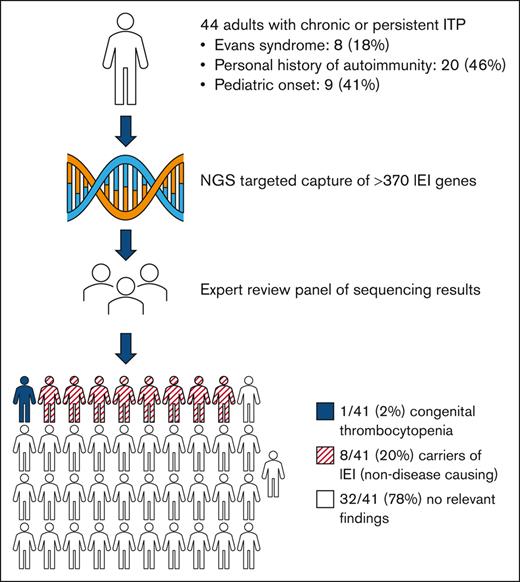
Key Points
Autoimmune cytopenias are a common manifestation of IEIs.
Although IEIs often underlie pediatric Evans syndrome, no cases of IEI were identified in this real-world study of adults with ITP and Evans syndrome.
Abstract
Inborn errors of immunity (IEIs) are monogenic disorders that predispose patients to immune dysregulation, autoimmunity, and infection. Autoimmune cytopenias, such as immune thrombocytopenia (ITP) and Evans syndrome (a combination of ITP and autoimmune hemolytic anemia), are increasingly recognized phenotypes of IEI. Although recent findings suggest that IEIs may commonly underlie pediatric ITP and Evans syndrome, its prevalence in adult patients with these disorders remains undefined. This study sought to estimate the prevalence of underlying IEIs among adults with persistent or chronic ITP or Evans syndrome using a next-generation sequencing panel encompassing >370 genes implicated in IEIs. Forty-four subjects were enrolled from an outpatient adult hematology clinic at a tertiary referral center in the United States, with a median age of 49 years (range, 20-83). Fourteen subjects (31.8%) had secondary ITP, including 8 (18.2%) with Evans syndrome. No cases of IEI were identified despite a high representation of subjects with a personal history of autoimmunity (45.5%) and early onset of disease (median age at diagnosis of 40 years [range, 2-77]), including 20.5% who were initially diagnosed as children. Eight subjects (18.2%) were found to be carriers of pathogenic IEI variants, which, in their heterozygous state, are not disease-causing. One case of TUBB1-related congenital thrombocytopenia was identified. Although systematic screening for IEI has been proposed for pediatric patients with Evans syndrome, findings from this real-world study suggest that inclusion of genetic testing for IEI in the routine work-up of adults with ITP and Evans syndrome has a low diagnostic yield.
Introduction
The inborn errors of immunity (IEIs) are a heterogenous group of genetic disorders characterized by immune deficiency and dysregulation, which lead to increased risk of infection, autoimmunity, and malignancy. The advent of comprehensive and affordable next-generation sequencing (NGS) has led to a rapid rise in the diagnosis and identification of IEIs, now numbering >450 monogenic defects.1
Immune thrombocytopenia (ITP) is a common manifestation of IEIs and can be the first presenting symptom of an underlying IEI in adults and children.2-5 Approximately 20% of patients with pediatric chronic ITP and 40% of patients with pediatric Evans syndrome (a combination of ITP and autoimmune hemolytic anemia) harbor an underlying IEI.6-9 In adults with immune cytopenias, the prevalence of underlying IEIs has not been defined, and IEIs may go unrecognized in these patients because of broad phenotypic heterogeneity and a lack of provider familiarity.10
Patients with autoimmune cytopenias are recommended to undergo quantitative immunoglobulin testing to assess the risk of immunosuppression.11 Formal diagnosis of an IEI may have additional utility as IEI-related autoimmune cytopenias are less responsive to standard therapies and often associate with other immunopathologic manifestations, such as immune-mediated gastrointestinal disease in children.8,12,13 Molecular diagnosis may also guide treatment given the mounting success of targeted therapies14-17 and hematopoietic stem cell transplantation for select patients with challenging IEI-associated sequelae, including refractory ITP.18
The purpose of this study was to estimate the prevalence of IEIs among a real-world cohort of adult patients from the United States with persistent or chronic ITP or Evans syndrome by applying an NGS panel encompassing >370 genes implicated in IEIs.10
Methods
Patient selection and study design
This was a prospective, single-center study conducted at the Fred Hutchinson Cancer Center from September 2021 to September 2022. The study protocol was approved by the University of Washington Institutional Review Board. All subjects provided written informed consent before enrollment.
Eligible patients were ≥18 years of age with a diagnosis of persistent or chronic ITP, defined as a platelet count of <100 × 109/L for 3 to 12 months or >12 months, respectively, in the absence of other causes.19 Subjects were excluded if they carried a diagnosis of a clonal lymphoproliferative disorder (eg, lymphoma or monoclonal B-cell lymphocytosis) or had prior genetic testing for IEIs. Potential subjects were recruited sequentially during clinic visits or approached for consent by phone or electronic mail in random order using a computerized number generator. Medical record review was conducted for relevant clinical data, including medical comorbidities, ITP history (ie, date of diagnosis, primary or secondary disease, prior lines of therapy, and history of splenectomy), the presence of persistent (≥3 months of) lymphadenopathy or splenomegaly on imaging, and laboratory results (ie, complete blood counts, mean platelet volume, immature platelet fraction, complement components 3 and 4, antinuclear antibodies, quantitative immunoglobulins, bone marrow aspirate and biopsy findings, and peripheral blood flow cytometry). Family history was obtained by interview, and a 3-generation pedigree was generated for each subject.
Genetic testing and analysis
DNA was isolated from peripheral blood or buccal swab samples. Targeted gene capture and massively parallel sequencing were performed at Invitae Corporation (San Francisco, CA), using an assay initially targeting mutations in 473 genes implicated in IEIs, congenital thrombocytopenia, inherited bone marrow failure, and other monogenic disorders (Table 1). In February 2022, halfway through enrollment, the panel was expanded to include 574 genes.
Targeted capture panel of genes associated with IEIs (n = 374), constitutional cytopenia (n = 21), and other monogenic disorders (n = 179)
| IEIs . | |
|---|---|
| Immunodeficiencies affecting cellular and humoral immunity | ADA, AK2, B2M, BCL10, CARD11, CD247, CD3D, CD3E, CD3G, CD40, CD40LG, CD8A, CIITA, CORO1A, DCLRE1C, DOCK2, DOCK8, FCHO1, ICOS, ICOSLG, IKBKB, IL21, IL21R, IL2RG, IL7R, IKZF1, ITK, JAK3, LAT, LCK, LIG4, MALT1, MAP3K14, MSN, NHEJ1, PAX1, POLD1, POLD2, PRKDC, PTPRC, RAG1, RAG2, REL, RELA, RELB, RFX5, RFXANK, RFXAP, RHOH, STK4, TAP1, TAP2, TAPBP, TFRC, TNFRSF4, and ZAP70 |
| Combined immunodeficiencies with associated or syndromic features | ARPC1B, ATM, BCL11B, BLM, CCBE1, CDCA7, CHD7, DNMT3B, EPG5, ERBIN, EXTL3, FAT4, FOXN1, GINS1, HELLS, IL6R, IL6ST, KDM6A, KMT2A, KMT2D, LIG1, MCM4, MTHFD1, MYSM1, NBN, NFE2L2, NFKBIA, NSMCE3, ORAI1, PGM3, PMS2, PNP, POLE, POLE2, RBCK1, RMRP, RNF168, RNF31, RNU4ATAC, SEMA3E, SKIV2L, SLC46A1, SMARCAL1, SP110, SPINK5, STAT3, STAT5B, STIM1, TBX1, TCN2, TGFBR1, TGFBR2, TTC37, TTC7A, WIPF1, ZBTB24, and ZNF341 |
| Predominantly antibody deficiencies | AICDA, ARHGEF1, ATP6AP1, BLNK, BTK, CD19, CD79A, CD79B, CD81, CR2, FNIP1, IGLL1, IRF2BP2, MOGS, MS4A1, MSH6, NFKB1, NFKB2, PIK3CD, PIK3R1, PTEN, SEC61A1, SH3KBP1, SLC39A7, TCF3, TNFRSF13B, TNFRSF13C, TNFSF12, TOP2B, TRNT1, and UNG |
| Diseases of immune dysregulation | AIRE, AP3B1, AP3D1, BACH2, CARMIL2, CASP10, CASP8, CD27, CEBPE, CTLA4, CTPS1, DEF6, FADD, FAS, FASLG, FERMT1, FOXP3, IL10, IL10RA, IL10RB, IL2RA, IL2RB, ITCH, LRBA, LYST, MAGT1, NFAT5, PEPD, PRF1, PRKCD, RAB27A, RASGRP1, RIPK1, SH2D1A, SLC7A7, STX11, STXBP2, TET2, TGFB1, TNFRSF9, TPP2, UNC13D, and XIAP |
| Congenital defects of phagocyte number or function | ACTB, CFTR, CLPB, CSF2RA, CSF2RB, CSF3R, CTSC, CXCR2, CYBA, CYBB, EFL1, ELANE, FERMT3, FPR1, G6PC3, G6PD, GATA2, GFI1, HAX1, HYOU1, ITGB2, JAGN1, LAMTOR2, MKL1, NCF2, NCF4, RAC2, SLC35C1, SLC37A4, SMARCD2, SRP54, TAZ, USB1, VPS13B, VPS45, WAS, and WDR1 |
| Defects in intrinsic and innate immunity | CARD9, CIB1, CLCN7, CXCR4, DBR1, HMOX1, IFIH1, IFNAR1, IFNAR2, IFNGR1, IFNGR2, IL12B, IL12RB1, IL12RB2, IL17F, IL17RA, IL17RC, IL23R, IL18BP, IRAK4, IRF4, IRF7, IRF8, IRF9, ISG15, JAK1, MYD88, NBAS, NCSTN, OSTM1, POLR3A, POLR3F, PSENEN, RANBP2, RORC, RPSA, SNX10, SPPL2A, STAT1, STAT2, TCIRG1, TICAM1, TLR3, TLR7, TMC6, TMC8, TNFRSF11A, TNFSF11, TRAF3, TRAF3IP2, TYK2, and UNC93B1 |
| Autoinflammatory disorders | ACP5, ADA2, ADAM17, ADAR, CARD14, CDC42, COPA, DNASE1L3, DNASE2, IL1RN, IL36RN, LPIN2, MEFV, MVK, NCKAP1L, NLRC4, NLRP1, NLRP12, NLRP3, NOD2, OAS1, OTULIN, PLCG2, POLA1, PSMB8, PSMG2, PSTPIP1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, SH3BP2, SLC29A3, TMEM173, TNFAIP3, TNFRSF1A, and TREX1 |
| Complement deficiencies | C1QA, C1QB, C1QC, C1S, C2, C3, C5, C6, C7, C8A, C8B, C9, CD46, CD55, CD59, CFB, CFD, CFH, CFI, CFP, SERPING1, and THBD |
| Bone marrow failure | ACD, BRCA1, BRCA2, BRIP1, CTC1, DKC1, DNAJC21, ERCC4, ERCC6L2, FANCA, FANCB, FANCC, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCL, FANCM, MAD2L2, NOP10, PALB2, PARN, RAD51, RAD51C, RFWD3, RTEL1, SAMD9, SAMD9L, SLX4, SRP72, STN1, TERC, TERT, TINF2, TP53, UBE2T, WRAP53, and XRCC2 |
| IEIs . | |
|---|---|
| Immunodeficiencies affecting cellular and humoral immunity | ADA, AK2, B2M, BCL10, CARD11, CD247, CD3D, CD3E, CD3G, CD40, CD40LG, CD8A, CIITA, CORO1A, DCLRE1C, DOCK2, DOCK8, FCHO1, ICOS, ICOSLG, IKBKB, IL21, IL21R, IL2RG, IL7R, IKZF1, ITK, JAK3, LAT, LCK, LIG4, MALT1, MAP3K14, MSN, NHEJ1, PAX1, POLD1, POLD2, PRKDC, PTPRC, RAG1, RAG2, REL, RELA, RELB, RFX5, RFXANK, RFXAP, RHOH, STK4, TAP1, TAP2, TAPBP, TFRC, TNFRSF4, and ZAP70 |
| Combined immunodeficiencies with associated or syndromic features | ARPC1B, ATM, BCL11B, BLM, CCBE1, CDCA7, CHD7, DNMT3B, EPG5, ERBIN, EXTL3, FAT4, FOXN1, GINS1, HELLS, IL6R, IL6ST, KDM6A, KMT2A, KMT2D, LIG1, MCM4, MTHFD1, MYSM1, NBN, NFE2L2, NFKBIA, NSMCE3, ORAI1, PGM3, PMS2, PNP, POLE, POLE2, RBCK1, RMRP, RNF168, RNF31, RNU4ATAC, SEMA3E, SKIV2L, SLC46A1, SMARCAL1, SP110, SPINK5, STAT3, STAT5B, STIM1, TBX1, TCN2, TGFBR1, TGFBR2, TTC37, TTC7A, WIPF1, ZBTB24, and ZNF341 |
| Predominantly antibody deficiencies | AICDA, ARHGEF1, ATP6AP1, BLNK, BTK, CD19, CD79A, CD79B, CD81, CR2, FNIP1, IGLL1, IRF2BP2, MOGS, MS4A1, MSH6, NFKB1, NFKB2, PIK3CD, PIK3R1, PTEN, SEC61A1, SH3KBP1, SLC39A7, TCF3, TNFRSF13B, TNFRSF13C, TNFSF12, TOP2B, TRNT1, and UNG |
| Diseases of immune dysregulation | AIRE, AP3B1, AP3D1, BACH2, CARMIL2, CASP10, CASP8, CD27, CEBPE, CTLA4, CTPS1, DEF6, FADD, FAS, FASLG, FERMT1, FOXP3, IL10, IL10RA, IL10RB, IL2RA, IL2RB, ITCH, LRBA, LYST, MAGT1, NFAT5, PEPD, PRF1, PRKCD, RAB27A, RASGRP1, RIPK1, SH2D1A, SLC7A7, STX11, STXBP2, TET2, TGFB1, TNFRSF9, TPP2, UNC13D, and XIAP |
| Congenital defects of phagocyte number or function | ACTB, CFTR, CLPB, CSF2RA, CSF2RB, CSF3R, CTSC, CXCR2, CYBA, CYBB, EFL1, ELANE, FERMT3, FPR1, G6PC3, G6PD, GATA2, GFI1, HAX1, HYOU1, ITGB2, JAGN1, LAMTOR2, MKL1, NCF2, NCF4, RAC2, SLC35C1, SLC37A4, SMARCD2, SRP54, TAZ, USB1, VPS13B, VPS45, WAS, and WDR1 |
| Defects in intrinsic and innate immunity | CARD9, CIB1, CLCN7, CXCR4, DBR1, HMOX1, IFIH1, IFNAR1, IFNAR2, IFNGR1, IFNGR2, IL12B, IL12RB1, IL12RB2, IL17F, IL17RA, IL17RC, IL23R, IL18BP, IRAK4, IRF4, IRF7, IRF8, IRF9, ISG15, JAK1, MYD88, NBAS, NCSTN, OSTM1, POLR3A, POLR3F, PSENEN, RANBP2, RORC, RPSA, SNX10, SPPL2A, STAT1, STAT2, TCIRG1, TICAM1, TLR3, TLR7, TMC6, TMC8, TNFRSF11A, TNFSF11, TRAF3, TRAF3IP2, TYK2, and UNC93B1 |
| Autoinflammatory disorders | ACP5, ADA2, ADAM17, ADAR, CARD14, CDC42, COPA, DNASE1L3, DNASE2, IL1RN, IL36RN, LPIN2, MEFV, MVK, NCKAP1L, NLRC4, NLRP1, NLRP12, NLRP3, NOD2, OAS1, OTULIN, PLCG2, POLA1, PSMB8, PSMG2, PSTPIP1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, SH3BP2, SLC29A3, TMEM173, TNFAIP3, TNFRSF1A, and TREX1 |
| Complement deficiencies | C1QA, C1QB, C1QC, C1S, C2, C3, C5, C6, C7, C8A, C8B, C9, CD46, CD55, CD59, CFB, CFD, CFH, CFI, CFP, SERPING1, and THBD |
| Bone marrow failure | ACD, BRCA1, BRCA2, BRIP1, CTC1, DKC1, DNAJC21, ERCC4, ERCC6L2, FANCA, FANCB, FANCC, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCL, FANCM, MAD2L2, NOP10, PALB2, PARN, RAD51, RAD51C, RFWD3, RTEL1, SAMD9, SAMD9L, SLX4, SRP72, STN1, TERC, TERT, TINF2, TP53, UBE2T, WRAP53, and XRCC2 |
| Constitutional cytopenia . |
|---|
| ABCG5, ABCG8, ACTN1, ANKRD26, BLOC1S6, CDAN1, CYCS, DIAPH1, ETV6, FLI1, GATA1, GP1BA, GP6, GP9, ITGA2B, ITGB3, MECOM, MPL, MYH9, RBM8A, RUNX1, THPO, TUBB1, VIPAS39, and VPS33B |
| Constitutional cytopenia . |
|---|
| ABCG5, ABCG8, ACTN1, ANKRD26, BLOC1S6, CDAN1, CYCS, DIAPH1, ETV6, FLI1, GATA1, GP1BA, GP6, GP9, ITGA2B, ITGB3, MECOM, MPL, MYH9, RBM8A, RUNX1, THPO, TUBB1, VIPAS39, and VPS33B |
| Miscellaneous . |
|---|
| ABCB7, ACAN, ADAMTS13, ADGRE2, AK7, ALAS2, ALG6, ALPK1, ANGPT1, ANKZF1, ANO6, ARMC4, ASAH1, ATR, BLOC1S3, C11orf70, C15orf41, C17orf62, CARD8, CBL, CCDC103, CCDC114, CCDC151, CCDC39, CCDC40, CCDC65, CCNO, CEP164, CFAP298, CHEK2, COL7A1, CYP27A1, DDX41, DDX58, DGAT1, DGKE, DNAAF1, DNAAF2, DNAAF3, DNAAF4, DNAAF5, DNAH1, DNAH11, DNAH5, DNAH8, DNAH9, DNAI1, DNAI2, DNAJB13, DNAL1, DRC1, DSG1, DTNBP1, DUOX2, EIF2AK3, EPCAM, ERCC2, ERCC3, FOXI3, G6PC, GAS8, GLRX5, GTF2H5, GTF2E2, GUCY2C, HPS1, HPS3, HPS4, HPS5, HPS6, HTRA2, ITGAM, JAK2, KAT6A, KDM1A, KIF23, KIT, KLF1, KLHDC8B, LARS2, LCT, LIPA, LRRC56, LRRC6, LRRC8A, LYN, MBD4, MCIDAS, MLH1, MPLKIP, MSH2, MYO5B, NAF1, NDUFB11, NEUROG3, NF1, NHP2, NME8, NPAT, NOTCH2, OFD1, P2RY12, PIH1D3, PLA2G4A, PLG, PLVAP, PMM2, PNLIP, POMP, POT1, PSMA3, PSMB4, PUS1, RASGRP2, RECQL4, RNF113A, RPGR, RPL11, RPL15, RPL18, RPL19, RPL23, RPL26, RPL27, RPL31, RPL35, RPL35A, RPL5, RPL9, RPS10, RPS15A, RPS19, RPS24, RPS26, RPS27, RPS28, RPS29, RPS7, RSPH1, RSPH3, RSPH4A, RSPH9, SAR1B, SBF2, SCO2, SEC23B, SGPL1, SI, SIAE, SLC10A2, SLC19A2, SLC25A38, SLC26A3, SLC5A1, SLC51B, SLC9A3, SPAG1, SPINT2, STAT4, STX3, TAOK2, TBXA2R, TERF2IP, TIMM50, TMPRSS15, TNFRSF6B, TONSL, TP63, TSR2, UNC45A, VAV1, WNT2B, YARS2, ZCCHC8, and ZMYND10 |
| Miscellaneous . |
|---|
| ABCB7, ACAN, ADAMTS13, ADGRE2, AK7, ALAS2, ALG6, ALPK1, ANGPT1, ANKZF1, ANO6, ARMC4, ASAH1, ATR, BLOC1S3, C11orf70, C15orf41, C17orf62, CARD8, CBL, CCDC103, CCDC114, CCDC151, CCDC39, CCDC40, CCDC65, CCNO, CEP164, CFAP298, CHEK2, COL7A1, CYP27A1, DDX41, DDX58, DGAT1, DGKE, DNAAF1, DNAAF2, DNAAF3, DNAAF4, DNAAF5, DNAH1, DNAH11, DNAH5, DNAH8, DNAH9, DNAI1, DNAI2, DNAJB13, DNAL1, DRC1, DSG1, DTNBP1, DUOX2, EIF2AK3, EPCAM, ERCC2, ERCC3, FOXI3, G6PC, GAS8, GLRX5, GTF2H5, GTF2E2, GUCY2C, HPS1, HPS3, HPS4, HPS5, HPS6, HTRA2, ITGAM, JAK2, KAT6A, KDM1A, KIF23, KIT, KLF1, KLHDC8B, LARS2, LCT, LIPA, LRRC56, LRRC6, LRRC8A, LYN, MBD4, MCIDAS, MLH1, MPLKIP, MSH2, MYO5B, NAF1, NDUFB11, NEUROG3, NF1, NHP2, NME8, NPAT, NOTCH2, OFD1, P2RY12, PIH1D3, PLA2G4A, PLG, PLVAP, PMM2, PNLIP, POMP, POT1, PSMA3, PSMB4, PUS1, RASGRP2, RECQL4, RNF113A, RPGR, RPL11, RPL15, RPL18, RPL19, RPL23, RPL26, RPL27, RPL31, RPL35, RPL35A, RPL5, RPL9, RPS10, RPS15A, RPS19, RPS24, RPS26, RPS27, RPS28, RPS29, RPS7, RSPH1, RSPH3, RSPH4A, RSPH9, SAR1B, SBF2, SCO2, SEC23B, SGPL1, SI, SIAE, SLC10A2, SLC19A2, SLC25A38, SLC26A3, SLC5A1, SLC51B, SLC9A3, SPAG1, SPINT2, STAT4, STX3, TAOK2, TBXA2R, TERF2IP, TIMM50, TMPRSS15, TNFRSF6B, TONSL, TP63, TSR2, UNC45A, VAV1, WNT2B, YARS2, ZCCHC8, and ZMYND10 |
The panel was expanded from 473 to 574 genes during the study, with the added genes denoted by underlining. Twenty-three patients (56%) underwent testing on the 574-gene panel.
Genetic analysis and variant curation were obtained via the commercial analytics pipeline. NGS data were also independently analyzed and interpreted by the research team. Raw NGS data files from each subject were obtained and aligned to the human reference genome (hg19) using the Burrows-Wheeler aligner,20 and single nucleotide and small insertion-deletion (indel) variants were identified with genome analysis toolkit,21 using best practice guidelines and annotated using an in-house pipeline, as previously described.22 Large indels and complex rearrangements were identified with Pindel23 and Delly.24 Copy number variants were called by read depth information using Confier,25 eXome-Hidden Markov Model,26 and in-house scripts developed in the laboratory. Variants with >5 variant reads and variant allele fraction consistent with heterozygosity (≥0.30) were retained. Somatic mutations defined by a variant allele fraction of <0.30 were excluded from the analysis. For known variants, we obtained allele frequencies from the Genome Aggregation Database (gnomAD) with 125 748 individuals and our in-house database of 2000 exomes from gene discovery projects that do not include hematologic disorders. Rare (minor allele frequency <0.001) nonsense, frameshift, splice site–altering variants, and predicted damaging missense variants were included. To assess the potential occurrence of a pathogenic allele in gnomAD, we conducted a comparative analysis of the frequencies of unequivocally pathogenic variants within specific subgroups of gnomAD, namely controls, individuals without cancer, and nonparticipants of the Trans-Omics for Precision Medicine program, which targeted subjects with underlying heart, lung, blood, and sleep disorders.
All in-house and commercially reported variants were independently reviewed by an expert review panel using the American College of Medical Genetics and Genomics (ACMG) standards and guidelines for variant classification.27 Commercially reported pathogenic or likely pathogenic variants were closely reviewed and reclassified as variants of unknown significance (VUSs) if there was insufficient evidence for pathogenicity as defined by ACMG.
To assess the impact of a splice site mutation on transcription, we designed reverse transcription polymerase chain reaction primers to amplify the aberrant transcript. Purified transcription polymerase chain reaction products were compared with transcripts from the unaffected allele and interpreted for predicted functional impact.
Statistical analysis
Differences in continuous and categorical values were assessed using t tests and Fisher exact tests, respectively. All analyses were conducted in R (version 4.1.0).
Results
Among 104 eligible participants, 61 were approached for consent and 44 were enrolled (supplemental Figure 1). The median age was 49 years (range, 20-83), and 56.8% were female (Table 2). Fourteen (31.8%) had secondary ITP, including 8 with Evans syndrome, 1 with common variable immunodeficiency, and 1 with an ill-defined inflammatory disorder, characterized by ≥5 years of persistent night sweats, diffuse fluorodeoxyglucose-avid lymphadenopathy, and repeated biopsies demonstrating only reactive changes (Table 2). An additional 8 subjects with primary ITP had a history of autoimmunity: 3 were diagnosed with a systemic autoimmune disease more than 1 year after the onset of ITP (systemic lupus erythematosus [n = 1], psoriatic arthritis [n = 1], and inflammatory bowel disease [n = 1]), and 5 had preexisting diagnoses of other autoimmune conditions (Raynaud disease [n = 2], Hashimoto thyroiditis [n = 1], neurosarcoidosis [n = 1], multiple sclerosis [n = 1], and autoimmune neutropenia [n = 1]). The median duration of ITP was 8.2 years (range, 0.3-32.9 years), and subjects had received a median of 4 lines of ITP-directed therapy (range, 0-13). At enrollment, 52% were receiving thrombopoietin receptor agonist therapy, and 14 participants (31.8%) had previously undergone splenectomy.
Baseline demographics and clinical characteristics of the study population
| Characteristics . | Subjects (n = 44) . |
|---|---|
| Median age (range), y | 49 (20-83) |
| Sex, n (%) | |
| Female | 25 (56.8) |
| Male | 19 (43.2) |
| Secondary ITP,∗n (%) | 14 (31.8) |
| Evans syndrome | 8 (18.2) |
| Systemic lupus erythematosus | 3 (6.8) |
| Inflammatory bowel disease | 2 (4.5) |
| Rheumatoid arthritis | 1 (2.3) |
| Antiphospholipid syndrome | 1 (2.3) |
| Common variable immunodeficiency | 1 (2.3) |
| Inflammatory disorder NOS | 1 (2.3) |
| Median duration of ITP (range), y | 8.2 (0.3 – 32.9) |
| Median age at diagnosis of ITP (range), y | 40 (2-77) |
| Mean baseline platelet count (range), x 109/L | 161 (16-646) |
| Median no. of previous ITP therapies (range) | 4 (0-13) |
| History of splenectomy, n (%) | 14 (31.8) |
| Prior rituximab, n (%) | 21 (47.7) |
| Most common current therapy, n (%) | |
| Thrombopoietin receptor agonists | 23 (52.3) |
| Not on therapy | 15 (34.1) |
| Corticosteroids | 11 (25.0) |
| Mycophenolate mofetil | 9 (20.5) |
| Lymphadenopathy or splenomegaly, n (%) | 9 (20.5) |
| Personal history of autoimmunity, n (%) | 20 (45.5) |
| Family history of autoimmunity | |
| In a first-degree relative, n (%) | 19 (43.2) |
| In any relative, n (%) | 25 (56.8) |
| Characteristics . | Subjects (n = 44) . |
|---|---|
| Median age (range), y | 49 (20-83) |
| Sex, n (%) | |
| Female | 25 (56.8) |
| Male | 19 (43.2) |
| Secondary ITP,∗n (%) | 14 (31.8) |
| Evans syndrome | 8 (18.2) |
| Systemic lupus erythematosus | 3 (6.8) |
| Inflammatory bowel disease | 2 (4.5) |
| Rheumatoid arthritis | 1 (2.3) |
| Antiphospholipid syndrome | 1 (2.3) |
| Common variable immunodeficiency | 1 (2.3) |
| Inflammatory disorder NOS | 1 (2.3) |
| Median duration of ITP (range), y | 8.2 (0.3 – 32.9) |
| Median age at diagnosis of ITP (range), y | 40 (2-77) |
| Mean baseline platelet count (range), x 109/L | 161 (16-646) |
| Median no. of previous ITP therapies (range) | 4 (0-13) |
| History of splenectomy, n (%) | 14 (31.8) |
| Prior rituximab, n (%) | 21 (47.7) |
| Most common current therapy, n (%) | |
| Thrombopoietin receptor agonists | 23 (52.3) |
| Not on therapy | 15 (34.1) |
| Corticosteroids | 11 (25.0) |
| Mycophenolate mofetil | 9 (20.5) |
| Lymphadenopathy or splenomegaly, n (%) | 9 (20.5) |
| Personal history of autoimmunity, n (%) | 20 (45.5) |
| Family history of autoimmunity | |
| In a first-degree relative, n (%) | 19 (43.2) |
| In any relative, n (%) | 25 (56.8) |
NOS, not otherwise specified.
One patient was concurrently diagnosed with Evans syndrome, systemic lupus erythematosus, and antiphospholipid syndrome; 1 was diagnosed with primary sclerosing cholangitis, ulcerative colitis, and rheumatoid arthritis.
Forty-one subjects completed genetic testing: 23 (56%) on the expanded 574-gene panel. There were no cases of IEI identified. Eight participants (18.2%) were found to be carriers of pathogenic IEI variants, including 1 who harbored 2 IEI-associated variants in different genes (Table 3). There were no significant clinical differences between carriers and noncarriers of a pathogenic IEI variant. An additional 6 subjects were carriers of pathogenic variants in genes associated with non-IEI monogenic disorders ranging from primary ciliary dyskinesia to inborn errors of metabolism (supplemental Table 1). One participant with a family history of thrombocytopenia was found to harbor a pathogenic variant in TUBB1 (p.Phe260Ser), consistent with a diagnosis of congenital thrombocytopenia.28
List of 9 pathogenic variants identified in genes implicated in IEIs with their associated phenotypic classification as outlined in 2022 by the International Union of Immunological Societies,1coding DNA reference sequence, protein-level amino acid sequences (if applicable), gene transcript, and variant-specific references denoted by PubMed Identifier
| IUIS phenotypic classification . | Gene . | cDNA (protein) . | Transcript . | Zygosity . | Disease . | Inheritance . | PMID . |
|---|---|---|---|---|---|---|---|
| Autoinflammatory disorders | MVK | c.1129G>A (p.Val377Ile) | NM_000433.3 | Het | Mevalonate kinase deficiency (hyper-IgD syndrome) | AR | 10369261 |
| Diseases of immune dysregulation | PRF1 | c.853_855del (p.Lys285del) | NM_001083116.1 | Het | Perforin deficiency | AR | |
| Combined immunodeficiencies with associated or syndromic features | PMS2 | c.746_753del (p.Asp249Valfs∗2) | NM_000535.5 | Het | PMS2 deficiency | AR | 20487569 and 27435373 |
| RMRP | n.239C>T | NR_003051.3 | Het | Cartilage hair hypoplasia | AR | 11940090 | |
| WAS | c.1203del (p.Pro402Argfs∗43) | NM000377.2 | Het (female) | Wiskott-Aldrich Syndrome | XL | ||
| Predominantly antibody deficiencies | TNFRSF13B | c.542C>A (p.Ala181Glu) | NM_012452.2 | Het | TACI deficiency | AR | 16007086; 16007087; and 20156508 |
| Congenital defects of phagocyte number or function | CFTR (2) | c.2052del (p.Lys684Asnfs∗38) | NM_000492.3 | Het | Cystic fibrosis | AR | 23974870 |
| c.1521_1523del (p.Phe508del) | Het | 2475911; 15371902; and 23974870 | |||||
| Bone marrow failure | FANCL | c.296_297del (p.Gln99Argfs∗17) | NM_018062.3 | Het | Fanconi anemia type L | AR |
| IUIS phenotypic classification . | Gene . | cDNA (protein) . | Transcript . | Zygosity . | Disease . | Inheritance . | PMID . |
|---|---|---|---|---|---|---|---|
| Autoinflammatory disorders | MVK | c.1129G>A (p.Val377Ile) | NM_000433.3 | Het | Mevalonate kinase deficiency (hyper-IgD syndrome) | AR | 10369261 |
| Diseases of immune dysregulation | PRF1 | c.853_855del (p.Lys285del) | NM_001083116.1 | Het | Perforin deficiency | AR | |
| Combined immunodeficiencies with associated or syndromic features | PMS2 | c.746_753del (p.Asp249Valfs∗2) | NM_000535.5 | Het | PMS2 deficiency | AR | 20487569 and 27435373 |
| RMRP | n.239C>T | NR_003051.3 | Het | Cartilage hair hypoplasia | AR | 11940090 | |
| WAS | c.1203del (p.Pro402Argfs∗43) | NM000377.2 | Het (female) | Wiskott-Aldrich Syndrome | XL | ||
| Predominantly antibody deficiencies | TNFRSF13B | c.542C>A (p.Ala181Glu) | NM_012452.2 | Het | TACI deficiency | AR | 16007086; 16007087; and 20156508 |
| Congenital defects of phagocyte number or function | CFTR (2) | c.2052del (p.Lys684Asnfs∗38) | NM_000492.3 | Het | Cystic fibrosis | AR | 23974870 |
| c.1521_1523del (p.Phe508del) | Het | 2475911; 15371902; and 23974870 | |||||
| Bone marrow failure | FANCL | c.296_297del (p.Gln99Argfs∗17) | NM_018062.3 | Het | Fanconi anemia type L | AR |
All variants in the heterozygous state are not associated with disease. One patient harbored 2 different pathogenic variants in IEI genes (CFTR and FANCL).
AR, autosomal recessive; cDNA, coding DNA reference sequence; Het, heterozygous; IgD, immunoglobulin D; IUIS, International Union of Immunological Societies; PMID, PubMed Identifier; XL, X-linked.
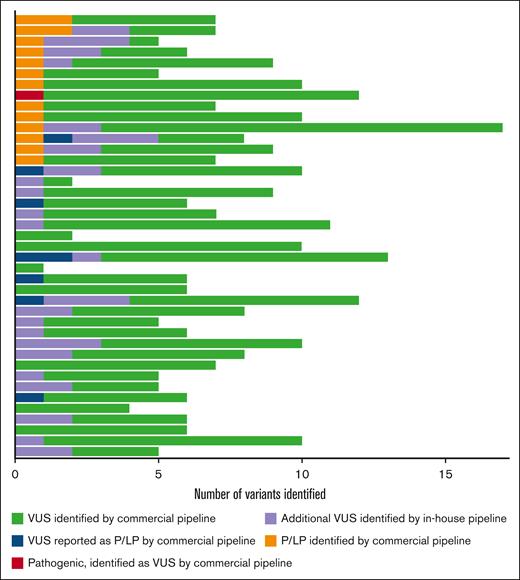
In total, 288 VUSs were identified (238 reported by the commercial analytics pipeline, 8 commercially reported pathogenic variants that were reclassified as VUSs by the in-house analytics pipeline (supplemental Table 2), and 42 VUSs identified by the in-house analytics pipeline), with a mean of 7 VUSs per subject (Figure 1). The TUBB1 p.Phe260Ser mutation was reported as a VUS by the commercial analytics pathway and was reclassified as pathogenic based on existing literature, which reports this variant in an affected family and provides functional data demonstrating the variant’s deleterious effect on platelet microtubule structure and proplatelet formation.28,29 The expert panel judged that 8 of 23 (34.8%) commercially reported pathogenic or likely pathogenic variants were VUSs based on insufficient data supporting pathogenicity according to ACMG guidelines. We performed RNA analysis on a VUS in DIAPH1 (c.3149-1 G>T) predicted to impact splicing and did not observe any effect.
Sequencing results for 41 patients with chronic or persistent ITP. Each row represents an individual subject. In total, there were 16 pathogenic variants (9 in genes implicated in IEIs, 1 variant associated with congenital thrombocytopenia, and 6 associated with miscellaneous disorders, such as primary ciliary dyskinesia and inborn errors of metabolism) and 288 VUSs. The commercial analytics pipeline reported 239 VUSs (green). Of the 23 commercially reported pathogenic or likely pathogenic variants, 8 were reclassified as VUSs by the research team (blue). The remaining 15 were assessed as pathogenic or likely pathogenic in agreement with the commercial interpretation (orange). The research team also identified 1 disease-causing pathogenic mutation associated with congenital thrombocytopenia (TUBB1) that was curated as a VUS by the commercial pipeline (red). An additional 42 VUSs (purple) were identified by the in-house analytics pipeline and not reported by the commercial analytics pipeline. P/LP, pathogenic or likely pathogenic.
Sequencing results for 41 patients with chronic or persistent ITP. Each row represents an individual subject. In total, there were 16 pathogenic variants (9 in genes implicated in IEIs, 1 variant associated with congenital thrombocytopenia, and 6 associated with miscellaneous disorders, such as primary ciliary dyskinesia and inborn errors of metabolism) and 288 VUSs. The commercial analytics pipeline reported 239 VUSs (green). Of the 23 commercially reported pathogenic or likely pathogenic variants, 8 were reclassified as VUSs by the research team (blue). The remaining 15 were assessed as pathogenic or likely pathogenic in agreement with the commercial interpretation (orange). The research team also identified 1 disease-causing pathogenic mutation associated with congenital thrombocytopenia (TUBB1) that was curated as a VUS by the commercial pipeline (red). An additional 42 VUSs (purple) were identified by the in-house analytics pipeline and not reported by the commercial analytics pipeline. P/LP, pathogenic or likely pathogenic.
Discussion
Despite estimates that IEIs commonly underlie pediatric ITP and Evans syndrome, no cases were identified by an NGS panel, encompassing >370 genes implicated in IEI in this real-world, single-center cohort of adult patients with persistent or chronic ITP. Although the prevalence was anticipated to be lower than in a pediatric population,9 the negative finding was surprising as the study cohort was unintentionally enriched for characteristics likely to be associated with an underlying IEI, including a high proportion of patients with heavily pretreated and longstanding ITP on par with contemporary interventional trials,30,31 personal and family history of autoimmunity, and early onset of disease, including 9 (20.5%) who presented in childhood, 2 of whom had pediatric Evans syndrome. Nearly one-third of the study population had ITP secondary to a systemic autoimmune disease, compared with a rate of ∼10% in population-based studies of adult ITP.32,33
Several issues surrounding the germ line genetic testing for IEIs might have affected our results. Among patients with a high pretest clinical suspicion for IEIs, the diagnostic yield of multigene panel testing ranges from 15% to 79%.34 This wide variability is explained partly by differences in methodology (eg, sequencing technique, number of genes evaluated, analytic and variant classification approaches) and study populations (eg, varying rates of carrier frequency and consanguinity).35 Compared with prior studies in pediatric Evans syndrome, this study had no cases of consanguinity8 and used an expert panel, which defined pathogenicity according to ACMG Standards and Guidelines, reclassifying more than one-third of commercially reported pathogenic or likely pathogenic variants as VUSs.8,9 Molecular diagnostic rates also vary among IEI phenotypic classifications. Less than 20% of those categorized as having autoinflammatory disease or predominantly antibody deficiency are molecularly diagnosed.36,37 Diagnostic yield is higher in early-onset highly penetrant disorders, such as severe combined immunodeficiency (100%), inherited bone marrow failure (55%), and syndromic IEIs (53%).36 We note that 6 patients who underwent screening for this study had previously been tested for IEIs, presumably based on a phenotype prompting a high pretest clinical suspicion. Two of the 6 were diagnosed with pathogenic variants in CTLA4 and FOXP3, consistent with a diagnosis of CTLA4 haploinsufficiency and immunodysregulation polyendocrinopathy enteropathy X-linked syndrome, respectively. The study population likely consisted of more subtle presentations. The negative findings in the study highlight the complexity underlying genetic determinants of autoinflammation, which may have greater variability in disease penetrance, expressivity, and polygenicity. Current knowledge of disease-causing variants might also be skewed toward early-onset, highly penetrant disorders and variants associated with subtle phenotypic presentation may be unrecognized at present. Further studies expanding on the clinical and genetic spectra of IEIs are critical. Whole-exome sequencing is underway for 1 subject with a strong family history of pediatric ITP and Evans syndrome (supplemental Figure 2).
Consistent with a prior study, we also observed a high rate of VUSs, with at least 1 VUS identified in each subject and an average of 7 per patient.35 The high rate may be partially attributed to the variant curation practices of commercial laboratories38 and reflect the paucity of data surrounding IEI-associated genes.
One commercially reported VUS (TUBB1 [p.Phe260Ser]) was reclassified as pathogenic. The variant was identified in a 23-year-old woman who carried a diagnosis of ITP after a 9-gene inherited thrombocytopenia panel was nondiagnostic.29 At least 1% to 2% of inherited thrombocytopenia can be misdiagnosed as ITP, although the actual prevalence may be higher.33
In summary, no cases of IEI were identified in this single-center cohort of adults with chronic ITP and Evans syndrome, despite high representation from patients with heavily pretreated, longstanding disease, underlying autoimmunity, and a young age of onset. As the phenotypic and genetic spectra of IEI continue to expand, disease-causing variants associated with adult immune cytopenias may be uncovered. At present, this work does not support implementation of routine testing for IEIs in adult patients with ITP, and sequencing is best reserved for cases where there is a high index of suspicion. Genetic testing for IEIs returns high rates of VUSs, and variant curation is complex, underscoring the importance of expert interpretation and efforts to advance disease-specific standardized variant curation.
Acknowledgments
The authors thank Megan Othus, Fred Hutchinson Cancer Center, for her guidance on biostatistical planning for this project.
This project was funded by the Barbara and Peter T. Pruitt, Jr ITP Research Award from Platelet Disorder Support Association. D.J. received support from an institutional training grant from the National Institutes of Health, National Heart, Lung, and Blood Institute (T32 HL007093).
Authorship
Contribution: D.J. contributed to the study concept and design, participant enrollment, data collection, analysis, and writing of the manuscript; S.B.K. contributed to the study concept and design, expert review of sequencing results, and writing of the manuscript; K.R. contributed to data collection; S.B. contributed to expert review of the sequencing results and performed RNA analysis; T.G. contributed to the study concept and design; S.G. analyzed the raw sequencing data and contributed to expert review of sequencing results; E.J.A. contributed to expert review of sequencing results; and all authors contributed to the critical revisions of the intellectual content and provided final approval of the manuscript.
Conflict-of-interest disclosure: T.G. is a member of the medical advisory board at Platelet Disorder Support Association; consults at Sanofi Corporation, Sobi Corporation, Alpine Immune Science: Bioproducts Laboratory, and Cellphire Corporation; and serves on the data monitoring board at Palisade Bio. The remaining authors declare no competing financial interests.
The current affiliation for S.B. is Sanofi Corporation, Paris, France.
Correspondence: Debbie Jiang, Division of Hematology, Massachusetts General Hospital, 40 Blossom St, Boston, MA, 02114; e-mail: dcjiang@mgh.harvard.edu; and Siobán B. Keel, Division of Hematology, University of Washington, 1705 NE Pacific St, Box 357710, Seattle, WA 98195; e-mail: sioban@uw.edu.
References
Author notes
The sequencing data for subjects who consented to genomic data sharing can be found in the Sequence Read Archive (SRA) database, accession number PRJNA1026521. For other forms of data sharing, please contact the corresponding author, Siobán B. Keel (sioban@uw.edu).
The full-text version of this article contains a data supplement.