPM1c mutation to induce high-penetrance myeloid leukemia in mice
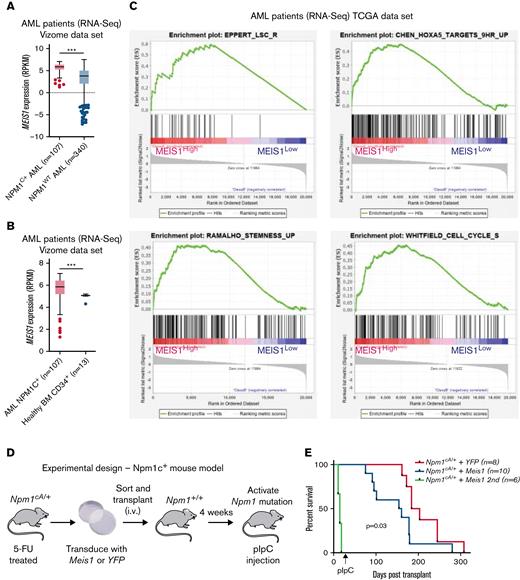
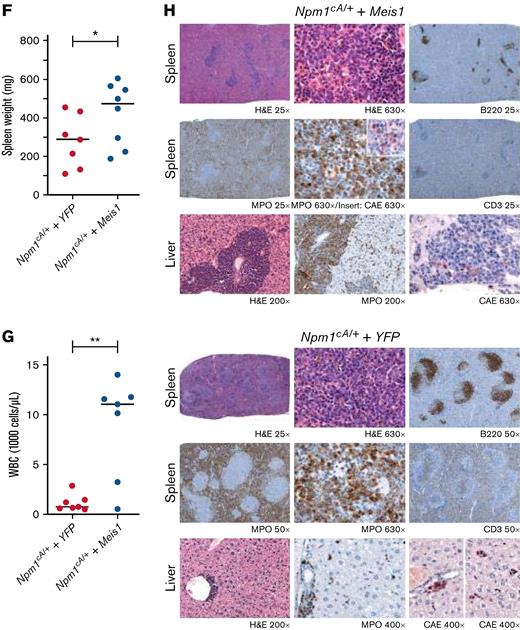
First, the examination of a large, previously published RNA-sequencing (RNA-seq) data set confirmed that MEIS1 exhibited significantly higher expression in mutant NPM1 (NPM1c) vs wild-type NPM1 (NPM1WT)-harboring AML patients (Figure 1A).26,27 Moreover, MEIS1 expression levels were higher in the majority of NPM1c AML patients compared with healthy CD34+ human stem progenitors (Figure 1B).26 The overexpression of MEIS1 was independent of the presence of frequently occurring collaborating mutations in NPM1c AML (supplemental Figure 1A-B).26 Furthermore, MEIS1 was expressed higher in relapsed NPM1c AML patients (supplemental Figure 1C).26 Notably, MEIS1 expression in NPM1c AML patients positively correlated with the expression of stem cell and cell cycle–associated pathways (Figure 1C). Because higher expression of stem cell signatures is associated with poor prognosis in AML patients, these data suggested that MEIS1 could be a determinant of disease aggressiveness in NPM1c AML.28,29 This notion is supported by data in the case of MLL leukemia, where Meis1 is an essential rate-limiting regulator of MLL leukemia stem cell potential.30 To test this hypothesis, we used mice harboring the humanized Npm1c mutation (Npm1cA/+) under the control of cre/lox recombinase,10 with the knowledge that Hox/Meis expression was endogenously higher in Npm1cA/+ vs Npm1wt–harboring murine hematopoietic stem progenitor cells (HSPCs).8Npm1cA/+containing BM-derived HSPCs, which had not yet undergone recombination to express the mutant allele, were retrovirally transduced with Meis1 or YFP and transplanted into lethally irradiated Npm1+/+ recipient mice. Four weeks posttransplant, expression of the Npm1c mutation was induced by intraperitoneal injection of polyinosinic:polycytidylic acid (Figure 1D). Meis1 overexpression exacerbated disease progression in NPM1c leukemia: all 10 mice transplanted with Npm1cA/+ + Meis1 cells died, with a significantly shorter median latency of 157 days, whereas Npm1cA/+ plus YFP succumbed to disease, with a longer latency of 194 days (P = .03, log-rank test). In retransplantation assays, all Npm1cA/+ plus Meis1 secondary mice succumbed to rapid myeloid leukemia, with a median latency of 14 days, whereas only 2 out of 5 Npm1cA/+ plus YFP secondary mice succumbed to disease, with a median latency of 77 days (data not shown) (Figure 1E). Second, Npm1cA/+ plus Meis1 mice succumbed to a disease resembling myeloid leukemia, with significantly higher WBC counts and spleen weights vs Npm1cA/+ plus YFP mice (P < .05, Student t test) (Figure 1F-G). Furthermore, histopathological analyses confirmed that, indeed, primary Npm1cA/+ plus Meis1 succumbed to AML with maturation, whereas primary Npm1cA/+ plus YFP mice exhibited only early signs of leukemia residing primarily in the spleen (Figure 1H). Immunophenotypically, the Npm1cA/+ + Meis1 leukemic cells showed a trend toward higher Gr1+, Mac1+, and cKit+ single-positive cells and lower Gr1+Mac1+ cells (supplemental Figure 1D). In sum, our data demonstrated that high Meis1 expression levels can collaborate with the NPM1c mutation to exacerbate leukemogenesis.
MEIS1 collaborates with NPM1c mutation to induce high-penetrance myeloid leukemia in mice. (A) MEIS1 expression in NPM1c de novo AML patients (n = 107) compared with NPM1WT AML patients (n = 340) from the Vizome data set.26 (B) MEIS1 expression in NPM1c de novo AML patients (n = 107) compared with CD34+ healthy BM (n = 13) from the Vizome data set.26 (C) GSEA analysis of 25% of highest MEIS1 (MEIS1High) expressing vs 25% lowest MEIS1 expressing (MEIS1Low) NPM1c de novo AML patients from the TCGA data set.27 (D) Schematic of the experimental design of in vivo experiments with Npm1cA/+ mouse model. (E) Kaplan-Meier plot of Npm1cA/+ mice transduced with a YFP empty vector (n = 8), Meis1 (n = 10), and secondary transplants of NPM1c+ Meis1 leukemic cells (n = 6). (F) Spleen weight of Npm1cA/+ mice (n = 7) transduced with an empty YFP control vector compared with Npm1cA/+ mice with Meis1 overexpression (n = 8). (G) WBC counts for Npm1cA/+ mice (n = 7) transduced with an empty YFP control vector compared with Npm1cA/+ mice with Meis1 overexpression (n = 7). (H) Histopathological analysis of a Npm1cA/+ plus YFP mouse compared with a Npm1cA/+ plus Meis1 mouse. ∗P < .05; ∗∗P < .001; ∗∗∗P < .0001. GSEA, gene set enrichment analysis; TCGA, the cancer genome atlas; WBC, white blood cell.
MEIS1 collaborates with NPM1c mutation to induce high-penetrance myeloid leukemia in mice. (A) MEIS1 expression in NPM1c de novo AML patients (n = 107) compared with NPM1WT AML patients (n = 340) from the Vizome data set.26 (B) MEIS1 expression in NPM1c de novo AML patients (n = 107) compared with CD34+ healthy BM (n = 13) from the Vizome data set.26 (C) GSEA analysis of 25% of highest MEIS1 (MEIS1High) expressing vs 25% lowest MEIS1 expressing (MEIS1Low) NPM1c de novo AML patients from the TCGA data set.27 (D) Schematic of the experimental design of in vivo experiments with Npm1cA/+ mouse model. (E) Kaplan-Meier plot of Npm1cA/+ mice transduced with a YFP empty vector (n = 8), Meis1 (n = 10), and secondary transplants of NPM1c+ Meis1 leukemic cells (n = 6). (F) Spleen weight of Npm1cA/+ mice (n = 7) transduced with an empty YFP control vector compared with Npm1cA/+ mice with Meis1 overexpression (n = 8). (G) WBC counts for Npm1cA/+ mice (n = 7) transduced with an empty YFP control vector compared with Npm1cA/+ mice with Meis1 overexpression (n = 7). (H) Histopathological analysis of a Npm1cA/+ plus YFP mouse compared with a Npm1cA/+ plus Meis1 mouse. ∗P < .05; ∗∗P < .001; ∗∗∗P < .0001. GSEA, gene set enrichment analysis; TCGA, the cancer genome atlas; WBC, white blood cell.
Npm1 haploinsufficiency and Meis1 overexpression collaborate independent of Npm1c mutation to induce AML
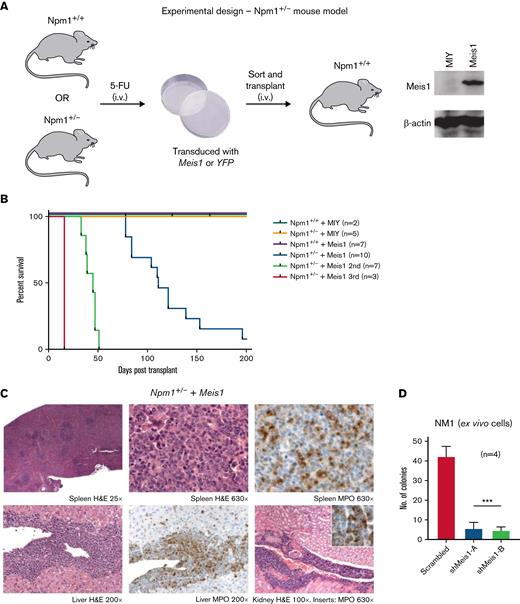
The Npm1cA/+ mouse model recapitulated the 2 main consequences of NPM1 mutations (ie, the increase of the NPM1c in the cytoplasm and the consequential insufficient levels of NPM1 in the nucleolus/nucleus). Of note, the insufficient levels of NPM1 do not cause AML in mouse models. We sought to test whether Meis1 can act as an oncogene collaboratively with the inadequate levels of NPM1, independent of the NPM1c mutation, for which we used a Npm1 haploinsufficient mouse model (Npm1+/−).11,24 In this model, the majority of the Hox/Meis genes were downregulated compared with Npm1+/+ HSPCs (supplemental Figure 2A). 5-FU enriched BM HSPCs from haploinsufficient (Npm1+/−) and wild-type (Npm1+/+) mice were transduced with Meis1 or YFP and transplanted into lethally irradiated Npm1+/+ mice (Figure 2A). The mice transplanted with Npm1+/+ stem progenitors overexpressing Meis1 or YFP and Npm1+/− HSPCs expressing YFP did not generate disease in mice (Figure 2B). However, 9 out of 10 mice transplanted with Npm1+/− cells overexpressing Meis1 (Npm1+/−+Meis1) generated a lethal hematopoietic disease, 7 of which were classified as AML, with a median latency of 111 days (Figure 2B). The leukemic mice displayed splenomegaly and elevated blasts and WBC counts and generated myeloid leukemia in secondary and tertiary recipients with median latencies of 45 and 16 days, respectively (Figure 2B; supplemental Figure 2C-D). Histopathology of mice from 3 independent cohorts indicated AML with maturation in all 3 cases tested, and 1 mouse also showed signs of T-cell lymphoblastic lymphoma in lymph nodes, lung, and spleen (Figure 2C; supplemental Figure 2D). Importantly, the phenotype observed in Npm1cA/++ Meis1 was similar to Npm1+/− plus Meis1 leukemic mice: both groups developed AML with maturation and infiltration of multiple organs (supplemental Figure 2E). Immunophenotypically, similar to Npm1cA/+ plus Meis1 leukemic mice, leukemic Npm1+/− plus Meis1 mice also showed a trend toward the presence of higher Gr1+, Mac1+, and c-kit+ single-positive cells in the BM (supplemental Figure 2F). In sum, this demonstrates that NPM1 haploinsufficiency in concert with overexpression of Meis1 is sufficient to induce AML in mice, independent of the NPM1c protein. However, cytogenetic analysis of a representative Npm1+/− plus Meis1 leukemic mouse BM grown ex vivo (referred to as NM1 cells) revealed that leukemia generated by Npm1+/− plus Meis1 was cytogenetically abnormal (supplemental Figure 2G). Therefore, we tested whether the leukemia was still reliant on MEIS1 or if it had become independent of MEIS1 due to the development of aneuploidy. For this, we suppressed Meis1 expression using 2 shRNA in this aneuploid cell population: shRNA-mediated depletion of Meis1 nearly eliminated clonogenic potential by inducing a >10-fold decrease in colony forming cell assay (CFC), demonstrating that Meis1 expression is not only crucial for leukemia initiation in the context of Npm1 haploinsufficiency but also for the maintenance of transformed leukemic cells (Figure 2D).
Npm1 haploinsufficiency and Meis1 overexpression collaborate to induce AML. (A) Schematic of the experimental design of in vivo experiments with Npm1. (B) Kaplan-Meier plot of NPM1+/+ mice transduced with a MIY empty vector (n = 2), NPM1+/− plus MIY (n = 5), NPM1+/+ plus Meis1 (n = 7), NPM1+/− plus Meis1 (n = 10), secondary transplants of NPM1+/− plus Meis1 leukemic BM (n = 7), and tertiary transplants of NPM1+/− plus Meis1 leukemic BM (n = 3). (C) Histopathological analysis of a NPM1+/− plus Meis1 mouse. (D) Colony-forming assay of NPM1+/− plus Meis1 ex vivo cells (NM1) transduced with scrambled control or shRNA against Meis1 (n = 4). indicates number of independent experiments. ∗P < .05; ∗∗P < .001; ∗∗∗P < .0001. MIY, MSCV IRES YFP plasmid; n, number of independent experiments.
Npm1 haploinsufficiency and Meis1 overexpression collaborate to induce AML. (A) Schematic of the experimental design of in vivo experiments with Npm1. (B) Kaplan-Meier plot of NPM1+/+ mice transduced with a MIY empty vector (n = 2), NPM1+/− plus MIY (n = 5), NPM1+/+ plus Meis1 (n = 7), NPM1+/− plus Meis1 (n = 10), secondary transplants of NPM1+/− plus Meis1 leukemic BM (n = 7), and tertiary transplants of NPM1+/− plus Meis1 leukemic BM (n = 3). (C) Histopathological analysis of a NPM1+/− plus Meis1 mouse. (D) Colony-forming assay of NPM1+/− plus Meis1 ex vivo cells (NM1) transduced with scrambled control or shRNA against Meis1 (n = 4). indicates number of independent experiments. ∗P < .05; ∗∗P < .001; ∗∗∗P < .0001. MIY, MSCV IRES YFP plasmid; n, number of independent experiments.
Meis1 collaborates with Npm1 haploinsufficiency to induce gene expression profile akin to NPM1cCN-AML patients
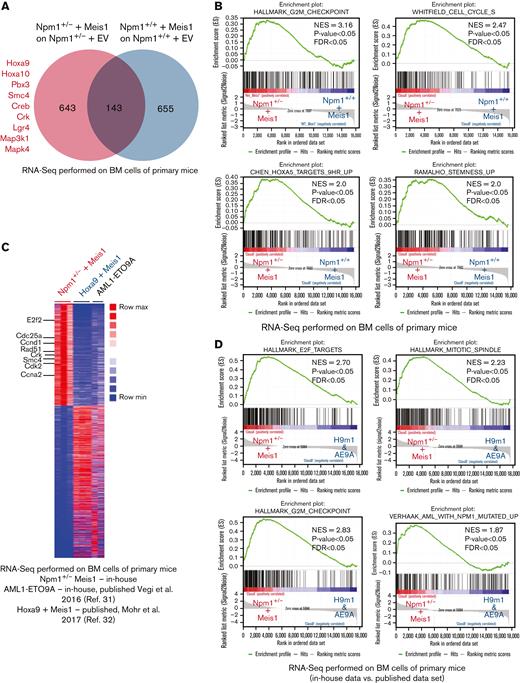
Next, we aimed to understand the molecular pathways governed by Meis1 in the context of Npm1 haploinsufficiency and assessed whether the Npm1+/− + Meis1 model recapitulated the transcriptional profile of human NPM1c AML. For this, we performed RNA-seq on Npm1+/− plus Meis1 vs Npm1+/+ plus Meis1 cells and empty vector controls. Our initial analysis revealed 643 genes differentially expressed genes in the Npm1+/− plus Meis1 arm, which included genes typically upregulated in NPM1c AML patients such as Hoxa cluster genes (Hoxa 5, 7, 9, and 10) and Pbx3 (Figure 3A; supplemental Table 1). Furthermore, oncogenes associated with proliferation and AML and AML-stem cell such as Crk, Lgr4, Smc4, and MAP kinases were also upregulated (Figure 3A). MEIS1 is required for the maintenance of an embryonic stem cell-like gene signature in MLL leukemia and is associated with the regulation of the cell cycle in multiple cell types.30 As mentioned previously, MEIS1 expression strongly correlated with stem cell and cell cycle gene expression signature in NPM1c AML patients. Concordantly, in GSEA, Npm1+/− plus Meis1 leukemic cells exhibited a significant enrichment for stem cell and cell cycle–associated gene expression signatures (Figure 3B).
Meis1 upregulates stem cell- and cell cycle–associated pathways in Npm1 haploinsufficient cells. (A) Venn diagram of RNA-seq data comparing differentially expressed genes in NPM1+/− plus Meis1 vs NPM1+/+ plus Meis1; both cohorts were first compared with their respective empty vector controls. (B) GSEA analysis for pathways upregulated in NPM1+/− plus Meis1 vs NPM1+/+ plus Meis1 cells. (C) Heatmap of differentially expressed genes in NPM1+/− plus Meis1, Hoxa9 plus Meis1 (H9M1), and AML1-ETO9a (AE9A) primary leukemic mice.31,32 (D) GSEA analysis for pathways enriched in NPM1+/− plus Meis1 vs H9M1 and AE9A primary leukemic mice. ∗P < .05; ∗∗P < .001; ∗∗∗P < .0001. FDR, false discovery rate; NES, normalized enrichment score.
Meis1 upregulates stem cell- and cell cycle–associated pathways in Npm1 haploinsufficient cells. (A) Venn diagram of RNA-seq data comparing differentially expressed genes in NPM1+/− plus Meis1 vs NPM1+/+ plus Meis1; both cohorts were first compared with their respective empty vector controls. (B) GSEA analysis for pathways upregulated in NPM1+/− plus Meis1 vs NPM1+/+ plus Meis1 cells. (C) Heatmap of differentially expressed genes in NPM1+/− plus Meis1, Hoxa9 plus Meis1 (H9M1), and AML1-ETO9a (AE9A) primary leukemic mice.31,32 (D) GSEA analysis for pathways enriched in NPM1+/− plus Meis1 vs H9M1 and AE9A primary leukemic mice. ∗P < .05; ∗∗P < .001; ∗∗∗P < .0001. FDR, false discovery rate; NES, normalized enrichment score.
Next, to further understand whether our Npm1+/− + Meis1 model recapitulated the transcriptional features of human NPM1c AML, we compared Npm1+/− plus Meis1 RNA-seq data against our own published RNA-seq data of AML1-ETO9A (AE9A)-transformed leukemic cells, which represents a nonhomeobox-driven leukemia,31 and a published data set of Hoxa9 plus Meis1 (H9M1)-transformed leukemia,32 exemplifying a Hox-driven Npm1wt cytogenetically normal AML. In comparison with AE9A, Npm1+/− plus Meis1 exhibited the key transcriptional feature of NPM1c AML, the upregulation of several Hoxa genes, which was supported by confirmatory real-time polymerase chain reaction (PCR) data (supplemental Figure 3A-B). Moreover, cell cycle- and stemness-associated oncogenes previously observed in our analysis, such as Smc4 and Crk, were again found upregulated in Npm1+/− plus Meis1 vs AE9A leukemic cells (supplemental Figure 3A). Importantly, GSEA revealed the enrichment of the human NPM1c AML gene signature in Npm1+/− plus Meis1 vs AE9A cells (supplemental Figure 3C). Next, both H9m1 and Npm1+/− plus Meis1 leukemic cells expectedly revealed upregulation of the Hoxa cluster and Pbx3 genes (supplemental Figure 3D). However, yet again, Npm1+/− plus Meis1 exhibited upregulation of oncogenes Smc4 and Crk. Overall, the transcriptional profile of Npm1+/− plus Meis1-driven leukemia was best revealed by collective comparison against AE9a and H9M1 using GSEA, which showed a clear enrichment of the human NPM1c AML gene signature and the overrepresentation of cell cycle–associated pathways in Npm1+/− plus Meis1 leukemic cells vs H9m1 and AE9A immortalized cells (Figure 3C-D; supplemental Figure 3E; supplemental Table 2). The overrepresentation of the cell cycle pathway in our RNA-seq data set was of particular interest as the NPM1c AML signature, which compares NPM1c-harboring patients to NPM1WT AML patients, also showed enrichment of cell cycle pathways, signifying the importance of the cell cycle pathway in NPM1c AML (supplemental Figure 3F-G). Taken together, these data indicated that NPM1 haploinsufficiency likely alters MEIS1 binding and allows it to upregulate several NPM1c AML–associated pathways, independent of the presence of NPM1c mutation.
Meis1 upregulates stem cell and cell cycle–associated oncogene SMC4 in the context of Npm1 haploinsufficiency
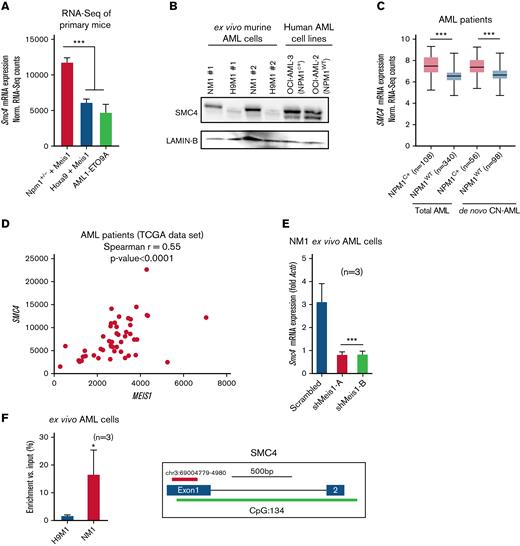
In line with our RNA-seq analysis, the cell cycle- and stemness-associated factor, small structural maintenance of chromosome protein 4 (SMC4), identified as an oncogene in multiple cancers, exhibited significantly higher expression in Npm1+/− plus Meis1 leukemic cells compared with H9M1 and AE9A leukemic cells in our RNA-seq analysis (Figure 4A). The SMC4 protein was considerably higher expressed in haploinsufficient NM1 cells and the NPM1c-harboring human AML cell line OCI-AML3 compared with Npm1wt-harboring H9M1 and OCI-AML2 cells (Figure 4B). SMC4 was significantly higher expressed in NPM1c vs NPM1WT AML patients (Figure 4C) and exhibited a strong positive correlation with MEIS1 expression in NPM1c AML patients (Figure 4D).27 Similar to MEIS1 expression, SMC4 expression levels were independent of collaborating mutations status in NPM1c AML patients (supplemental Figure 4A).26 Furthermore, knockdown of Meis1 in NM1 cells lead to a significant decrease in mRNA expression levels of Smc4 (Figure 4E). MEIS1 cleavage under targets and release using nuclease followed by quantitative real-time PCR (CUT&RUN qPCR) revealed that MEIS1 was enriched at the Smc4 promoter in NM1 cells but exhibited negligible enrichment in H9M1 cells, whereas a known binding site of MEIS1, the Maf1 promoter, was comparably enriched in both (Figure 4F; supplemental Figure 4C). Lastly, the depletion of SMC4 via shRNA impeded growth and colony formation more effectively in NM1 and OCI-AML3 cells than in Npm1wt-harboring OCI-AML2 and H9M1 cells, indicating that Npm1 haploinsufficient and NPM1c+ cells are more vulnerable to SMC4 depletion (supplemental Figure 4D-E).
The cell cycle protein SMC4 is a downstream target of MEIS1 in NPM1c AML. (A) Smc4 mRNA expression in NPM1+/− plus Meis1, Hoxa9 plus Meis1, and AML1-ETO9A leukemic mice; values are shown as normalized RNA-seq counts. (B) Western blot analysis on protein extracts from NM1, H9M1 murine ex vivo leukemic cells, and human AML cell lines OCI-AML3 and OCI-AML2 probed with antibody against SMC4 and LAMIN-B as endogenous control. (C) SMC4 mRNA expression in NPM1c vs NPM1WT AML patients in total human AML and de novo CN-AML patient cohorts from the Vizome data set; values are shown as normalized RNA-seq counts.26 (D) Spearman correlation of SMC4 and MEIS1 expression in NPM1c AML patients (r = 0.55, P < .0001). (E) Smc4 mRNA expression in NM1 cells transduced with scrambled control or shRNA against Meis1 (n = 3); values are shown as fold change to housekeeping gene control, Actb. (F) CUT&RUN of MEIS1 in H9M1 and NM1 cells analyzed for enrichment of Smc4 promoter (n = 3); values are shown as percentage Smc4 enrichment in input control. ∗P < .05; ∗∗P < .001; ∗∗∗P < .0001. mRNA, messenger RNA. Actb, Actin Beta; n, number of experimental replicates.
The cell cycle protein SMC4 is a downstream target of MEIS1 in NPM1c AML. (A) Smc4 mRNA expression in NPM1+/− plus Meis1, Hoxa9 plus Meis1, and AML1-ETO9A leukemic mice; values are shown as normalized RNA-seq counts. (B) Western blot analysis on protein extracts from NM1, H9M1 murine ex vivo leukemic cells, and human AML cell lines OCI-AML3 and OCI-AML2 probed with antibody against SMC4 and LAMIN-B as endogenous control. (C) SMC4 mRNA expression in NPM1c vs NPM1WT AML patients in total human AML and de novo CN-AML patient cohorts from the Vizome data set; values are shown as normalized RNA-seq counts.26 (D) Spearman correlation of SMC4 and MEIS1 expression in NPM1c AML patients (r = 0.55, P < .0001). (E) Smc4 mRNA expression in NM1 cells transduced with scrambled control or shRNA against Meis1 (n = 3); values are shown as fold change to housekeeping gene control, Actb. (F) CUT&RUN of MEIS1 in H9M1 and NM1 cells analyzed for enrichment of Smc4 promoter (n = 3); values are shown as percentage Smc4 enrichment in input control. ∗P < .05; ∗∗P < .001; ∗∗∗P < .0001. mRNA, messenger RNA. Actb, Actin Beta; n, number of experimental replicates.
Menin-MLL1 interaction inhibitor MI-3454 downregulates MEIS1 and SMC4, and Meis1 expression is a determinant of sensitivity to MI-3454
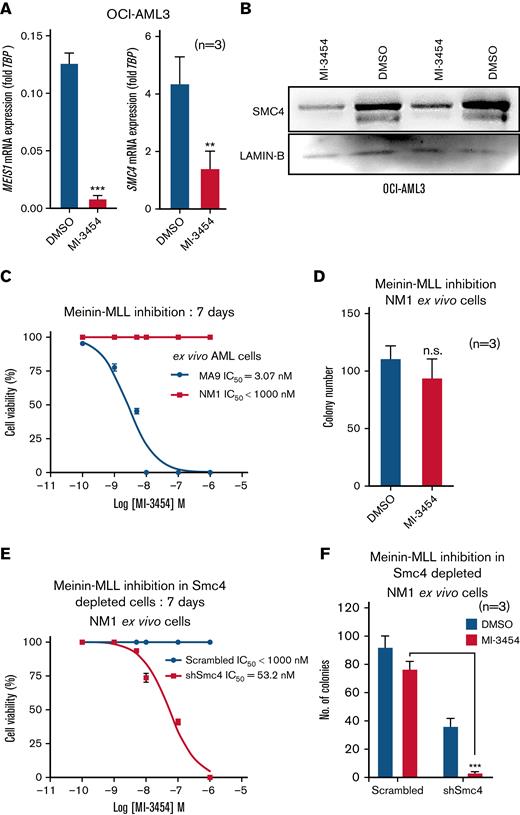
Recent studies have shown that the Menin-MLL interaction inhibitors successfully abrogate growth of NPM1c AML cells and dramatically downregulate MEIS1 expression in vivo.20,22,23 In line with these findings, we observed that the OCI-AML3 cells were sensitive to the Menin-MLL interaction inhibitor MI-3454 (half-maximal inhibitory concentratio (IC50) = 25 nM) and induced a drastic decrease in MEIS1 expression (Figure 5A; supplemental Figure 5A). Consistent with our observation of a close correlation between Meis1 and SMC4 expression, SMC4 mRNA and protein were also downregulated in MI-3454–treated OCI-AML3 cells (Figure 5A-B). Moreover, published RNA-seq data of OCI-AML3 cells treated with the Menin-MLL interaction inhibitor MI-503 also showed MEIS1 and SMC4 downregulation vs control (supplemental Figure 5B).22 In NPM1c AML, high MEIS1 expression is associated with treatment response to the pharmacological inhibition of the Menin-MLL interaction.22,23 In this regard, NM1 cells exhibited very high tolerance for MI-3454 compared with endogenous Meis1 expressing MLL-AF9 cells (IC50 MLL-AF9 = 3.07 nM vs IC50 NM1 > 1000nM; P < .0001) (Figure 5C) and exhibited only a trend toward reduced colony-forming ability vs DMSO (P = .2, Student t test) (Figure 5D). Notably, the depletion of Smc4 was sufficient to sensitize Meis1 overexpressing NM1 cells to MI-3454, as it reduced the IC50 concentration of MI-3454 by several folds (>1000 nM to 53.2 nM, P < .0001) and significantly reduced colony-forming ability vs DMSO and scrambled controls (P < .0001) (Figure 5E-F). These data suggest that in addition to Meis1, its downstream target, Smc4 is also a determinant of the efficacy of Menin-MLL interaction inhibitors, characterizing SMC4 inhibitors as an attractive therapeutic avenue for NPM1c AML.
MEIS1 and SMC4 expression levels affect response to MI-3454 treatment. (A) MEIS1 and SMC4 mRNA expression in OCI-AML3, treated with DMSO control or MI-3454 (25 nM, n = 3) for 7 days; values are shown as fold change to housekeeping gene control, TBP. (B) Western blot analysis showing 2 experimental replicates of SMC4 protein expression in OCI-AML3 cells treated with 25 nM MI-3454 for 7 days vs DMSO control. LAMIN-B was used as endogenous control. (C) IC50 calculation of MI-3454 inhibitor on Meis1 overexpressing NM1 cells vs endogenous Meis1 expressing ex vivo MLL-AF9 cells. (D) Colony number in the colony forming assay of NM1 cells treated with DMSO and MI-3454. (E) IC50 calculation of MI-3454 inhibitor on NM1 cells transduced with scrambled control or shRNA against Smc4. (F) Colony forming assay of NM1 cells transduced with scrambled control and shRNA-mediated Smc4 knockdown, treated with DMSO and MI-3454 (n = 3). ∗P < .05; ∗∗P < .001; ∗∗∗P < .0001. DMSO, dimethyl sulfoxide; n, number of independent experiments; TBP, TATA-binding protein.
MEIS1 and SMC4 expression levels affect response to MI-3454 treatment. (A) MEIS1 and SMC4 mRNA expression in OCI-AML3, treated with DMSO control or MI-3454 (25 nM, n = 3) for 7 days; values are shown as fold change to housekeeping gene control, TBP. (B) Western blot analysis showing 2 experimental replicates of SMC4 protein expression in OCI-AML3 cells treated with 25 nM MI-3454 for 7 days vs DMSO control. LAMIN-B was used as endogenous control. (C) IC50 calculation of MI-3454 inhibitor on Meis1 overexpressing NM1 cells vs endogenous Meis1 expressing ex vivo MLL-AF9 cells. (D) Colony number in the colony forming assay of NM1 cells treated with DMSO and MI-3454. (E) IC50 calculation of MI-3454 inhibitor on NM1 cells transduced with scrambled control or shRNA against Smc4. (F) Colony forming assay of NM1 cells transduced with scrambled control and shRNA-mediated Smc4 knockdown, treated with DMSO and MI-3454 (n = 3). ∗P < .05; ∗∗P < .001; ∗∗∗P < .0001. DMSO, dimethyl sulfoxide; n, number of independent experiments; TBP, TATA-binding protein.
NPM1 haploinsufficiency is a targetable vulnerability of NPM1c AML cells and can be combined with MI-3454 to specifically antagonize NPM1c AML growth
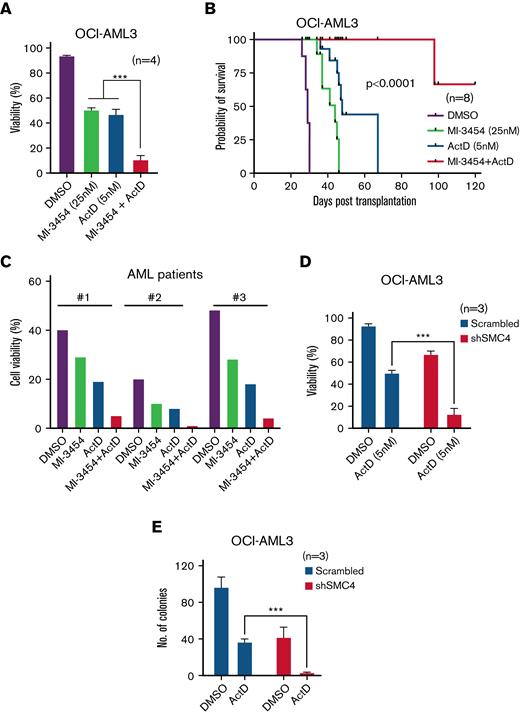
Recent studies strongly indicate that inadequate levels of NPM1 render NPM1c AML cells unable to regulate nucleolar stress efficiently and leave them vulnerable to chemically induced nucleolar stress via drugs such as actinomycin d (ActD).33 Consistent with these findings, NPM1c harboring OCI-AML3 human AML cells and Npm1 haploinsufficient NM1 murine AML cells were more sensitive to ActD compared with NPM1WT-harboring OCI-AML2 human AML cells and H9M1 murine AML cells (supplemental Figure 6A-B).9,33 With the aim to capitalize on NPM1c-induced haploinsufficiency and simultaneously antagonize the crucial Meis1-Smc4 axis, in a second step we combined ActD treatment with MI-3454. This combination, applying the IC50 concentrations of ActD (5 nM) and MI-3454 (25 nM), induced a marked reduction in the viability of OCI-AML3 cells, decreased colony-forming potential, and significantly enhanced survival of xenografts vs DMSO and single-drug treatment (Figure 6A-B; supplemental Figure 6C). Primary NPM1c AML patient cells treated with ActD and MI-3454 exhibited enhanced apoptosis vs DMSO and single-drug therapy in vitro (Figure 6C; supplemental Table 3). Lastly, MI-3454 treatment could be substituted by the knockdown of SMC4 in OCI-AML3 cells as SMC4 depletion combined with ActD treatment–induced cell death and reduction in the colony-forming ability of OCI-AML3 cells (Figure 6D-E). Taken together, our data demonstrate that the growth of NPM1c AML cells can be abolished by capitalizing on NPM1 haploinsufficiency and simultaneously targeting the oncogenic MEIS1-SMC4 axis.
Effect of MI-3454 in combination with actinomycin D. (A) Viability of OCI-AML3 treated with DMSO, MI-3454 (25nM) or Act-D (5nM) as single agent or with the combination therapy of MI-3454 (25nM) and Act-D (5nM) (n=4). n indicates number of independent experiments. (B) Kaplan Maier plot of NSG mice injected with OCI-AML3 treated with DMSO, MI-3454, Act-D or the combination of MI-3454 and Act-D (n=8). n indicates number of independent NSG mice used per experimental arm. (C) Cell viability of BM cells originating from three primary NPM1c AML patients treated with DMSO, MI-3454, Act-D or the combination of MI-3454 and Act-D. (D) The difference in cell viability of SMC4 depleted OCI-AML3 cells versus scrambled control, measured 48h post-treatment with DSMO or Act-D. n indicates number of independent experiments. (E) No. of colonies in the colony forming assay of OCI-AML3 transduced with scrambled control and shRNA mediated SMC4 knockdown, treated with DMSO or Act-D (n=3). n indicates number of independent experiments. ∗P < 0.05; ∗∗P < 0.001; ∗∗∗P < 0.0001.
Effect of MI-3454 in combination with actinomycin D. (A) Viability of OCI-AML3 treated with DMSO, MI-3454 (25nM) or Act-D (5nM) as single agent or with the combination therapy of MI-3454 (25nM) and Act-D (5nM) (n=4). n indicates number of independent experiments. (B) Kaplan Maier plot of NSG mice injected with OCI-AML3 treated with DMSO, MI-3454, Act-D or the combination of MI-3454 and Act-D (n=8). n indicates number of independent NSG mice used per experimental arm. (C) Cell viability of BM cells originating from three primary NPM1c AML patients treated with DMSO, MI-3454, Act-D or the combination of MI-3454 and Act-D. (D) The difference in cell viability of SMC4 depleted OCI-AML3 cells versus scrambled control, measured 48h post-treatment with DSMO or Act-D. n indicates number of independent experiments. (E) No. of colonies in the colony forming assay of OCI-AML3 transduced with scrambled control and shRNA mediated SMC4 knockdown, treated with DMSO or Act-D (n=3). n indicates number of independent experiments. ∗P < 0.05; ∗∗P < 0.001; ∗∗∗P < 0.0001.
Discussion
In recent years, the molecular mechanisms by which NPM1c initiates and maintains AML have been well studied.6-9 However, the contribution of NPM1 haploinsufficiency remains relatively less explored. Using the Npm1+/− mouse model, we demonstrate as a proof of principle that even in the absence of NPM1c, NPM1 haploinsufficiency can provide a fertile ground for myeloid leukemogenesis if appropriate collaboration partners are highly expressed, such as MEIS1, a hallmark of human NPM1c AML.
MEIS1 has a well-established but complex and context-dependent role in cell cycle regulation in normal hematopoietic stem cells as well as leukemia.30,34-38 It was shown in murine embryonic fibroblasts that the tumor-promoting signature of Meis1 is associated with the regulation of the centrosome cycle, and inhibition of Meis1 transforming activity suppresses cell cycle pathways.39 Furthermore, the MEIS family member MEIS2 prevents mitotic catastrophe in neuroblastoma by regulating the cell cycle.40 Our studies in the Npm1 haploinsufficient mouse model demonstrate that inadequate levels of NPM1 abet the oncogenic function of MEIS1, among others, by its regulation of the cell cycle and stem cell–associated pathways.
Our and published RNA-seq data sets elucidates that compared with NPM1wt-harboring AML, AML generated by the combination of NPM1 haploinsufficiency and MEIS1 overexpression or that observed in NPM1c AML patients is characterized by the upregulation of several cell cycle pathway genes. Interestingly, in NPM1 haploinsufficient AML cells, MEIS1 exhibits increased occupancy on the promoter of one such cell cycle–associated oncogene, Smc4, but not in NPM1wt AML cells. Moreover, Meis1 knockdown decreases Smc4 expression in haploinsufficient AML cells. Collectively, these data illustrate that MEIS1 regulates Smc4 by displaying a binding affinity at its promoter in the context of NPM1 haploinsufficiency that it does not demonstrate in AML cells harboring NPM1wt. These data indicate that Npm1 haploinsufficiency alters MEIS1 binding, which in turn upregulates a set of cell cycle- and stemness-associated genes, such as SMC4, that allow Npm1 haploinsufficient cells to overcome a myelodysplastic syndrome–like phenotype and instead develop a fully penetrant AML. Furthermore, these data also suggest that NPM1 haploinsufficiency could contribute to MEIS1 function in human AML.
The upstream regulator and binding partner of MEIS1, the leucine zipper transcription factor cAMP response element-binding protein (CREB) was consistently transcriptionally upregulated in haploinsufficient Npm1+/− plus Meis1 murine AML cells vs Npm1wt harboring Npm1+/+ plus Meis1 and Hoxa9 plus Meis1 AML cells (supplemental Tables 1 and 2). As CREB interacts with MEIS1 to facilitate its binding at cis-regulatory elements and the expression of its target genes, higher levels of CREB could explain the altered MEIS1 binding.41 The fact that Smc4 and several other cell cycle genes upregulated in haploinsufficient AML cells and identified as signature genes in NPM1c AML harbor CRE sites on their promoter supports this theory.42 Nevertheless, in the future, understanding the underlying mechanisms that alter MEIS1 binding in haploinsufficient cells necessitates detailed investigations into the posttranslational modifications and binding partners of MEIS1 in haploinsufficient vs NPM1wt leukemic cells.
Our data indicate that MEIS1 performs its oncogenic function in association with SMC4.43 SMC4 levels peak at mitosis and play a key role in faithful chromosome condensation and segregation during mitosis.44 In cancer, SMC4 is assigned an oncogenic role in multiple solid tumors and MLL-AF9 leukemia.45-47 SMC4 likely plays a role in NPM1WT AML as well. However, our findings suggest that MEIS1 exhibits considerably stronger binding at the SMC4 promoter in the context of NPM1 haploinsufficiency, leading to increased expression levels in NPM1c AML patients vs NPM1WT AML patients. This might contribute to our observation that NPM1c AML cells are more reliant on SMC4 expression for leukemic growth compared with NPM1wt AML cells. Taking this into account, targeting MEIS1 and SMC4 would be a potential approach to counteract AML growth, although at the moment, there are no chemical inhibitors of SMC4 readily available. Because it has been previously shown that inhibiting Menin-MLL interaction induces downregulation of MEIS1 expression in vivo, inhibiting the MEIS1-SMC4 axis using the Menin-MLL inhibitors in NPM1c AML is a logical choice.20-23
Here, we demonstrate that the efficacy of the Menin-MLL interaction inhibitor MI-3454 depends on the expression of MEIS1 and its downstream target SMC4. Constitutive Meis1 expressing NM1 cells were resistant to MI-3454, whereas SMC4 depletion potentiated the sensitivity of NM1 cells to MI-3454. This suggests that the MEIS1-SMC4–driven cell cycle program contributes to the resistance against MI-3454. Thus, the efficacy of MLL1-Menin inhibitors on NPM1c AML cells would likely improve if combined with SMC4 or cell cycle inhibitors. Lastly, recent studies have elucidated that ActD, an RNA polymerase I inhibitor, is particularly effective at targeting NPM1c AML cells by inducing nucleolar stress and promyelocytic leukemia protein nuclear body formation.9,33 We could show that NPM1 haploinsufficient NM1 cells are vulnerable to ActD, and its combination with MEIS1 downregulation via MI-3454 abrogates NPM1c AML leukemic growth in vitro and in vivo. These data suggest that exploiting NPM1 haploinsufficiency in combination with abrogation of the MEIS1-SMC4 axis could be a viable avenue for treating NPM1c AML patients.
Acknowledgments
The authors would like to thank all members of the animal facility and genomics and flow cytometry core facilities of the University of Ulm, Germany, and at the animal facility at Wellcome Trust Sanger Institute, United Kingdom. They thank all members of the Institute of Experimental Cancer Research for technical support such as patient sample preparation, cell sorting, irradiation of mice, or injection of cells into mice. They thank K. Döhner and H. Döhner (Department of Internal Medicine III, University Hospital Ulm) for providing primary patient samples.
C.B. was funded by a grant from the DFG (SFB 1074 project A4 to C.B.). S.B. was funded by the Baustein program 3.2, University of Ulm, Germany.
Authorship
Contribution: C.B, S.B., and A. Muranyi designed the project; A. Muranyi, T.A., A.K., A. Mupo, V.P.S.R., and S.B. performed the experiments; S.B. and T.A. performed the RNA-seq data analysis in collaboration with A.S. and Basepair; I.G.-M. and L.Q.-M. performed histopathology; A.A. and C.G. performed and analyzed the cytogenetic analyses; G.V. provided the Npm1cA/+ mouse model, and experiments pertaining to it were performed in G.V.'s laboratory in Cambridge with the support of G.V.; and C.B. and S.B. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christian Buske, Institute of Experimental Cancer Research, University Hospital Ulm, Albert Einstein Allee-11, Ulm 89081, Germany; e-mail: christian.buske@uni-ulm.de; and Shiva Bamezai, Institute of Experimental Cancer Research, University Hospital Ulm, Albert Einstein Allee-11, Ulm 89081, Germany; e-mail: shiva.bamezai@uni-ulm.de.
References
Author notes
Data can be accessed by e-mail to the corresponding author, Christian Buske (christian.buske@uni-ulm.de).
The full-text version of this article contains a data supplement.
A. Muranyi and T.A. contributed equally to the manuscript.