TO THE EDITOR:
Translocations involving the mixed lineage leukemia (MLL/KMT2A) gene generally confer poor prognosis in acute myeloid leukemia (AML) and display a large intertumor and intratumor heterogeneity.1 By conducting a single-cell RNA sequencing (scRNA-seq) analysis, the different developmental stages along the hematopoietic stem cell (HSC) to myeloid trajectory can be resolved, which is relevant for self-renewal, interactions of leukemic cells with nonmalignant cells in the microenvironment, and therapy resistance.2-5 However, information on MLL-rearranged (MLL-r) cases of AML is scarce as previous scRNA-seq studies of AML by van Galen et al2 and Shlush et al3 include only 1 patient with MLL-r each. In our previous work, we have described a novel MLL fusion with the enhancer of messenger RNA decapping 4 gene (MLL-EDC4),6 for which recently another case has been reported.7
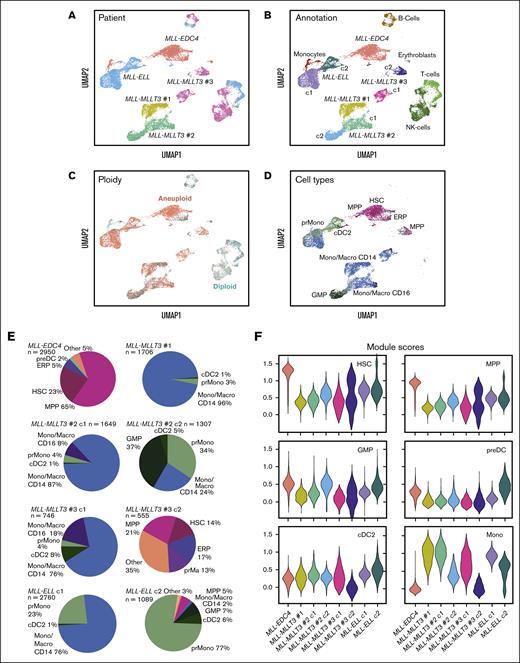
Here, we dissected cell types and developmental stages in 5 patients with AML by scRNA-seq to compare the novel MLL-EDC4 translocation with MLL-MLLT3 and MLL-ELL fusions (supplemental Table 1). Mononuclear cells were collected from peripheral blood or bone marrow and subjected to scRNA-seq to yield 17 600 cells as described in further detail in the supplemental information. Transcriptome features of the merged scRNA-seq data obtained from the 5 patients were visualized by uniform manifold approximation and projection (UMAP) and clustering (Figure 1A-B; supplemental Figure 1A-B). We then annotated leukemic vs nonmalignant cells according to marker gene expression profiles and validated the results with the chromosome ploidy computed from the scRNA-seq data (Figure 1C). The scRNA-seq analysis revealed a significant intratumor heterogeneity of the MLL-MLLT3 #2, MLL-MLLT3 #3, and MLL-ELL patient samples with 2 distinctive clusters (c1 and c2) of leukemic cells. In contrast, the MLL-EDC4 and MLL-MLLT3 #1 samples showed a more homogeneous phenotype. Nonmalignant cells determined by marker gene expression were clustered per cell type across all patients without further batch correction, whereas leukemic cells from each patient sample were clustered individually.
Intratumor heterogeneity and cell type assignment of MLL-r samples. (A) UMAP embedding of all AML samples colored by patient. (B) UMAP embedding colored by cell types determined from marker gene expression. AML cells form separate clusters for each patient whereas nonmalignant cell types from different samples cluster together. (C) UMAP embedding colored by ploidy with AML cells annotated as aneuploid (red) and microenvironment cells as diploid (cyan). (D) UMAP embedding of AML cells colored by cell type prediction with the SingleR annotation software package against the Human Cell Atlas as reference data set. (E) Pie charts of predicted cell type composition for AML cell clusters. (F) Violin plots of myeloid cell signature module scores according to supplemental Table 2 for AML cell clusters. c1, cluster1; and c2, cluster2.
Intratumor heterogeneity and cell type assignment of MLL-r samples. (A) UMAP embedding of all AML samples colored by patient. (B) UMAP embedding colored by cell types determined from marker gene expression. AML cells form separate clusters for each patient whereas nonmalignant cell types from different samples cluster together. (C) UMAP embedding colored by ploidy with AML cells annotated as aneuploid (red) and microenvironment cells as diploid (cyan). (D) UMAP embedding of AML cells colored by cell type prediction with the SingleR annotation software package against the Human Cell Atlas as reference data set. (E) Pie charts of predicted cell type composition for AML cell clusters. (F) Violin plots of myeloid cell signature module scores according to supplemental Table 2 for AML cell clusters. c1, cluster1; and c2, cluster2.
We characterized the differentiation state of leukemic cells with an automated cell type prediction approach using the Human Cell Atlas8 bone marrow data set from 8 healthy donors as training data set. Genes signatures and scores for the different cell types were assigned based on the most expressed cell type markers from the Human Cell Atlas data (Figure 1D-F; supplemental Figure 1C-E, supplemental Table 2). Leukemic cells with MLL-EDC4 translocation represented a distinct leukemic cell cluster and were almost exclusively classified as HSCs, multipotent progenitors (MPPs), or erythroblasts (ERPs), which is in line with their CD34+/CD14– signature from fluorescence-activated cell sorting (FACS) (supplemental Table 3). In contrast, malignant cells from the common MLL fusions presented a more differentiated phenotype that unveiled a trajectory from myeloid progenitors to monocyte-like cells from cluster 2 to 1 for MLL-MLLT3 #2, MLL-MLLT3 #3, and MLL-ELL (Figure 1D; supplemental Figure 1F). Interestingly, a fraction of cells from cluster 2 of MLL-MLLT3 #3 stood out because it displayed signatures of MPP (21%) and HSC (14%) cells (Figure 1E-F). The MLL-EDC4 patient showed elevated module scores for HSC- and MPP-genes, whereas no upregulation in monocytic CD14+related genes was evident (Figure 1E-F). This phenotype was also partly present in cluster 2 of MLL-MLLT3 #3 as apparent from the bimodal distribution of the violin plot and the low monocyte score of the whole cluster and in a minor fraction of cluster 2 from MLL-ELL. In contrast, leukemic cells from patients MLL-MLLT3 #1, MLL-MLLT3 #2 and cluster 1 of MLL-MLLT3 #3 and MLL-ELL showed an almost opposite pattern. The analysis of the microenvironment revealed monocytes with an unusual gene expression signature in MLL-EDC4 that was characterized by expression of CD36, cathepsins, and C-type lectin (CLEC) receptors (supplemental Figure 1G).9
Next, we performed a differential gene expression analysis of gene sets and pathways for the different MLL-r cases. Gene set enrichment analysis showed a downregulation of myeloid leukocyte mediated immunity and activation and a dampened immune response in the MLL-EDC4+ leukemic cells. Pathways associated with MYC targets, interferon alpha response, eukaryotic translation initiation or elongation, and reactive oxygen species were upregulated (Figure 2A). The upregulation of various ribosomal proteins in MLL-EDC4+ AML may be linked to the malignant transformation of cells.10 Furthermore, the upregulation of reactive oxygen species pathways has been shown to interfere with hematopoiesis because of an increase in oxidative stress causing genomic instability.11 Transcriptomes of leukemic cells from the MLL-MLLT3 and MLL-ELL patients displayed an upregulation of classical monocyte markers in contrast to MLL-EDC4 (Figure 2B). Interestingly, the most differentially expressed gene in MLL-EDC4 was lactate dehydrogenase B (LDHB), which mediates the switch on the anaerobic glycolysis and lactate production that could reflect a high proliferation rate of leukemic cells (Warburg effect) and/or adaption to hypoxia.12
Gene expression and transcription factor activity in MLL-EDC4 compared with other MLL-r cases. (A) Enriched gene sets in upregulated and downregulated genes of MLL-EDC4 AML cells compared with all other AML cells visualized as dot plots. Gene sets from the Hallmark, Reactome, and Gene Ontology Biological Processes (GO:BP) databases were used. (B) Clustered single-cell transcriptomic heat map of the most differentially expressed genes between AML cell clusters. (C) Heat map of transcription factor activities for AML cells based on scRNA-seq data. (D) Transcription factor network colored by transcription factor activity.
Gene expression and transcription factor activity in MLL-EDC4 compared with other MLL-r cases. (A) Enriched gene sets in upregulated and downregulated genes of MLL-EDC4 AML cells compared with all other AML cells visualized as dot plots. Gene sets from the Hallmark, Reactome, and Gene Ontology Biological Processes (GO:BP) databases were used. (B) Clustered single-cell transcriptomic heat map of the most differentially expressed genes between AML cell clusters. (C) Heat map of transcription factor activities for AML cells based on scRNA-seq data. (D) Transcription factor network colored by transcription factor activity.
The MLL-EDC4 fusion showed a distinctive upregulation of genes known to have an impact on cell-fate decision and cellular differentiation in hematopoiesis and endothelial-to-hematopoietic transition (NPM1, CDK6, SOX4, GATA2, MYC, and DACH1) or leukemic stem cell activation (FLT3, HOPX, HOXA9, and RUNX1)13-19 (Figure 2B). It is noted that transcription factors (TFs) such as SOX4, GATA2, MYC, and RUNX1 are well established master regulators of stem cell programs. These findings prompted us to systematically evaluate TF expression and activity based on target gene expression. Compared with other fusions (Figure 2C), MLL-EDC4 displayed an increased activity of interferon-related TFs such as STAT2 and IRF9, of oncogenes MYC and MYB as well as other TFs such as E2F4, ETS1, GATA1, NFYA, POU2F1, SPI1, and TAL1 that have been linked to stemness in hematopoietic cells.20-22 Based on these data, a network of interacting TFs was generated (Figure 2D). Unsupervised clustering highlighted MYC as a central node in the network of regulatory factors that is linked to many TFs as first or second edge. MYC is known to play a crucial role in cell growth, proliferation, and tumorigenesis.23 In addition, TF activity showed an upregulation of POU2F1 in MLL-EDC4 leukemic cells, which can function in cell growth control, cellular stress response, stem cell identity, and immune regulation.24 Finally, activity of hematopoietic key regulator RUNX1 was high as inferred from the aberrant expression of its downstream targets UBB, PSNE1, ARID1B, and KIAA0125 involved in differentiation of myeloid cells.25
In summary, our scRNA-seq analysis of MLL fusions in AML revealed variable degrees of intratumor heterogeneity and differentiation stages. The MLL-EDC4 AML case was associated with a more primitive cell differentiation state than MLL-MLLT3 or MLL-ELL. The unique hematological progenitor-like cell type in MLL-EDC4 is evident from an extensive upregulation of a network of TFs that are known to be crucial for differentiation block and leukemic development. Furthermore, a fraction of leukemic cells with an HSC/progenitor-like cell type in 1 cluster of the MLL-MLLT3 #3 sample was detected, which points to a complex interplay of MLL fusion partners and the cell type that develops the AML initiating translocation. It is well established that a more stem cell like phenotype is highly relevant for prognosis and therapy response.2-5 Accordingly, it will be important to extend the approach described here to a larger patient cohort to reveal the relation between the developmental stage along the myeloid trajectory and clinical parameters for different MLL fusions.
Informed consent was obtained from all participants involved in the research reported in the manuscript at the Department of Medicine I of University Freiburg Medical Center.
Acknowledgments: The research was supported by the Deutsche Forschungsgemeinschaft through grants RI 1283/15-2 (K.R.), LU 429/16-2 (M.L.), MA 7792/1-2 (J.-P.M.), and BE 6461/1-2 (H.B.) within Research Group FOR2674 and through subproject Z1 within SFB1074.
Contribution: M.L. and K.R. designed and coordinated the study; H.B., T.M., J.D.-A., and S.O. acquired patient samples; L.C.S. and J.-P.M. acquired the data; L.C.S., A.P.S., S.M.T., S.S., I.S., J.-P.M., and K.R analyzed and curated the data; L.C.S. and K.R. drafted the manuscript; all authors reviewed and edited the manuscript; and K.R. and M.L. provided supervision.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Karsten Rippe, German Cancer Research Center (DKFZ), Division of Chromatin Networks, Im Neuenheimer Feld 280, 69120 Heidelberg, Germany; e-mail: karsten.rippe@dkfz.de; and Michael Lübbert, Department of Medicine I, University Freiburg Medical Center, Hugstetter Straße 55, 79106 Freiburg, Germany; e-mail: michael.luebbert@uniklinik-freiburg.de.
References
Author notes
The single-cell RNA sequencing data are available via the Zenodo open repository at https://doi.org/10.5281/zenodo.7832875. Analysis scripts are provided at Github from the link https://github.com/RippeLab/MLL-EDC4. Other data are available on request from the corresponding authors, Karsten Rippe (karsten.rippe@dkfz.de) and Michael Lübbert (michael.luebbert@uniklinik-freiburg.de).
The full-text version of this article contains a data supplement.