Key Points
T-ALL cells are exquisitely sensitive to the inhibition of DHODH.
DHODH represents a promising metabolic target for the treatment of patients with T-ALL.
Abstract
Patients with relapsed or refractory T-cell acute lymphoblastic leukemia (T-ALL) have a poor prognosis with few therapeutic options. With the goal of identifying novel therapeutic targets, we used data from the Dependency Map project to identify dihydroorotate dehydrogenase (DHODH) as one of the top metabolic dependencies in T-ALL. DHODH catalyzes the fourth step of de novo pyrimidine nucleotide synthesis. Small molecule inhibition of DHODH rapidly leads to the depletion of intracellular pyrimidine pools and forces cells to rely on extracellular salvage. In the absence of sufficient salvage, this intracellular nucleotide starvation results in the inhibition of DNA and RNA synthesis, cell cycle arrest, and, ultimately, death. T lymphoblasts appear to be specifically and exquisitely sensitive to nucleotide starvation after DHODH inhibition. We have confirmed this sensitivity in vitro and in vivo in 3 murine models of T-ALL. We identified that certain subsets of T-ALL seem to have an increased reliance on oxidative phosphorylation when treated with DHODH inhibitors. Through a series of metabolic assays, we show that leukemia cells, in the setting of nucleotide starvation, undergo changes in their mitochondrial membrane potential and may be more highly dependent on alternative fuel sources. The effect on normal T-cell development in young mice was also examined to show that DHODH inhibition does not permanently damage the developing thymus. These changes suggest a new metabolic vulnerability that may distinguish these cells from normal T cells and other normal hematopoietic cells and offer an exploitable therapeutic opportunity. The availability of clinical-grade DHODH inhibitors currently in human clinical trials suggests a potential for rapidly advancing this work into the clinic.
Introduction
Leukemia remains the most common childhood cancer and the second leading cause of cancer-related deaths in children.1 The subset of T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive disease affecting both children and young adults. This leukemia has high rates of relapse as well as primary refractory disease, with survival rates of <15% in both instances.2-5 Adult patients with T-ALL have similarly poor outcomes and survival rates.6-8 Although patients with B-cell ALL (B-ALL) have benefited greatly from the addition of tyrosine-kinase inhibitors as well as chimeric antigen receptor T-cell therapy and other immune-based therapies to treatment regimens, these therapies are not available for patients with T-ALL, making new and more effective treatments urgently needed.9,10
T-ALL is known to be sensitive to inhibitors of nucleotide synthesis. Indeed, some of the mainstays of current therapy, methotrexate and 6-mercaptopurine, take advantage of this vulnerability. Inhibition of the 1-carbon folate pathway via serine hydroxymethyltransferase (SHMT) 1 and SHMT2 is also a known metabolic dependency in T-ALL.11 In addition, activated lymphocytes have been shown to have higher resting pools of intracellular pyrimidines at baseline than other white blood cells.12 Activated T cells have dramatic upregulation of the enzymes involved in the de novo pyrimidine synthesis pathway.13
Dihydroorotate dehydrogenase (DHODH) catalyzes the conversion of dihydroorotate to orotate during the de novo synthesis of pyrimidines.14 In eukaryotic cells, it is located in the inner mitochondrial membrane between complex I and complex II and is the only step in the pyrimidine synthesis pathway that occurs inside the mitochondrion.15 DHODH is a ubiquitously expressed and essential enzyme; without it, a cell cannot make uridine, cytidine, or thymidine ribonucleotides and deoxyribonucleotides. Inhibiting DHODH rapidly leads to a state of nucleotide starvation and forces cells to rely on extracellular nucleotide salvage via nucleoside transporters in the cell membrane. Some malignant cells, including T lymphoblasts, are relatively deficient in their ability to carry out extracellular salvage of nucleosides from their environment.16,17
The sensitivity of T cells, particularly malignant T cells, to DHODH inhibition, is likely due to fundamental differences in their nucleotide metabolism. In fact, this likely explains the clinical activity of the Food and Drug Administration–approved, low potency DHODH inhibitors leflunomide and teriflunomide in autoimmune disease. In this context, DHODH inhibition is clinically beneficial, suppressing the abnormal T-cell activation in the treatment of rheumatoid arthritis and multiple sclerosis.18-20
Our previous work demonstrated that inhibiting DHODH using the highly potent and bioavailable small molecule brequinar (BRQ) leads to the differentiation of myeloid cells and produces a marked antileukemic effect in preclinical models of acute myeloid leukemia (AML).21-23 Subsequently, multiple small molecule DHODH inhibitors have been used in clinical trials of patients with myeloid malignancies.24,25 These high potency inhibitors have been safe when used in humans, though their efficacy as single agents in myeloid malignancies remains unproven. We expanded our investigations to lymphoid disease, given the need for improved therapeutic options as well as the unique aspects of lymphoid biology.
In this study, we demonstrate that lymphoid cells, specifically malignant T lymphoblasts, are extremely sensitive to DHODH inhibition using both small molecule inhibitors as well as genetic approaches. We demonstrate that DHODH inhibition is a potent, effective therapy in multiple preclinical models of T-ALL.
Methods
Cell culture and cell viability assays
The human T-ALL cell lines used in this study (Jurkat, PF-382, CUTLL-1, RPMI-8402, DND-41, KOPTK-1, CCRF-CEM, and MOLT-16) were purchased from the American Type Culture Collection or the Leibniz-Institute Deutsche Sammlung von Mikroorganismen und Zellkulturen. All lines were tested and confirmed to be negative for mycoplasma.
Cell lines were maintained in RPMI 1640 (Corning) supplemented with 1% penicillin/streptomycin (Gibco) and 10% fetal bovine serum (Gibco) in a 37°C incubator with 5% CO2. Cells were counted using acridine orange. Cell viability was measured using the CellTiter-Glo Luminescent Cell Viability Assay (Promega) following the recommended incubation/concentration. Luminescence was measured using a SynergyHTX plate reader (BioTEK). The values of 50% inhibitory concentration were determined using Prism GraphPad version 9 software.
Compounds
BRQ sodium was synthesized by the Broad Institute.
Plasmids and CRISPR constructs
The intracellular domain of Notch1 was cloned into the MigR1 as previously described.26 This construct was packaged into ecotropic retrovirus and transduced into adult murine bone marrow progenitors. Animals that underwent transplantation were subjected to bleeding 4 weeks after transplant to assess the presence of CD4+CD8+ double-positive T cells in the peripheral blood, an aberrant cell population created by Notch1-induced, ectopic T-cell development in the bone marrow.27 Leukemic cells from the bone marrow and spleen were harvested, purified by Ficoll gradient, and frozen down for subsequent transplantation.
Single guide RNAs targeting the human DHODH sequence as well as control nontargeting guides were constructed based on previously published design rules.28 These guides were cloned into the LentiCRISPRv2 green fluorescent protein (GFP) backbone (plasmid no. 82416, Addgene). For virus production, 5μg of the backbone plasmid was used, together with 4μg psPAX2 and 1μg VSVG. These were transfected into 293 T cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocols. Lentiviral supernatant was harvested, and viral titers determined as described.29
Flow cytometry
Antibodies (human: CD45, CD7, TdT, CD34, CD1a, CD99, CD10, CD3, CD4, and CD8; mouse: CD45, CD8, CD4, T-cell receptor ß, CD3, CD5, CD25, CD44, CD62L, CD117, CD24, NK1.1, Ter-119, T-cell receptor gamma/delta, B220, and CD11b) were purchased from BioLegend. Cells were suspended in fluorescence-activated cell sorting buffer (phosphate-buffered saline + 2% fetal bovine serum + 1 mM EDTA) and stained for 30 minutess at 4°C in the dark. 7-Aminoactinomycin D, 4′,6-diamidino-2-phenylindole, or Live/Dead Fixable Near-IR Dead Cell Stain (Invitrogen) was included as a viability dye to help identify dead cells. Flow cytometry data were collected on an LSR2 flow cytometer (BD Biosciences) and analyzed using FlowJo software. Fluorescence-activated cell sorting was performed on a BD Aria 2, with the support of the Harvard Stem Cell Institute–Center for Regenerative Medicine Flow Cytometry Facility. Cell cycle staining and analysis were performed as previously described.22,30-33
Measuring intracellular purines and pyrimidines
Incubation of cells with drug
Cells were incubated with the indicated concentration of drug for different time periods. Cultures were maintained, and aliquots (1 × 107 cells) were extracted at the indicated times. After being washed with phosphate-buffered saline, cells were processed for nucleotide extraction. Nucleotides were extracted using perchloric acid, and the extracts were neutralized with potassium hydroxide and stored at −20°C until analyzed.34
HPLC quantification of nucleoside 5′-triphosphate levels
Perchloric acid extracts were analyzed using either a Waters 2695e high-performance liquid chromatography (HPLC) with a Waters 2489 UV/visible detector or a Waters 2695 HPLC with a Waters 2487 Dual λ Absorbance detector. A Partisil 10 SAX column was used to separate nucleoside triphosphates using a 50-minute concave gradient curve (curve 8) at a flow rate of 1.5 mL/min starting from 60% 0.005 M ammonium dihydrogen phosphate (NH4H2PO4; pH 2.8) and 40% 0.75 M NH4H2PO4 (pH 3.8) to 100% 0.75 M NH4H2PO4 (pH 3.8). The column eluate was monitored at 262 nm. Standard ribonucleoside triphosphates were used to create a standard curve, which was used to quantitate nucleotide pools.22
Measuring oxidative phosphorylation via Seahorse assay
The Agilent Seahorse XF Cell Mito Stress Test was performed according to the manufacturer’s instructions. Briefly, human T-ALL cell lines were grown in suspension culture, treated or untreated with BRQ, and then transferred into the 96-well poly-D-lysine-coated microplate at a density of 1 × 105 cells per well. The plates were centrifuged at 200 g for 1 minute without a brake to adhere the cells to the bottom of the well in an even distribution. Then the remainder of the Cell Mito Stress Test protocol was followed according to the instructions. All assays were performed on a Seahorse XFe96 Analyzer. The following drug concentrations were used: oligomycin 1.5 μM, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone 1.0 μM, and rotenone/antimycin A 0.5 μM. Analysis was performed using Agilent Analytics software as well as GraphPad Prism version 9 software.
Gene expression analysis by RNA sequencing
Total RNA was harvested from 5 × 106 Jurkat cells either treated or untreated with 1 μM BRQ over differing time periods using an RNeasy-Plus Mini Kit (Qiagen). After RNA isolation, samples were sent to Novogene for library preparation and sequencing.
RNA sequencing analysis
RNA sequencing data analysis was performed in alignment with the Encyclopedia of DNA Elements Consortium standards (https://www.encodeproject.org/chip-seq/). Quality control tests for unmapped reads were performed based on the FastQC version 0.11.9 software (Babraham Bioinformatics; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and summarized with multiQC version 1.9.35 The human reads were mapped to hg19/gencode v19 genome using STAR version 2.7.2b36 with standard parameters: --outSAMtype BAM SortedByCoordinate--outSAMunmapped None--outSAMattributes NH HI NM MD AS XS--outReadsUnmapped Fastx--outSAMstrandField intronMotif --quantMode TranscriptomeSAM GeneCounts--quantTranscriptomeBan IndelSoftclipSingleend. Quality control for the mapped reads and for replicate reproducibility were performed using SARTools version 1.7.3.37 Gene level reads were summarized by counting the reads that overlapped the hg19/gencode v19 gene exons by using the featureCounts version 1.6.3 method implemented in the Subread version 2.0.0 package (http://subread.sourceforge.net).38 Gene counts were normalized and used to quantify the differential genes between the experimental and control conditions using DESeq2 version 1.32.0.39 Gene expression was estimated based on the log2(transcripts per million [TPM] + 1) scores for normalized reads.40 The expressed genes were identified as the genes with maximum log2(TPM + 1) expression ≥ 1 across all conditions. The gene differentiability was assessed with DESeq2 based on the robust shrunken log2 fold change scores and the approximate posterior estimation for generalized linear model coefficients (apeglm version 1.6) method for effect size.39 The cutoffs for gene differentiability were |fold change expression| ≥ 1.5 and adjusted P value ≤ .10. Hea tmaps for the visualization of transcriptional changes induced by treatment were created by using the Morpheus software platform (https://software.broadinstitute.org/morpheus/).
Dependency data analysis
The CRISPR 22Q2 public data from the screens published by Broad Achilles and Sanger Score projects41 were downloaded from the DepMap portal https://depmap.org/portal/download/. The CRISPR 22Q2 screening was performed for 31 tumor lineages on 1086 cell lines, of which 5 were annotated as T-ALL and 12 as B-ALL. The gene effect scores summarizing the guide depletion were determined based on the Chronos algorithm.42 Negative Chronos dependency scores below −0.3 estimate cell growth inhibition and/or death after gene knockout (KO). Common essential genes have a median Chronos score of −1.
The genetic differential dependencies enriched in the T-ALL cell lines vs all other non–T-ALL cell lines were identified based on the Two-class Comparison method available from the DepMap Data Explorer interactive platform www.depmap.org, with a cutoff ≤ 0.10 for the P value corrected for multiple hypothesis testing using the false discovery rate.43
Gene set enrichment analyses
Gene set enrichment analysis (GSEA) version 4.2.0 software44,45 was used to identify functional associations of the molecular phenotypes induced by treatment vs control with the MSigDB version 7.4 collections of gene sets,46 including the hallmark gene sets (h) and the Kyoto Encyclopedia of Genes and Genomes and Reactome curated pathways. For the experimental comparison, the hg19/gencode v19 expressed genes were ranked based on the expression fold change in treated vs control phenotypes. The goal of GSEA was to identify the gene sets that are distributed at the top or at the bottom of the ranked list of genes based on the Kolmogorov-Smirnov enrichment test. Gene sets with absolute normalized enrichment score (NES) ≥ 1.3, a nominal P value ≤ 0.05 and an false discovery rate ≤ 0.25 for the Kolmogorov-Smirnov test were considered significant hits. The results were visualized on volcano plots for the NES vs –log10(P) and on GSEA plots.
Mice and animal housing
Mice were maintained under pathogen-free conditions, and experiments were approved by the Massachusetts General Hospital Institutional Animal Care and Use Committee. C57BL/6J and NOD scid gamma (NSG) mice were purchased from The Jackson Laboratories.
In vivo studies
For the studies with the genetically engineered mouse models, adult bone marrow progenitor cells from wild-type CD45.1STEM mice described previously47 were infected with retrovirus containing the intracellular domain of Notch1 construct as detailed earlier, and cells were selected. After this, 5 × 106 cells were injected retro-orbitally into each 8 to 12-week-old female C57BL/6J recipient mouse. Disease burden was tracked using peripheral white blood cell (WBC) counts and bone marrow GFP measurements. Upon detection of at least 10% leukemia in the bone marrow, treatment with BRQ was initiated, and serial assessments were conducted.
For the studies with the patient-derived xenograft (PDX) models, vials of P1 passage cells from 2 different established T-ALL PDX models were purchased from Proxe,48 and currently available from Dana-Farber Cancer Institute Center for Patient Derived Models, https://www.dana-farber.org/research/departments-centers-and-labs/integrative-research-centers/center-for-patient-derived-models/hematologic-pdx-models/.
After thawing, 1×106 cells were injected retro-orbitally into 8-week-old female recipient NSG mice that were sublethally irradiated with 200 cGy. Approximately 6 weeks after injection, the mice were moribund and euthanized. P2 leukemic cells were harvested from the bone marrow and spleen, purified by Ficoll gradient, and frozen. For subsequent larger-scale experiments, 1 × 106 P2 cells were injected retro-orbitally into 8-week-old female recipient NSG mice which were sublethally irradiated with 200 cGy. Disease burden was tracked using human CD45 antibody staining of peripheral blood and bone marrow. When there was at least 1% leukemia present in the bone marrow, treatment with BRQ was initiated, and serial assessments were conducted.
Statistical analyses
GraphPad Prism version 9 software was used to perform statistical analyses. The Wilcoxon-Mann-Whitney test was used for pairwise comparisons of significance. The log-rank (Mantel-Cox) test was used for the survival curves analyses. The analyses of gene expression datasets have been described earlier. Statistical methods as well as the number of animals or independent experiments are listed in the figures or figure legends.
Results
T-ALL cell lines are highly dependent on de novo pyrimidine synthesis
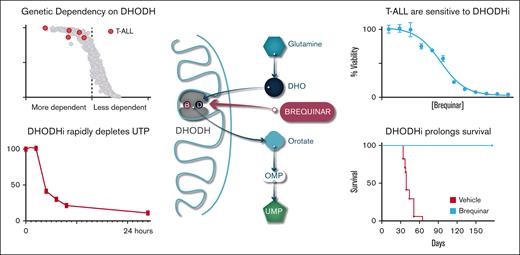
Our goal was to identify new metabolic dependencies in T-ALL that could be targeted. We used the Cancer Dependency Map project data and specifically interrogated the T-ALL cell lines that had undergone whole-genome CRISPR screening to identify dependencies.49 We noted that DHODH was one of their most significant dependencies (Figure 1A; The Cancer Dependency Map portal [RRID: SCR_017655]). We then chose to look at DHODH across all cell lines. It is well established that AML blasts are dependent on the machinery of pyrimidine synthesis and particularly dependent on the activity of DHODH.22,45,50,51 However, T-ALL and B-ALL cell lines ranked among the most highly dependent on DHODH, even more so than most of the AML cell lines included in the database (Figure 1B).
T-ALL cell lines are highly dependent on de novo pyrimidine synthesis. (A) Volcano plot depicting the 22Q2 CRISPR (DepMap Public+Score, Chronos) genome-wide differential dependency (effect size) vs -log10(false discovery rate [FDR] q value) for T-ALL vs non–T-ALL cell lines. Dots correspond to genes. The DHODH gene is highlighted red. Differential dependency is estimated based on the 2-class comparison method implemented in DepMap (limma empirical Bayes; significance cutoff FDR q ≤ 0.10). (B) Scatter dot plot demonstrating the dependency on DHODH of the T-ALL, B-ALL, and AML cell lines in the CRISPR (DepMap 22Q2 Public+Score, Chronos) screen for 1086 cell lines. Dots represent cell lines. The 5 T-ALL cell lines in the screen are highlighted red, the 12 B-ALL cell lines are highlighted black, and the 26 AML cell lines are highlighted blue. The x-axis shows the DHODH Chronos dependency score (gene effect) per cell line. The y-axis shows the probability of dependency on DHODH per cell line. Dependency on DHODH is estimated by low negative (≤−0.5) Chronos gene effect scores. (C) Schematic outline of the de novo pyrimidine synthesis pathway and the role of DHODH, which is located in the inner mitochondrial membrane. Also shown is the location where uridine feeds into the pathway. (D) CRISPR KO of DHODH was performed in the Jurkat cell line. Parental and KO cells were seeded at equivalent densities, and supplemental uridine was withdrawn, at the time when DHODH-KO cells rapidly died while the parental cells continued to grow rapidly. dCTP, deoxycytosine triphosphate; dTMP, deoxythymidine 5′-monophosphate; dTTP, deoxythymidine 5′-triphosphate; dUDP, deoxyuridine diphosphate; dUMP, deoxyuridine monophosphate; dUTP, deoxyuridine triphosphate; GlcNAc, N-acetylglucosamine; OMP, orotidylate monophosphate.
T-ALL cell lines are highly dependent on de novo pyrimidine synthesis. (A) Volcano plot depicting the 22Q2 CRISPR (DepMap Public+Score, Chronos) genome-wide differential dependency (effect size) vs -log10(false discovery rate [FDR] q value) for T-ALL vs non–T-ALL cell lines. Dots correspond to genes. The DHODH gene is highlighted red. Differential dependency is estimated based on the 2-class comparison method implemented in DepMap (limma empirical Bayes; significance cutoff FDR q ≤ 0.10). (B) Scatter dot plot demonstrating the dependency on DHODH of the T-ALL, B-ALL, and AML cell lines in the CRISPR (DepMap 22Q2 Public+Score, Chronos) screen for 1086 cell lines. Dots represent cell lines. The 5 T-ALL cell lines in the screen are highlighted red, the 12 B-ALL cell lines are highlighted black, and the 26 AML cell lines are highlighted blue. The x-axis shows the DHODH Chronos dependency score (gene effect) per cell line. The y-axis shows the probability of dependency on DHODH per cell line. Dependency on DHODH is estimated by low negative (≤−0.5) Chronos gene effect scores. (C) Schematic outline of the de novo pyrimidine synthesis pathway and the role of DHODH, which is located in the inner mitochondrial membrane. Also shown is the location where uridine feeds into the pathway. (D) CRISPR KO of DHODH was performed in the Jurkat cell line. Parental and KO cells were seeded at equivalent densities, and supplemental uridine was withdrawn, at the time when DHODH-KO cells rapidly died while the parental cells continued to grow rapidly. dCTP, deoxycytosine triphosphate; dTMP, deoxythymidine 5′-monophosphate; dTTP, deoxythymidine 5′-triphosphate; dUDP, deoxyuridine diphosphate; dUMP, deoxyuridine monophosphate; dUTP, deoxyuridine triphosphate; GlcNAc, N-acetylglucosamine; OMP, orotidylate monophosphate.
In eukaryotes, the reduction of dihydroorotate to orotate by DHODH occurs in the inner mitochondrial membrane, the only step in de novo pyrimidine biosynthesis that does not occur in the cytoplasm. Glutamine is the key metabolite that feeds the pathway, ultimately resulting in the production of uridine monophosphate. Supplementing cells with uridine bypass a cell’s requirement of de novo pyrimidine synthesis and can rescue cells from the effects of DHODH inhibition or the genetic loss of DHODH enzyme activity (Figure 1C).
To validate that T-ALL is indeed dependent on the activity of DHODH, we performed CRISPR-Cas9 KO of DHODH in vitro in the Jurkat cell line, a T-lymphoblast cell line established from a 14-year-old male patient. Cells that have lost DHODH enzyme activity, either genetically or chemically, can survive in the presence of supplemental uridine. Removal of exogenous uridine will then be used to assess a cell’s dependency on DHODH activity. In a competition experiment in which wild-type Jurkat cells were mixed in equal ratio with Jurkat cells that had undergone CRISPR KO, there was a rapid selection against DHODH-KO cells, demonstrating the dependency on DHODH (Figure 1D). Additionally shown is the western blot demonstrating the knockdown of DHODH in the Jurkat cells (supplemental Figure 1A). When quantified, normalization to the actin loading controls demonstrated a ∼50% reduction in DHODH protein expression compared with that of the parental Jurkat cell line. This is consistent with a heterozygous KO phenotype (supplemental Figure 1B). Interestingly, none of the clones tested demonstrated complete KO of DHODH, even in the presence of supplemental uridine. This suggested an absolute dependence, to some degree, on de novo pyrimidine synthesis in this cell line. In the Jurkat DHODH-HET cell line, even the loss of a single copy of DHODH performed like a complete KO in our functional growth assay (Figure 1). Without uridine, the DHODH-HET Jurkat line demonstrated a complete loss of viability. This haploinsufficiency phenotype that we observed in the Jurkat cells highlights an absolute dependence on DHODH for survival.
Inhibition of DHODH leads to cell cycle arrest and apoptosis in T lymphoblasts
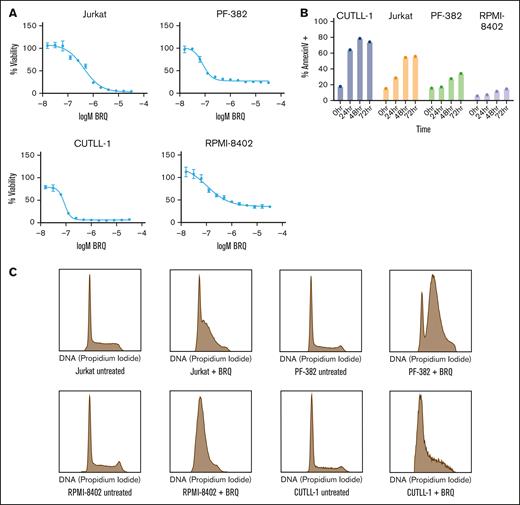
BRQ is a small molecular inhibitor of DHODH originally developed by the Dupont company in the 1980s, which has been studied in multiple clinical trials.14,22,52-54 We screened a panel of T-ALL cell lines for their sensitivity to BRQ. All cell lines demonstrated sensitivity to DHODH inhibition, with 50% inhibitory concentration values in the nanomolar range over a treatment range from 72 to 120 hours (Figure 2A; supplemental Figure 2A).
Inhibition of DHODH leads to cell cycle arrest and apoptosis in T lymphoblasts. (A) Treatment of T-ALL cell lines with BRQ, showing dose-dependent decreases in cell viability, as measured by CellTiter-Glo. Shown are the mean ± standard deviation of 3 replicates. (B) T-ALL cell lines were treated with 1 μM BRQ. Graph showing a time-dependent increase in annexin V staining measured by flow cytometry. Shown are data from a representative experiment, which was repeated 3 times. (C) Treatment of T-ALL cell lines in vitro with 1 μM BRQ for 72 hours resulted in an S-phase arrest. Shown are data from a representative experiment, which was repeated 3 times.
Inhibition of DHODH leads to cell cycle arrest and apoptosis in T lymphoblasts. (A) Treatment of T-ALL cell lines with BRQ, showing dose-dependent decreases in cell viability, as measured by CellTiter-Glo. Shown are the mean ± standard deviation of 3 replicates. (B) T-ALL cell lines were treated with 1 μM BRQ. Graph showing a time-dependent increase in annexin V staining measured by flow cytometry. Shown are data from a representative experiment, which was repeated 3 times. (C) Treatment of T-ALL cell lines in vitro with 1 μM BRQ for 72 hours resulted in an S-phase arrest. Shown are data from a representative experiment, which was repeated 3 times.
Having observed sensitivity across multiple models of T-ALL, we determined the impact of BRQ treatment on alteration of cell cycle and induction of cell death. Cells treated with BRQ showed increased apoptosis over 24 to 72 hours (Figure 2B). Although all cell lines demonstrated a time-dependent increase in annexin V–positive staining, there were clear differences in the apoptotic sensitivity to BRQ treatment. We hypothesized that cells that displayed a lower degree of apoptosis might instead be undergoing cell cycle arrest. Indeed, we observed an S-phase arrest in treated cell lines (Figure 2C).
DHODH inhibition is specific to the pyrimidine synthesis pathway and is rescued by supplemental uridine
Although DHODH inhibitors have been well studied in the setting of myeloid malignancies,22,23,25,55 little is known about their effect in lymphoid malignancies. We wanted to study the metabolic effects of BRQ treatment in T-ALL. T-ALL cell lines were treated with BRQ for 24 hours, and metabolites were extracted at multiple time points using perchloric acid for the quantification of intracellular adenosine triphosphate (ATP), guanosine triphosphate, uridine triphosphate (UTP), and cytosine triphosphate. The intracellular pyrimidines (UTP and cytosine triphosphate) were rapidly depleted within the first 8 hours, whereas the purines (ATP and guanosine triphosphate) remained relatively unchanged (Figure 3A; supplemental Figure 3A).
DHODH inhibition is specific to the pyrimidine synthesis pathway and is rescued by supplemental uridine. (A) Graphs showing intracellular nucleotide concentrations in cells treated with 1 μM BRQ over a time course. Concentrations were normalized to those of untreated controls. Shown are the mean ± standard deviation of 3 replicates. (B) Treatment of Jurkat cells with BRQ showed dose-dependent decreases in cell viability, which is rescued by the addition of supplemental uridine, as measured by CellTiter-Glo. Shown are the mean ± standard deviation of 3 replicates. CTP, cytosine triphosphate; GTP, guanosine triphosphate.
DHODH inhibition is specific to the pyrimidine synthesis pathway and is rescued by supplemental uridine. (A) Graphs showing intracellular nucleotide concentrations in cells treated with 1 μM BRQ over a time course. Concentrations were normalized to those of untreated controls. Shown are the mean ± standard deviation of 3 replicates. (B) Treatment of Jurkat cells with BRQ showed dose-dependent decreases in cell viability, which is rescued by the addition of supplemental uridine, as measured by CellTiter-Glo. Shown are the mean ± standard deviation of 3 replicates. CTP, cytosine triphosphate; GTP, guanosine triphosphate.
Because DHODH and the de novo pyrimidine synthesis pathway are ultimately responsible for the generation of UTP (Figure 1C), treating T-ALL cells with supplemental uridine should overcome the effects of BRQ and nucleotide starvation. Indeed, the inclusion of exogenous uridine (100 μM) did completely rescue the growth inhibitory effects of BRQ (Figure 3B; supplemental Figure 3B), confirming that the effects of BRQ and DHODH inhibition are specific to pyrimidine biosynthesis in T lymphoblasts.
DHODH inhibition upregulates a transcriptional signature of oxidative phosphorylation
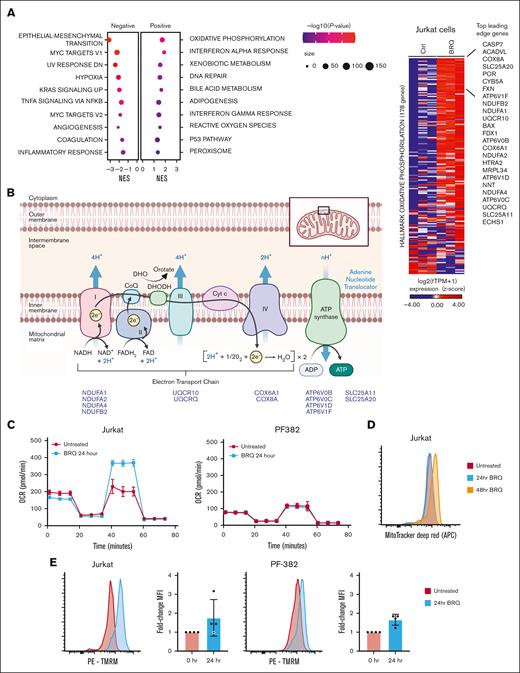
To understand the potential transcriptional effects of DHODH inhibition in T-ALL, triplicate samples of Jurkat cells were treated with 1 μM BRQ for 24 hours. Gene set enrichment analysis demonstrated the top hallmark gene sets up- or down-regulated after BRQ treatment in Jurkat cells (Figure 4A; supplemental Figure 4A). Although some of the altered pathways were similar to those observed in other malignant cell lines, such as interferon gamma signaling and translation regulation,21,22 we were particularly intrigued by the enrichment of gene sets associated with oxidative phosphorylation.
DHODH inhibition upregulates a transcriptional signature of oxidative phosphorylation. (A) Bubble dot plot depicting the GSEA NES for top hallmark gene sets enriched in the genome-wide transcriptional changes induced by BRQ treatment over 24 hours in Jurkat cells. Hallmark gene sets are presented as bubble dots. The dot size depicts the number of differentially expressed genes that are within the specified hallmark pathway. The dot color depicts the significance range of the P value. Significance cutoffs for GSEA: |NES| > 1.3, -log10(P value) > 1, and FDR < 0.25. (B) Schematic representation of the electron transport chain in the inner mitochondrial membrane and the location of DHODH. Highlighted are 14 genes and their relative positions within the electron transport chain, which were among the leading-edge genes driving the oxidative phosphorylation signature in Jurkat cells treated with BRQ. Additionally shown is a heat map depicting the log2(TPM + 1) expression for the replicate samples treated with BRQ vs dimethyl sulfoxide on the hallmark oxidative phosphorylation genes. Genes are ranked based on the log2 fold-change in expression induced by BRQ. Top 25 up-regulated leading-edge genes are annotated. (C) The oxygen consumption rate (OCR) was measured using the Seahorse XF Cell Mito Stress Test assay. Cells were treated with BRQ for 24 hours preceding the assay. OCR was measured over 74 minutes. This experiment was performed twice. (D) Jurkat cells were treated with 1 μM BRQ over a time course. Cells were then stained with MitoTracker Deep Red and fluorescence was measured using flow cytometry. This experiment was performed 3 times. (E) Jurkat and PF-382 cells were treated with 1 μM BRQ for 24 hours. Cells were then stained with tetramethyl rhodamine methyl ester (TMRM) and fluorescence was measured using flow cytometry. This experiment was performed 4 times. APC, allophycocyanin; MFI, mean fluorescence intensity; PE, phycoerythrin. Panel B was adapted from “Electron Transport Chain,” by BioRender.com (2020).
DHODH inhibition upregulates a transcriptional signature of oxidative phosphorylation. (A) Bubble dot plot depicting the GSEA NES for top hallmark gene sets enriched in the genome-wide transcriptional changes induced by BRQ treatment over 24 hours in Jurkat cells. Hallmark gene sets are presented as bubble dots. The dot size depicts the number of differentially expressed genes that are within the specified hallmark pathway. The dot color depicts the significance range of the P value. Significance cutoffs for GSEA: |NES| > 1.3, -log10(P value) > 1, and FDR < 0.25. (B) Schematic representation of the electron transport chain in the inner mitochondrial membrane and the location of DHODH. Highlighted are 14 genes and their relative positions within the electron transport chain, which were among the leading-edge genes driving the oxidative phosphorylation signature in Jurkat cells treated with BRQ. Additionally shown is a heat map depicting the log2(TPM + 1) expression for the replicate samples treated with BRQ vs dimethyl sulfoxide on the hallmark oxidative phosphorylation genes. Genes are ranked based on the log2 fold-change in expression induced by BRQ. Top 25 up-regulated leading-edge genes are annotated. (C) The oxygen consumption rate (OCR) was measured using the Seahorse XF Cell Mito Stress Test assay. Cells were treated with BRQ for 24 hours preceding the assay. OCR was measured over 74 minutes. This experiment was performed twice. (D) Jurkat cells were treated with 1 μM BRQ over a time course. Cells were then stained with MitoTracker Deep Red and fluorescence was measured using flow cytometry. This experiment was performed 3 times. (E) Jurkat and PF-382 cells were treated with 1 μM BRQ for 24 hours. Cells were then stained with tetramethyl rhodamine methyl ester (TMRM) and fluorescence was measured using flow cytometry. This experiment was performed 4 times. APC, allophycocyanin; MFI, mean fluorescence intensity; PE, phycoerythrin. Panel B was adapted from “Electron Transport Chain,” by BioRender.com (2020).
Oxidative phosphorylation is the process of redox reactions involving the flow of electrons along complexes of inner mitochondrial membrane–bound proteins, resulting in the generation of ATP. We analyzed the leading-edge genes to determine what was driving this oxidative phosphorylation signature and noticed that 14 of the top 25 leading-edge genes were components of the electron transport chain (Figure 4B).
To functionally evaluate changes in oxidative phosphorylation in the Jurkat cell line, we directly measured the oxidative phosphorylation after treatment with BRQ. Cells were treated with BRQ for 24 hours, live cells were plated, and oxygen consumption was measured over time using the Seahorse XF platform after the addition of standard mitochondrial inhibitors. The rate of basal respiration was similar between untreated and treated cells (Figure 4C). However, compared with untreated cells, the maximal respiratory capacity in BRQ-treated cells (24 hours) was significantly higher (Figure 4C), suggesting that treated cells have a greater spare capacity or a greater capacity to respond to increased energy demands. This increase in oxidative phosphorylation in BRQ-treated cells supports the findings from the RNA sequencing.
In addition, we profiled another T-ALL cell line, PF-382, which showed minimal differences in oxygen consumption between untreated and treated cells, suggesting that T-ALL is a metabolically heterogenous disease that does not respond uniformly to nucleotide starvation and other forms of metabolic stress.
To further interrogate mitochondrial function, flow cytometry was used to quantify mitochondrial mass and mitochondrial membrane potential (Figure 4D). Mitochondrial mass was assessed using MitoTracker Deep Red, a dye that binds to thiol groups in mitochondria. After 24 to 48 hours of treatment with BRQ in multiple cell lines, there was a consistent increase (1.64-fold) in mitochondrial mass (representative data shown in Figure 4D). BRQ led to an S-phase arrest, an accumulation of cells that were unable to complete cytokinesis and expectedly larger in size, as evidenced by an increase in their forward scatter measurement (supplemental Figure 4B). Although the MitoTracker dye did not allow for a precise measurement of mitochondrial number, it does seem most plausible that the increase in mitochondrial mass was due to the concurrent increase in average cell size and average amount of available cytoplasm, rather than due to an actual increase in the average number of mitochondria per cell. Notably, previous work done in AML cell lines that show resistance to chemotherapy also show increased mitochondrial mass and increased OxPhos.56
Mitochondrial membrane potential was quantified using the dye tetramethylrhodamine methyl ester. Tetramethylrhodamine methyl ester normally accumulates in healthy mitochondria with intact membrane potentials and fluoresces brightly, but when the membrane potential is lost, the fluorescence dims. There was an increase in the mitochondrial membrane potential after BRQ treatment in all cell lines at 24 hours (Figure 4E). This is consistent with the Jurkat cell Seahorse assay data and, again, suggests that T-ALL cells treated with BRQ have increased capacity to tolerate increased energy demands or metabolic stress.
DHODH inhibition is curative in a genetically engineered mouse model of T-ALL
Based on the consistent and robust in vitro response of T-ALL cell lines to DHODH inhibition, we tested BRQ in preclinical mouse models of T-ALL. NOTCH1 activation is a hallmark of T-ALL, occurring in >70% of cases.57,58 We used a syngeneic retroviral transduction model of leukemia driven by a constitutively activated intracellular NOTCH59 to test the efficacy of BRQ. This model was transplanted into immunocompetent C57/BL6J recipient mice, demonstrated a high penetrance and short latency, and had a GFP marker for disease tracking.
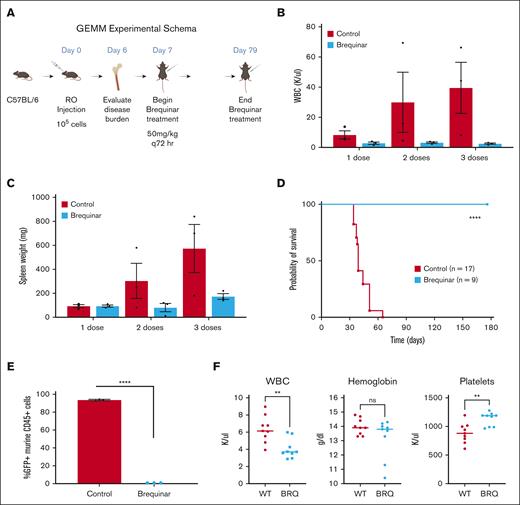
After leukemia was established, mice were randomized to treatment with BRQ (Figure 5A). The mice in the treatment group received BRQ at a dose of 50 mg/kg every 72 hours, which had previously been optimized by our laboratory as the maximal tolerated dose.22 The peripheral WBC count rapidly increased in the untreated animals but remained in the normal range in the mice treated with BRQ (Figure 5B). Spleen weight followed the same pattern (Figure 5C).
DHODH inhibition is curative in a genetically engineered mouse model of T-ALL. (A) Schema outline of the experiment, with dosing and treatment time points. (B) Peripheral WBC counts measured in mice with T-ALL that either received no treatment (untreated) or treatment with BRQ 50 mg/kg every 3 days (brequinar). Each group included 3 animals. Measurements were recorded after 1, 2, or 3 doses of BRQ, with matched untreated controls. (C) Spleen weights measured untreated or brequinar mice. Each group included 3 animals. Measurements were recorded after 1, 2, or 3 doses of BRQ, with matched untreated controls. (D) Kaplan-Meier survival curve comparing untreated brequinar mice. n = 17 in the untreated group; n = 9 in the brequinar group; P < .0001. (E) Disease burden in the bone marrow as measured by the percentage of GFP+ murine CD45+ cells detected by flow cytometry after 5 doses of BRQ. Each group included 3 animals. (F) WBC counts, hemoglobin, and platelet counts from both wild-type (n = 9) and BRQ-treated mice (n = 9). GEMM, genetically engineered mouse model; RO, retroorbital. Created with BioRender.com.
DHODH inhibition is curative in a genetically engineered mouse model of T-ALL. (A) Schema outline of the experiment, with dosing and treatment time points. (B) Peripheral WBC counts measured in mice with T-ALL that either received no treatment (untreated) or treatment with BRQ 50 mg/kg every 3 days (brequinar). Each group included 3 animals. Measurements were recorded after 1, 2, or 3 doses of BRQ, with matched untreated controls. (C) Spleen weights measured untreated or brequinar mice. Each group included 3 animals. Measurements were recorded after 1, 2, or 3 doses of BRQ, with matched untreated controls. (D) Kaplan-Meier survival curve comparing untreated brequinar mice. n = 17 in the untreated group; n = 9 in the brequinar group; P < .0001. (E) Disease burden in the bone marrow as measured by the percentage of GFP+ murine CD45+ cells detected by flow cytometry after 5 doses of BRQ. Each group included 3 animals. (F) WBC counts, hemoglobin, and platelet counts from both wild-type (n = 9) and BRQ-treated mice (n = 9). GEMM, genetically engineered mouse model; RO, retroorbital. Created with BioRender.com.
The mice were treated for 79 days, until all mice in the untreated group (n = 17) had succumbed to disease. All mice in the treatment group (n = 9) were alive at this time. BRQ was well tolerated, and there were no treatment-related toxicities. The mice maintained stable weights throughout. Treatment with BRQ dramatically improved survival (Figure 5D).
After discontinuation of treatment, we anticipated that the leukemia would relapse, based on previous experience in models of myeloid disease.60 The mice were monitored for signs of relapse for an additional 98 days after the end of the treatment. On day 175 of the experiment, the remaining animals were euthanized for bone marrow evaluation of leukemia burden. None of the BRQ-treated mice had any evidence of leukemia (GFP+ cells) in their bone marrow (Figure 5E). These data provide robust evidence that DHODH inhibition led to rapid and complete disease response in a genetically engineered NOTCH-activated model of T-ALL.
To provide further evidence that BRQ treatment does not have broad untargeted effects on hematopoiesis, we also examined complete blood count data from the mice in the treatment group, and compared these with those of healthy wild-type mice that were not exposed to the effects of nucleotide starvation (Figure 5F). Although there are statistically significant differences between the WBC and platelet counts, it is important to note that in both groups (wild-type and BRQ-treated), both WBC and platelet count values were in the range considered normal for adult mice. This supports that there is no meaningful nonspecific effect of BRQ on general hematopoiesis or lymphopoiesis.
DHODH inhibition results in disease response in 2 PDX models of T-ALL
Given the dramatic disease response in the syngeneic genetically engineered mouse model model of T-ALL, we then used 2 PDX models to evaluate the efficacy of BRQ (Figure 6A). One PDX (MAAT-43298) was originally derived from a 5-year-old male with relapsed leukemia after chemotherapy. This PDX harbored an FBXW7p.R465H mutation that leads to activated NOTCH1 signaling.
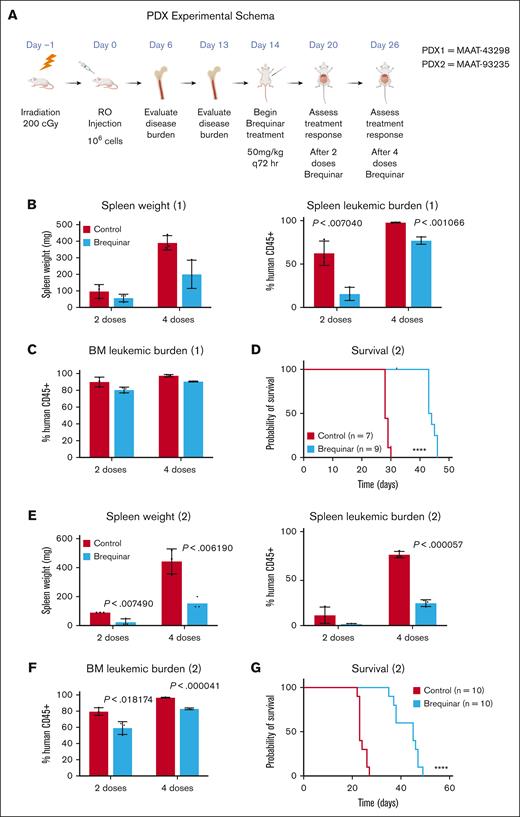
DHODH inhibition results in disease response in a PDX model of T-ALL. (A) Schematic outline of the experiment, with dosing and treatment time points. (B) Spleen weights measured in mice after the engraftment of the T-ALL PDX (MAAT-43298), which were either untreated or designated as brequinar. Each group included 3 animals. Measurements were recorded fter 2 or 4 doses of BRQ, with matched untreated controls. (C) Disease burden in the bone marrow as measured by the percentage of human CD45+ cells detected by flow cytometry following either 2 or 4 doses of BRQ. Each group included 3 animals. (D) Kaplan-Meier survival curve comparing mice after the engraftment of the T-ALL PDX (MAAT-43298), which were either untreated or designated as brequinar; n = 7 in the untreated group; n = 9 in the brequinar group; P < .0001. (E) Spleen weights measured in mice after the engraftment of the T-ALL PDX (MAAT-93235) that were either untreated or designated as brequinar. Each group included 3 animals. Measurements were recorded after 2 or 4 doses of BRQ, with matched untreated controls. (F) Disease burden in the bone marrow as measured by the percentage of human CD45+ cells detected by flow cytometry after either 2 or 4 doses of BRQ. Each group included 3 animals. (G) Kaplan-Meier survival curve comparing mice after the engraftment of the T-ALL PDX (MAAT-93235), which were either untreated or designated as brequinar. n = 10 in the untreated group; n =10 in the brequinar group; P < .0001. Created with BioRender.com.
DHODH inhibition results in disease response in a PDX model of T-ALL. (A) Schematic outline of the experiment, with dosing and treatment time points. (B) Spleen weights measured in mice after the engraftment of the T-ALL PDX (MAAT-43298), which were either untreated or designated as brequinar. Each group included 3 animals. Measurements were recorded fter 2 or 4 doses of BRQ, with matched untreated controls. (C) Disease burden in the bone marrow as measured by the percentage of human CD45+ cells detected by flow cytometry following either 2 or 4 doses of BRQ. Each group included 3 animals. (D) Kaplan-Meier survival curve comparing mice after the engraftment of the T-ALL PDX (MAAT-43298), which were either untreated or designated as brequinar; n = 7 in the untreated group; n = 9 in the brequinar group; P < .0001. (E) Spleen weights measured in mice after the engraftment of the T-ALL PDX (MAAT-93235) that were either untreated or designated as brequinar. Each group included 3 animals. Measurements were recorded after 2 or 4 doses of BRQ, with matched untreated controls. (F) Disease burden in the bone marrow as measured by the percentage of human CD45+ cells detected by flow cytometry after either 2 or 4 doses of BRQ. Each group included 3 animals. (G) Kaplan-Meier survival curve comparing mice after the engraftment of the T-ALL PDX (MAAT-93235), which were either untreated or designated as brequinar. n = 10 in the untreated group; n =10 in the brequinar group; P < .0001. Created with BioRender.com.
The PDX was engrafted and established in immunodeficient NSG mice before the initiation of BRQ treatment, and cohorts of mice were analyzed at predetermined end points. Treated animals showed a decrease in spleen size and leukemia burden (Figure 6B), though without an associated decrease in bone marrow disease (Figure 6C). There was a significant survival benefit in the BRQ-treated mice as compared with that in the untreated mice (Figure 6D).
The second PDX (MAAT-93235) was originally derived from a 6-year-old female with relapsed leukemia. This PDX harbored multiple mutations in a number of different oncogenes, including KMT2C, CREBBP, NOTCH2, and PDGFRA among others. Disease was established in the same manner, and animals were analyzed. This model, again, showed a decrease in spleen size and leukemia burden (Figure 6E) but also showed a significant decrease in bone marrow disease (Figure 6F). There was, again, a significant survival benefit in the BRQ-treated mice compared with that in the untreated mice (Figure 6G).
DHODH inhibition affects the developing thymus, although changes rapidly reverse when nucleotide starvation is removed
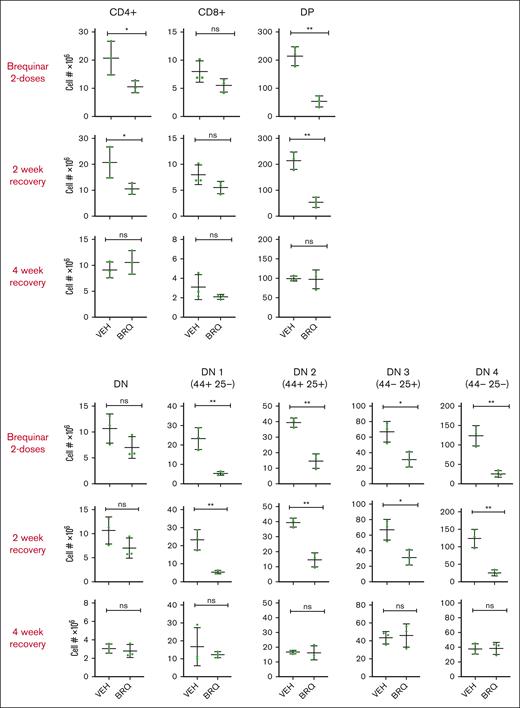
Because there is interest in the use of DHODH inhibitors as a potential therapy for pediatric and adult leukemias, we turned our attention to the effects of DHODH inhibition on development and specifically focused on the developing thymus. This compartment would be of particular interest in pediatric patients, whose thymi have not yet completed developing. To model this, we used juvenile mice (3 weeks) whose thymi were just reaching the peak of their size and growth rate. These mice were treated with BRQ 50 mg/kg every 72 hours and were paired with age- and sex-matched controls at each time point of analysis. A subset of mice was euthanized after the completion of 2 doses of BRQ, and the thymus, bone marrow, spleen, and peripheral blood were analyzed for T-cell populations, both relative and absolute numbers. Another cohort was allowed to recover for 2 weeks after treatment before analysis, and a third cohort was allowed a 4-week recovery period.
Treatment with 2 doses of BRQ significantly affects T cells across all stages of development, as shown in Figure 7. This is expected, because developing T cells are rapidly proliferative (often dividing as quickly as every 6 hours) and, therefore, highly dependent on their supply of nucleotides for DNA replication as well as transcription. However, after both the 2- and 4-week recovery periods after BRQ treatment, these differences were no longer detectable, demonstrating the resilience of the developing thymus and that the effects of nucleotide starvation were temporary and reversible. These data are encouraging, because this may also suggest that the developing human thymus, which is similarly rapidly proliferative in nature would, also recover on the removal of nucleotide starvation. We are hopeful these findings will pave the way for additional preclinical studies and eventually clinical trials for pediatric patients with T-cell disease.
The developing thymus recovers rapidly after exposure to DHODH inhibition. T-cell populations were quantified by flow cytometric analysis. T cells were isolated from the thymi of juvenile mice treated with vehicle (VEH) or BRQ 50 mg/kg every 3 days for 2 doses, followed by a 2-week or a 4-week recovery period. Shown are double-negative (DN) as well as DN1-4 subgroups, single positive (CD4+ and CD8+), and double-positive (DP) groups. n = 3 mice for each interval time point analysis. ns, not significant.
The developing thymus recovers rapidly after exposure to DHODH inhibition. T-cell populations were quantified by flow cytometric analysis. T cells were isolated from the thymi of juvenile mice treated with vehicle (VEH) or BRQ 50 mg/kg every 3 days for 2 doses, followed by a 2-week or a 4-week recovery period. Shown are double-negative (DN) as well as DN1-4 subgroups, single positive (CD4+ and CD8+), and double-positive (DP) groups. n = 3 mice for each interval time point analysis. ns, not significant.
Discussion
Metabolic reprogramming is now known to be a hallmark of cancer cells in general and of T-ALL specifically.61,62 Targeting metabolic pathways has been of great interest in the treatment of cancer; indeed, some of the earliest chemotherapy agents used (eg, methotrexate and mercaptopurine) take advantage of cancer cells’ dependence on nucleotide synthesis and continue to be a backbone of therapy today. Targeting the 1-carbon folate pathway using SHMT1 and SHMT2 inhibitors is another promising approach.11 In this study, we demonstrated that BRQ specifically inhibits the pyrimidine synthesis pathway in T lymphoblasts and rapidly depletes concentrations of intracellular pyrimidines but not purines, which can be rescued by the addition of supplemental uridine. Given the metabolic similarities between lymphocytic populations, B-ALL may also be sensitive to DHODH inhibition and is the subject of ongoing efforts. The understanding of metabolic remodeling of cancer cells continues to expand, and as it does, approaches such as targeting an essential enzyme, such as DHODH, may not only be effective but also offer a larger therapeutic window and/or prove less apt to generate resistance.
We also demonstrated that DHODH is a promising target in the treatment of T-ALL. T-ALL cell lines are differentially sensitive to genetic loss of DHODH. Additionally, we showed numerous T-ALL cell lines to be sensitive to chemical inhibition of DHODH in vitro. BRQ is a highly specific and potent inhibitor of DHODH.22,63 Given the interest in the development of clinical grade DHODH inhibitors and ongoing clinical trials, it is imperative to define diseases in which these compounds may be active. Based on their well-studied, on-target effects, and in vitro and in vivo activity, we suspect that the effects we observe with BRQ would be consistent across other DHODH inhibitors.64-66
The specific mechanism by which T lymphoblasts are sensitive to DHODH inhibition remains incompletely understood. In myeloid malignancies, DHODH inhibitors were found to be potent inducers of differentiation, which occurred over a period of several days.22,23,25,55 However, a different mechanism is likely to be at play in T-ALL, in which in vitro or in vivo differentiation during the course of treatment has not been a feature of this disease.
The role of oxidative phosphorylation in the response of T lymphoblasts to DHODH inhibition also remains incompletely understood. The physical location of DHODH in the inner mitochondrial membrane and the fact that the reduction of DHO to orotate is the only step in pyrimidine biosynthesis to take place inside the mitochondria (in eukaryotes), and not in the cytosol, is unique.67 Our data demonstrated an increase in transcription of genes involved in oxidative phosphorylation in the setting of DHODH inhibition. Furthermore, the genes driving this effect seemed primarily concentrated in the electron transport chain itself. Some proteins, including NDUFA1, NDUFA2, NDUFA4, and NDUFB2, have specific oxidoreductase activity and work to create the electron gradient across the inner mitochondrial membrane. Others (UQCRQ) have ubiquinone-binding protein activity, ATPase activity (ATP6V0B, ATP6V0C, ATP6V1D, and ATP6V1F), or comprise parts of the cytochrome c oxidase complex (COX6A1 and COX8A). These findings suggest that upon inhibition of DHODH, T lymphoblasts exhibit a compensatory increase in the expression of these and other genes in the electron transport chain to maintain the gradient across the membrane and preserve ATP production.
Although the Jurkat cell line is one of the most commonly used and well-studied T-ALL cell lines in literature, it cannot be representative of all T-ALLs. Future studies will include evaluating the impact of BRQ on gene expression in both additional cell lines and primary patient samples. Moreover, although the increase in spare capacity was seen only in the Jurkat cells, and not the PF-382 cells (or in other cell lines tested), it is notable that in all cell lines studied, an increase in mitochondrial membrane potential was observed after treatment with BRQ over 24 hours. This contrasts to some previously published literature; however, this may be explained by the fact that other studies have used a different class of compounds to inhibit DHODH, which binds the active site of the enzyme.68 We hypothesize that inhibition of DHODH causes cells to upregulate the expression of other genes, which allow the cell to increase its mitochondrial membrane potential to try to maximize mitochondrial energy production because DHODH-dependent respiration is inhibited. Additional experiments with BRQ in combination with specific mitochondrial complex inhibitors are underway to better elucidate the role of DHODH inhibition in mitochondrial respiration.
We demonstrated in 3 different preclinical animal models that DHODH inhibition was well tolerated and led to rapid disease response and improvement in survival. Of course, in most cases of cancer treatment, combination therapy is essential for cure. One of the paradigms of combination therapy is to select drugs with nonoverlapping patterns of toxicity. This is due to the observation that although combination therapy is almost always more effective, it is almost always more toxic. Thus, the additive or synergistic efficacy of a combination needs to exceed its additive or synergistic side-effects. For this reason, the treatment of lymphoid malignancies is particularly attractive, given that many of the standard-of-care approved agents are not nucleoside analogs. Additional combination studies are needed to determine how to best leverage the therapeutic potential of DHODH inhibition.
Finally, we also show pilot studies in juvenile, healthy mice, demonstrating that although there are significant impacts on the developing thymus and T-cell populations across the spectrum of maturation in response to DHODH inhibition, these changes are short lived and reversible when the DHODH inhibition is stopped. This is encouraging as we continue to think about the possibility of brequinar and other related agents as potential new therapies for children with T-cell malignancies, whose T-cell repertoires are still rapidly expanding and may be susceptible to similar impacts but who are similarly likely to quickly recover after the stimulus is removed.
Together, our encouraging preclinical data, combined with the knowledge gained from active clinical trials of DHODH inhibitors in other malignancies, will hopefully pave the way for additional investigations into DHODH as a promising therapeutic target in T-ALL.
Acknowledgments
The authors thank Jon Aster for generously sharing plasmid constructs and for helpful discussions. The authors also thank Warren Pear for sharing plasmids, Giulia Schiroli for her guidance in generating the CRISPR-KO cell lines, and Thale Olsen for helpful discussions regarding dependency data. They thank Maris Handley and the staff of the Harvard Stem Cell Institute–Center for Regenerative Medicine Flow Cytometry Core for assistance with flow cytometry. A.N.S. was supported by research funding from the National Institutes of Health (National Heart, Lung, and Blood Institute 5T32HL007574). K.G. was supported by a Swedish Research Council International postdoc grant. J.M. was supported by an international postdoc grant from the Swedish Research Council (2018-06724). Y.P. was supported by the National Institutes of Health (National Cancer Institute K08CA222684) and a Hyundai Hope on Wheels grant. K.S. was supported by National Institutes of Health (R35 CA210030). D.B.S. was supported by research funding from an American Society of Hematology scholar grant and by the St. Baldrick's Foundation.
Authorship
Contribution: A.N.S., K.S., and D.B.S. undertook study conception and design; A.N.S., E.Z., M.A., J.M., and K.G. collected data; A.N.S., G.A., Y.P., V.G., K.S., and D.B.S. undertook analysis and interpretation of the results; A.N.S. wrote the original draft of the manuscript; Y.P., K.S., and D.B.S. reviewed and edited the manuscript; and all authors reviewed the results and approved the final draft of the manuscript.
Conflict-of-interest disclosure: V.G. has a Sponsored Research Agreement from Clear Creek Bio. K.S. is on the scientific advisory board and has stock options in Auron Therapeutics; receives grant funding from Novartis and KronosBio; and has previously consulted for AstraZeneca and KronosBio on topics unrelated to this manuscript. D.B.S. is a cofounder and holds equity in Clear Creek Bio. The remaining authors declare no competing financial interests.
Correspondence: Amy N. Sexauer, Department of Pediatric Oncology, Dana-Farber Cancer Institute, 450 Brookline Ave, Boston, MA 02215; e-mail: amy_sexauer@dfci.harvard.edu; and David B. Sykes, Center for Regenerative Medicine, Massachusetts General Hospital, 185 Cambridge St, CPZN 4-4238, Boston, MA, 02114; e-mail: dbsykes@mgh.harvard.edu.
References
Author notes
RNA sequencing data are available at Gene Expresssion Omnibus database (accession number GSE216753).
Original data, the syngeneic murine GEMM model of leukemia, information on resources and reagents are available on request from the corresponding authors, Amy Sexauer (amy_sexauer@dfci.harvard.edu) and David Sykes (dbsykes@mgh.harvard.edu).
The full-text version of this article contains a data supplement.

![T-ALL cell lines are highly dependent on de novo pyrimidine synthesis. (A) Volcano plot depicting the 22Q2 CRISPR (DepMap Public+Score, Chronos) genome-wide differential dependency (effect size) vs -log10(false discovery rate [FDR] q value) for T-ALL vs non–T-ALL cell lines. Dots correspond to genes. The DHODH gene is highlighted red. Differential dependency is estimated based on the 2-class comparison method implemented in DepMap (limma empirical Bayes; significance cutoff FDR q ≤ 0.10). (B) Scatter dot plot demonstrating the dependency on DHODH of the T-ALL, B-ALL, and AML cell lines in the CRISPR (DepMap 22Q2 Public+Score, Chronos) screen for 1086 cell lines. Dots represent cell lines. The 5 T-ALL cell lines in the screen are highlighted red, the 12 B-ALL cell lines are highlighted black, and the 26 AML cell lines are highlighted blue. The x-axis shows the DHODH Chronos dependency score (gene effect) per cell line. The y-axis shows the probability of dependency on DHODH per cell line. Dependency on DHODH is estimated by low negative (≤−0.5) Chronos gene effect scores. (C) Schematic outline of the de novo pyrimidine synthesis pathway and the role of DHODH, which is located in the inner mitochondrial membrane. Also shown is the location where uridine feeds into the pathway. (D) CRISPR KO of DHODH was performed in the Jurkat cell line. Parental and KO cells were seeded at equivalent densities, and supplemental uridine was withdrawn, at the time when DHODH-KO cells rapidly died while the parental cells continued to grow rapidly. dCTP, deoxycytosine triphosphate; dTMP, deoxythymidine 5′-monophosphate; dTTP, deoxythymidine 5′-triphosphate; dUDP, deoxyuridine diphosphate; dUMP, deoxyuridine monophosphate; dUTP, deoxyuridine triphosphate; GlcNAc, N-acetylglucosamine; OMP, orotidylate monophosphate.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/21/10.1182_bloodadvances.2023010337/2/m_blooda_adv-2023-010337-gr1.jpeg?Expires=1769831719&Signature=DT57W9FrAnO1sVnO~ckzDaqtS-rqq9Yks5Ek1rqUaeZUePyXEWVIr-g6gjONoVArvHmMjhy~39LquYBy0iMNGpOqg~mVYXgH4jo8UJ7t1Sxr80r~mD5PhFZ8uKZb-gszUJ7ozg1TLoPFRHuQ08rIfgvBP9YQbQWdu~0amHxarfkQyK~ukpIGh3zQIVfVFU7OnqYfPM-uWHionkp1C2byEGqk-qIW02h8K6f8pee-KCzcPTKzb3bV-jyKtMfUGHOwEPJTn2otwqpqnmDywFvfWZ7Vwa-gT8cZQ-KOD~elEGcA43gWmFNFzwJfxgwIxzGQ34-RS03pulP0m6otHXxMpA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)