Key Points
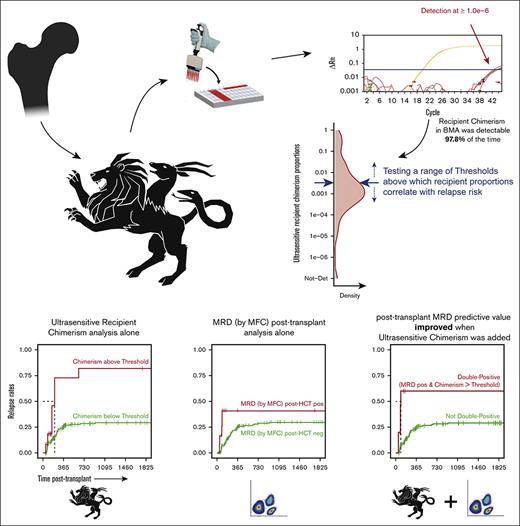
We developed an ultrasensitive method to quantify chimerism after allo-HCT, leveraging its sensitivity to predict relapse more accurately.
Our assay significantly enhanced flow-based MRD data, revealing a previously unappreciated clinical utility of posttransplant chimerism.
Abstract
Increasing mixed chimerism (reemerging recipient cells) after allogeneic hematopoietic cell transplant (allo-HCT) can indicate relapse, the leading factor determining mortality in blood malignancies. Most clinical chimerism tests have limited sensitivity and are primarily designed to monitor engraftment. We developed a panel of quantitative polymerase chain reaction assays using TaqMan chemistry capable of quantifying chimerism in the order of 1 in a million. At such analytic sensitivity, we hypothesized that it could inform on relapse risk. As a proof-of-concept, we applied our panel to a retrospective cohort of patients with acute leukemia who underwent allo-HCT with known outcomes. Recipient cells in bone marrow aspirates (BMAs) remained detectable in 97.8% of tested samples. Absolute recipient chimerism proportions and rates at which these proportions increased in BMAs in the first 540 days after allo-HCT were associated with relapse. Detectable measurable residual disease (MRD) via flow cytometry in BMAs after allo-HCT showed limited correlation with relapse. This correlation noticeably strengthened when combined with increased recipient chimerism in BMAs, demonstrating the ability of our ultrasensitive chimerism assay to augment MRD data. Our technology reveals an underappreciated usefulness of clinical chimerism. Used side by side with MRD assays, it promises to improve identification of patients with the highest risk of disease reoccurrence for a chance of early intervention.
Introduction
Chimerism in an individual refers to the presence of allogeneic cells/genomic DNA (gDNA) and is commonly a natural phenomenon, a legacy from maternal–fetal exchange during pregnancy.1,2 The origin of chimerism can also be iatrogenic, acquired from a blood transfusion or from liquid or solid organ transplant.
Allogeneic hematopoietic cell transplant (allo-HCT) from a related (family) or unrelated donor source is a procedure resulting in acquired chimerism. Allo-HCT is currently the best curative option for ∼50% of adult patients with acute leukemias, those categorized with intermediate-/high-risk disease, or those with resistant or relapsed disease after induction therapy.3-5 Despite its association with graft-versus-host disease (GVHD), allo-HCT is preferred for its graft-versus-leukemia effect in treating leukemia.6 Currently, ∼75% of allo-HCT worldwide are used to treat leukemia, 94% of which are acute lymphoblastic and myeloid leukemias and myelodysplastic syndrome (MDS).7,8
After allo-HCT, complete donor chimerism resulting in full donor hematopoiesis indicates successful engraftment. However, posttransplant chimerism is dynamic. Donor and recipient hematopoiesis could coexist in the recipient, resulting in mixed chimerism or even a return to complete autologous reconstitution if engraftment fails. Split chimerism can also occur when chimerism is in 1 cell subpopulation but not others.9 Mixed chimerism may indicate the survival of leukemic or healthy host hematopoietic cells or a combination of both and, thus, can be indicative of relapse. A probable mechanism is decreased immunocompetence of donor effector cells and reduced donor-derived graft-versus-leukemia, facilitating both healthy host and leukemic cell reemergence.10 In patients with acute leukemias and MDS, a growing body of data shows that increasing mixed chimerism is associated with the highest risk of relapse.11-20 This underscores the predictive usefulness of chimerism analysis as a surrogate marker for minimal/measurable residual disease (MRD), especially when direct MRD measurement becomes difficult because of the wide heterogeneity of many subtypes of acute leukemias.21,22
Sensitive and quantitative monitoring of chimerism after transplant is key to acquiring information on graft maintenance/rejection and, at the same time, on leukemia reappearance by associating it with MRD. We designed a chimerism panel composed of polymorphism-specific primers and fluorogenic probes used to detect and quantify nonshared polymorphisms in highly sensitive real-time TaqMan quantitative polymerase chain reaction (qPCR). The limit-of-detection (LOD) is in the order of 1 in a million, at least an order of magnitude below commercially available tests,23 and multiple orders of magnitude below the current gold standard in clinical chimerism analysis, the widely used PCR-based short/variable tandem repeat (STR) characterization (LOD ≥ 1:100). We applied this panel to a cohort of patients with leukemia with known outcomes after allo-HCT. We hypothesized that by leveraging the high resolution of our technique, reemerging recipient cells (ie, increasing mixed chimerism) could be detected/quantified with sufficient accuracy to inform the patient about the risk of relapse.
Methods
Patients
We included 298 consecutive patients with a diagnosis of acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), or MDS, who consented to master protocol #999.209 for data collection and specimen storage at the Fred Hutchinson Cancer Center and underwent their first transplant and had chimerism their follow-up data measured via the STR methodology in whole bone marrow aspirates (BMAs) and in leukemia-relevant peripheral blood (PB)-derived subsets (CD33+, CD3+, and CD19+ cells). We excluded 38 patients with incomplete baseline specimen inventory (ie, not all donors and recipients of pretransplant samples had leftover gDNA available), 35 whose recipient DNA could not be uniquely targeted, and 2 additional patients because of chimerism assay failure. Patient clinical characteristics are summarized in Table 1.
Patient characteristics included in chimerism analyses
| Characteristic . | Relapse status . | |
|---|---|---|
| No . | Yes . | |
| Numbers, n | 155 | 68 |
| Chimerocyte type of assay run, n (%) | ||
| HLA-specific assay(s) (with or without non-HLA specificities) | 31 (20.0) | 15 (22.1) |
| Non-HLA–specific assay(s) only | 124 (80.0) | 53 (77.9) |
| Age at transplant (y), median (IQR) | 52 [40 - 62] | 52 [39 - 60] |
| Sex, n (%) | ||
| Female | 63 (40.6) | 35 (51.5) |
| Male | 92 (59.4) | 33 (48.5) |
| Ancestral background, n (%) | ||
| Caucasian | 87 (75.0) | 46 (83.6) |
| Asian | 14 (12.1) | 2 (3.6) |
| American Native (including Alaska, Hawaii, and Pacific Islands) | 7 (6.0) | 4 (7.3) |
| Mixed/multiple | 3 (2.6) | 3 (5.5) |
| Sub-Saharan African | 5 (4.3) | 0 (0.0) |
| Unknown/missing data | 39 | 13 |
| Transplant type, n (%) | ||
| Related (haplo or HLA matched) | 15 (9.7) | 16 (23.5) |
| Unrelated cord blood (single or combined) | 57 (36.8) | 15 (22.1) |
| Unrelated (HLA matched or mismatched) | 83 (53.5) | 37 (54.4) |
| Conditioning, n (%) | ||
| TBI 1200-1320; FLU, CY | 40 (25.8) | 14 (20.6) |
| TBI 300-400; CY with or without FLU | 27 (17.4) | 7 (10.3) |
| TBI 200; FLU based (with CY, ATG, RAB, and/or TREO) | 47 (30.3) | 27 (39.7) |
| No TBI; BU based (with CY, FLU, and/or ATG) or FLU based (with either L-PAM or TREO) | 41 (26.5) | 20 (29.4) |
| GVHD prophylaxis, n (%) | ||
| CSP + MMF (with or without RAPA, CY, or FK506) | 88 (56.8) | 22 (32.4) |
| CSP + MTX (with or without CY or FK506) | 8 (5.2) | 6 (8.8) |
| FK506 + MMF (with or without CY or RAPA) | 15 (9.7) | 16 (23.5) |
| FK506 + MTX (with or without steroids or ABA) | 44 (28.4) | 24 (35.3) |
| Diagnosis, n (%) | ||
| ALL | 30 (19.4) | 7 (10.3) |
| AML | 84 (54.2) | 49 (72.1) |
| MDS | 41 (26.5) | 12 (17.6) |
| MRD based on MFC status before transplant, n (%) | ||
| Positive (>0%) | 41 (27.3) | 31 (48.4) |
| Negative (=0%) | 109 (72.7) | 33 (51.6) |
| Missing data | 5 | 4 |
| Disease status at transplant, n (%) | ||
| Active disease | 13 (8.4) | 15 (22.1) |
| CR1 | 70 (45.2) | 28 (41.2) |
| CR2 | 30 (19.4) | 13 (19.1) |
| CR3 | 5 (3.2) | 1 (1.5) |
| Undefined (mostly including patients with MDS) | 37 (23.9) | 11 (16.2) |
| Characteristic . | Relapse status . | |
|---|---|---|
| No . | Yes . | |
| Numbers, n | 155 | 68 |
| Chimerocyte type of assay run, n (%) | ||
| HLA-specific assay(s) (with or without non-HLA specificities) | 31 (20.0) | 15 (22.1) |
| Non-HLA–specific assay(s) only | 124 (80.0) | 53 (77.9) |
| Age at transplant (y), median (IQR) | 52 [40 - 62] | 52 [39 - 60] |
| Sex, n (%) | ||
| Female | 63 (40.6) | 35 (51.5) |
| Male | 92 (59.4) | 33 (48.5) |
| Ancestral background, n (%) | ||
| Caucasian | 87 (75.0) | 46 (83.6) |
| Asian | 14 (12.1) | 2 (3.6) |
| American Native (including Alaska, Hawaii, and Pacific Islands) | 7 (6.0) | 4 (7.3) |
| Mixed/multiple | 3 (2.6) | 3 (5.5) |
| Sub-Saharan African | 5 (4.3) | 0 (0.0) |
| Unknown/missing data | 39 | 13 |
| Transplant type, n (%) | ||
| Related (haplo or HLA matched) | 15 (9.7) | 16 (23.5) |
| Unrelated cord blood (single or combined) | 57 (36.8) | 15 (22.1) |
| Unrelated (HLA matched or mismatched) | 83 (53.5) | 37 (54.4) |
| Conditioning, n (%) | ||
| TBI 1200-1320; FLU, CY | 40 (25.8) | 14 (20.6) |
| TBI 300-400; CY with or without FLU | 27 (17.4) | 7 (10.3) |
| TBI 200; FLU based (with CY, ATG, RAB, and/or TREO) | 47 (30.3) | 27 (39.7) |
| No TBI; BU based (with CY, FLU, and/or ATG) or FLU based (with either L-PAM or TREO) | 41 (26.5) | 20 (29.4) |
| GVHD prophylaxis, n (%) | ||
| CSP + MMF (with or without RAPA, CY, or FK506) | 88 (56.8) | 22 (32.4) |
| CSP + MTX (with or without CY or FK506) | 8 (5.2) | 6 (8.8) |
| FK506 + MMF (with or without CY or RAPA) | 15 (9.7) | 16 (23.5) |
| FK506 + MTX (with or without steroids or ABA) | 44 (28.4) | 24 (35.3) |
| Diagnosis, n (%) | ||
| ALL | 30 (19.4) | 7 (10.3) |
| AML | 84 (54.2) | 49 (72.1) |
| MDS | 41 (26.5) | 12 (17.6) |
| MRD based on MFC status before transplant, n (%) | ||
| Positive (>0%) | 41 (27.3) | 31 (48.4) |
| Negative (=0%) | 109 (72.7) | 33 (51.6) |
| Missing data | 5 | 4 |
| Disease status at transplant, n (%) | ||
| Active disease | 13 (8.4) | 15 (22.1) |
| CR1 | 70 (45.2) | 28 (41.2) |
| CR2 | 30 (19.4) | 13 (19.1) |
| CR3 | 5 (3.2) | 1 (1.5) |
| Undefined (mostly including patients with MDS) | 37 (23.9) | 11 (16.2) |
ABA, abatacept; ATG, antithymocyte globulin; BU, busulfan; CSP, cyclosporine; CR1/2/3, first/second/third complete remission; CY, cyclophosphamide; FK506, tacrolimus; FLU, fludarabine; haplo, haploidentical; IQR, interquartile range; L-PAM, melphalan; MMF, mycophenolate mofetil; RAB, radiolabeled antibodies; RAPA, rapamycin; TBI, total body irradiation (in cGy); TREO, treosulfan.
In the drawn BMAs, red blood cells were lysed to collect nucleated cells. Drawn PB was sorted via fluorescence-activated cell sorting (FACS) to collect cell subsets (≤30 000 cells per subset). gDNA was extracted, and chimerism via STR was assessed using the Promega Powerplex 16 system (Promega, Madison, WI).
MFC to detect residual leukemia
Pretransplant MRD was assessed via a standardized 10-color multiparameter flow cytometry (MFC) approach,24 using red blood cell–lysed BMAs, on an average of 24 days before allo-HCT (range, 7-64 days). MRD was identified using different-from-normal and/or leukemia-associated immunophenotype approaches, with any level of residual disease considered MRD positive.25 MFC was also conducted after allo-HCT, and whenever such information was available for the same time point as for a chimerism measurement, MRD after allo-HCT was analyzed as an orthogonal approach to assess relapse risk via chimerism analysis.
Ultrasensitive chimerism
Donor–recipient genetic mismatches were used to measure chimerism. Our panel of highly sensitive, polymorphism-specific TaqMan qPCR assays consisted of 40 assays targeting polymorphism in HLA (n = 23) and non-HLA regions (n = 17)26-33 (supplemental Table 1). Part of the genomic regions and molecular assay oligonucleotides are available in the patent US 10 604 805 B2. The assays were rigorously optimized for high analytic specificity (no amplification of unintended polymorphisms; supplemental Table 1; supplemental Figure 1) and high analytic sensitivity (could detect a single target in the equivalent of a million background copies). Additional quality control metrics were assessed, including assay linearity, reproducibility, precision, and reportable range (supplemental Methods; supplemental Figure 2).
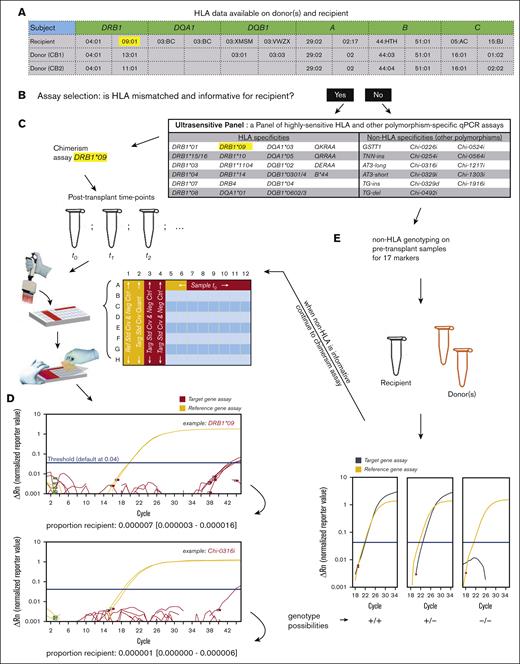
To identify donor–recipient mismatches, HLA-specific assays were chosen based on readily available patient records (example in Figure 1A). When an HLA mismatch was unavailable, genotyping for non-HLA markers was performed using a multiplexed qPCR method modified from a previously described method.34,35 Pretransplant genomic material stored for each patient and their candidate donor(s) was used (Figure 1E). A reference and a target set of primer/probe oligos were mixed per the reaction. Quantification was per the respective standard curve (absolute quantification method), and the target-to-reference quantity ratio was calculated for each sample, with values at ∼1, ∼0.5, or 0 if homozygous, heterozygous, or null for the target locus, respectively (Figure 1E; supplemental Figure 3).
Chimerism assay workflow. (A) HLA data readily available for donor(s) and recipient are analyzed. (B) Assay selection is performed. (C) If an informative HLA marker is available, chimerism assays are planned and run using recipient gDNA samples taken after transplant, typically in 2 reference gene replicates and 6 target replicates (all scalable to accommodate more DNA mass) in plate columns from 5 to 12. (D) Actual examples of an HLA-specific and a non-HLA–specific chimerism are provided; chimerism is quantified based on the ratio of a target to a reference quantity, accompanied by a 95% CI of the measurement. (E) If an informative HLA marker is not available, genotyping for 17 non-HLA markers is conducted on donor(s) and recipient samples taken before transplant; our genotyping assays can distinguish the homozygous (+/+) or heterozygous (+/–) presence of an allele based on quantity comparison vs a reference gene; a non-HLA marker is informative if it is positive in the recipient and negative in the donor(s).
Chimerism assay workflow. (A) HLA data readily available for donor(s) and recipient are analyzed. (B) Assay selection is performed. (C) If an informative HLA marker is available, chimerism assays are planned and run using recipient gDNA samples taken after transplant, typically in 2 reference gene replicates and 6 target replicates (all scalable to accommodate more DNA mass) in plate columns from 5 to 12. (D) Actual examples of an HLA-specific and a non-HLA–specific chimerism are provided; chimerism is quantified based on the ratio of a target to a reference quantity, accompanied by a 95% CI of the measurement. (E) If an informative HLA marker is not available, genotyping for 17 non-HLA markers is conducted on donor(s) and recipient samples taken before transplant; our genotyping assays can distinguish the homozygous (+/+) or heterozygous (+/–) presence of an allele based on quantity comparison vs a reference gene; a non-HLA marker is informative if it is positive in the recipient and negative in the donor(s).
Ultrasensitive chimerism detection was performed via real-time qPCR with TaqMan chemistry, using the absolute quantification method. Each sample was measured in sextuplicates for the target assay and in duplicates for the reference assay, in separate wells (Figure 1C). The total gDNA quantity (in genome equivalents [gEqs]) was counted with the reference standard curve, whereas chimeric DNA was counted with the target standard curve. Chimerism proportion was calculated as the ratio of these 2 quantities. Accuracy was evaluated using a 95% confidence interval (95% CI), calculated with the Wilson score without continuity correction.36,37
Statistical analyses
To assess the association of highly sensitive chimerism testing with relapse, 2 approaches were used for analyses in each material tested (BMAs and CD33+, CD3+, and CD19+ cells): high resolution chimerism values at each time point after allo-HCT, and high resolution chimerism dynamics (representing the rate of change in chimerism values, calculated between 2 time points) when multiple time points per patient were available. The first 540 days after allo-HCT was set as a predetermined window of analysis, chosen because it contained the most relapse events (88.2%). Because chimerism measurement was used to predict the outcome, chimerism time points within less than a week of a relapse diagnosis were excluded to avoid the self-fulfilling prophecy of a bad effect of chimerism at relapse. Chimerism measurements after relapse were also excluded. All BMA chimerism measurements (except n = 9) and all subset-derived chimerism measurements eligible in the outcome analyses were performed in the first 540 days after transplant. Chimerism values with relapse were assessed using the Cox proportional hazard regression, with chimerism modeled as a time-dependent covariate. In the multivariate models, covariates shown to correlate with outcome were included: pretransplant MRD, disease status at transplant, leukemia diagnosis, GVHD prophylaxis, and transplant type (see “Results”). Kaplan-Meier survival curves for time-dependent covariates (interval censored) were computed using the “survfit” function of the R package “survival,”38 whereas the corresponding Cox regression hazard ratios (HRs) were computed using the “coxph” function for interval-censored data.39
Results
Performance characteristics of the assay
We created a panel of highly sensitive and specific assays for ultrasensitive chimerism testing. These assays targeted HLA polymorphisms on chromosome 6 as well as other chromosomes.26-32,34,35
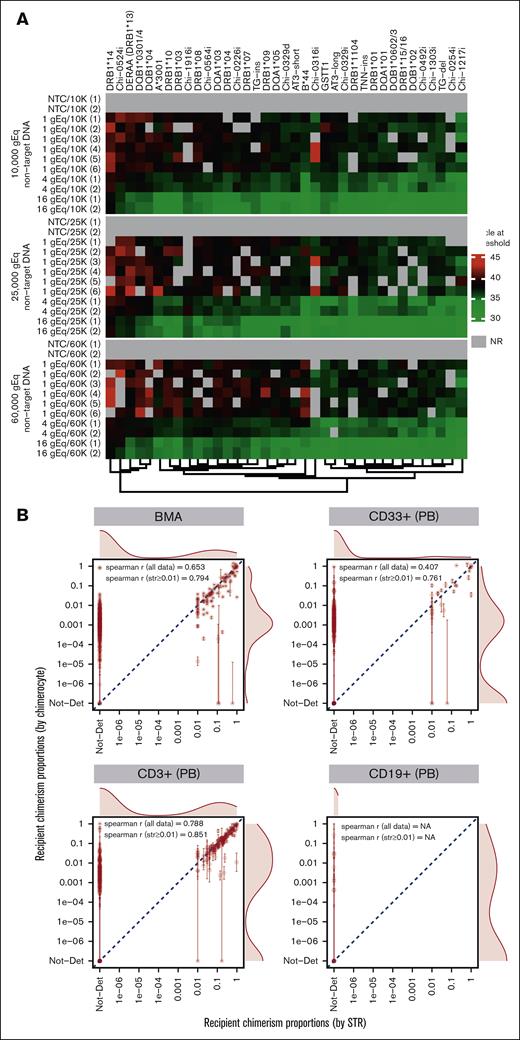
All the assays in the panel had high analytic sensitivity, quantifying down to a single target gDNA copy in a reaction with high concentrations of background gDNA, and high analytic specificity, that is, no amplification of background DNA when it was the only DNA present in the reaction (Figure 2A). Background DNA was tested up to 60 000 gEqs per reaction well (as allowed based on the concentration of the DNA purchased from the International Histocompatibility Working Group [IHWG]). The amplification patterns of target DNA at low levels did not appear to deteriorate with increasing background DNA (Figure 2A), and the nominal LOD per reaction well was set at 1 over twice the maximum background gEq tested, resulting in an effective LOD of 1.38e−6 per typical 6-replicate assay run.
Chimerocyte panel detects low target copies in high background levels (highly sensitive), does not amplify background (highly specific), and correlates with measurements from a gold standard method. (A) Two, 2, and 6 replicates, respectively, of 16, 4, and 1 target gEq were run with 3 different nontarget (background) levels amplifying at an acceptable cycle at a threshold ranging from 31.4 to 44.6. The amplifications were comparable per the background level and self-consistent per the individual assay. The 1 gEq dilution sometimes yielded no amplification (threshold, NR); this is statistically expected when pipetting such low target copy numbers. When targets were absent (nontemplate controls) the background was never amplified. (B) Comparison of recipient chimerism quantification using ultrasensitive chimerism (Chimerocyte) technique vs standard-of-care STR technique conducted using the gDNA from BMAs and PB-derived cells of patients with leukemia. Below a proportion of 0.01, chimerism was not detected (Not-Det) using STR. Error bars are 95% CIs of the measurements. Agreement between the methods was high and extreme outliers (ie, >0.1 in 1 assay vs Not-Det in the other) remained sparse and often with reduced accuracy (ie, large 95% CIs). Not-Det, not detected; NR, not reached; NA, missing or not available.
Chimerocyte panel detects low target copies in high background levels (highly sensitive), does not amplify background (highly specific), and correlates with measurements from a gold standard method. (A) Two, 2, and 6 replicates, respectively, of 16, 4, and 1 target gEq were run with 3 different nontarget (background) levels amplifying at an acceptable cycle at a threshold ranging from 31.4 to 44.6. The amplifications were comparable per the background level and self-consistent per the individual assay. The 1 gEq dilution sometimes yielded no amplification (threshold, NR); this is statistically expected when pipetting such low target copy numbers. When targets were absent (nontemplate controls) the background was never amplified. (B) Comparison of recipient chimerism quantification using ultrasensitive chimerism (Chimerocyte) technique vs standard-of-care STR technique conducted using the gDNA from BMAs and PB-derived cells of patients with leukemia. Below a proportion of 0.01, chimerism was not detected (Not-Det) using STR. Error bars are 95% CIs of the measurements. Agreement between the methods was high and extreme outliers (ie, >0.1 in 1 assay vs Not-Det in the other) remained sparse and often with reduced accuracy (ie, large 95% CIs). Not-Det, not detected; NR, not reached; NA, missing or not available.
Chimerism detection in whole BMAs and cell lineage subsets
Of the 298 consecutive patients who underwent a first-time transplant and had long-term follow-up, 260 had available specimens at baseline (before the transplant) and after the transplant, along with baseline specimens of all their donors. Of those, 225 (87%) were informative for at least 1 assay in our panel. This percentage was close to theoretical panel informativeness (94%) and surprisingly high, considering that 36% of patients (93 of 260) were recipients of double cord blood transplant (multiple donors tend to reduce the probability of an informative assay). Chimerism assessment failed for 2 patients, so 223 patients were included for further analyses (Table 1).
Of the 223 patients, 207 had BMAs, 166 had PB-derived CD33+ cells, and 168 had PB-derived CD3+ cells, but only 10 had PB-derived CD19+ cells available to test (supplemental Table 2). The BMAs were not subjected to FACS, and chimerism testing was performed using extracted gDNA directly from the processed specimen. Samples taken at multiple time points after allo-HCT were available for ∼50% of patients (supplemental Figure 4), whereas the remaining patients had samples available from only 1 time point.
We compared our chimerism testing data with those of STR testing routinely performed at our institution for post–allo-HCT follow-up. The agreement between the 2 measurements was high, as shown from the correlation coefficients (undefined correlation coefficient for the CD19+ subset because all STR values were 0; Figure 2B).
Although the goal of an allo-HCT is to replace the hematopoietic cells of the recipient with those of the donor, a complete replacement is usually not achieved. Using our technique, recipient chimerism remained detectable in 97.8% of measurements in the BM (vs 29.1% observed via STR) and detectable recipient chimerism proportions ranged from 1.0e–6 to 1.4. Detectability in CD33+ cells was 56.8% (vs 9.6% using STR) and detectable proportions ranging from 3.9e–5 to 1.4. Detectability in CD3+ cells was 75.0% (vs 36.7% using STR), with detectable proportions ranging from 2.8e–4 to 1.0 (Figures 2B and 3A). Finally, detectability in CD19+ cells was 45.0% (vs 0.0% using STR), with detectable proportions ranging from 4.0e–4 to 0.27 (Figure 2B; supplemental Figure 5). The detectability in the sorted subsets was lower than in BMAs, unsurprisingly, given the lower cell number recovery from FACS. When chimerism was undetectable, the upper limit of the Wilson 95% CI had a mean of 0.0014, 0.0073, 0.011, and 0.11 for measurements in BMAs, CD33+ cells, CD3+ cells, and CD19+ cells, respectively, suggesting a high probability of true recipient chimerism proportions anywhere from 0 to these values, given the opportunity of assaying enough gDNA.
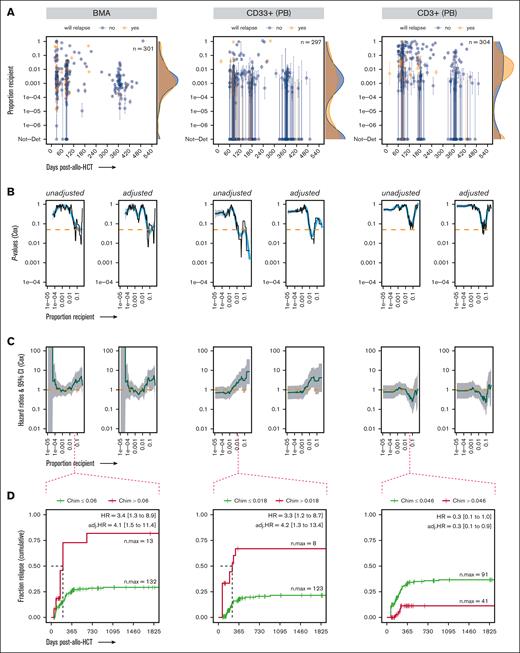
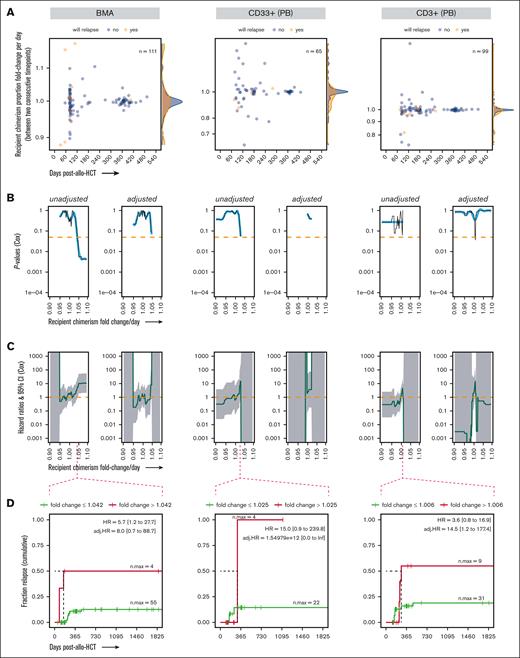
Evaluating recipient chimerism levels to identify a threshold above which patients with leukemia are at high risk of relapse after allo-HCT. (A) Highly sensitive chimerism measurements in BMA, PB-derived CD33+ myeloid cells, and PB-derived CD3+ T cells in the first 540 days after allo-HCT, excluding time points at which relapsed disease was already present and/or time points that self-fulfilled the relapse “prophecy” (see “Methods”). P values (B) and HRs (C) from the Cox proportional hazard regression (unadjusted and adjusted for confounding covariates), with chimerism as a time-dependent covariate, computed over a range of thresholds of chimerism proportions incrementally increasing between 1.0e–5 and 0.5. (D) Probability (cumulative incidence) of relapse with follow-up in a 5-year window (1825 days) after allo-HCT for a representative chimerism proportion threshold. Ticks represent individuals who were censored using the interval censoring approach. A generalized additive model was used to smooth fit the P values (blue line), which were truncated when HRs gave infinite CIs; the HRs gray ribbon represents the 95% CI; Chim, chimerism proportion (recipient); n.max, the maximum number of patients at risk; Not-Det, not detected; adj, adjusted.
Evaluating recipient chimerism levels to identify a threshold above which patients with leukemia are at high risk of relapse after allo-HCT. (A) Highly sensitive chimerism measurements in BMA, PB-derived CD33+ myeloid cells, and PB-derived CD3+ T cells in the first 540 days after allo-HCT, excluding time points at which relapsed disease was already present and/or time points that self-fulfilled the relapse “prophecy” (see “Methods”). P values (B) and HRs (C) from the Cox proportional hazard regression (unadjusted and adjusted for confounding covariates), with chimerism as a time-dependent covariate, computed over a range of thresholds of chimerism proportions incrementally increasing between 1.0e–5 and 0.5. (D) Probability (cumulative incidence) of relapse with follow-up in a 5-year window (1825 days) after allo-HCT for a representative chimerism proportion threshold. Ticks represent individuals who were censored using the interval censoring approach. A generalized additive model was used to smooth fit the P values (blue line), which were truncated when HRs gave infinite CIs; the HRs gray ribbon represents the 95% CI; Chim, chimerism proportion (recipient); n.max, the maximum number of patients at risk; Not-Det, not detected; adj, adjusted.
Risk of leukemia relapse as a posttransplant outcome of interest
Overall, the 5-year leukemia relapse rate was 32.5%, and 88.2% of relapse events occurred within the first 540 days after HCT (supplemental Figure 6). To test the prognostic value of ultrasensitive chimerism testing on leukemia relapse, we conducted univariate and multivariate analyses using Cox regression, with chimerism modeled as a time-dependent covariate. Although chimerism alone was included in the univariate model, covariates that were suspected to influence relapse outcomes were included in addition to chimerism in the multivariate model. Disease status (active vs in remission)40 and MRD41,42 at the time of transplant are known to be strongly associated with relapse outcomes after transplant and should be considered as covariates. Similarly, the source of donor cells can influence post–allo-HCT outcomes. In particular, umbilical cord blood transplant is known to be associated with lower relapse rates than those with other graft sources.43,44 In our cohort of patients, these 3 covariates were associated with relapse risk after allo-HCT (supplemental Figure 7). This was in addition to the leukemia diagnosis, in which patients with AML fared worse than those with ALL or MDS, and to the GVHD prophylaxis regimen, in which patients receiving cyclosporine and mycophenolate mofetil–based prophylaxis regimens had more favorable relapse outcomes (supplemental Figure 7). Those covariates were, thus, included in the adjusted Cox models in the subsequent analyses.
From 1037 ultrasensitive chimerism tests (361 in BMAs, 324 in CD33+ cells, 332 in CD3+ cells, and 20 in CD19+ cells), 115 (11.1%) measurements were excluded from the relapse risk analyses. The exclusions comprised measurements within less than a week of relapse (n = 44), after relapse (n = 62), and beyond 540 days after transplant (for patients without relapse; n = 9; see “Methods”).
Association of chimerism proportions with relapse risk
The association of chimerism with posttransplant relapse was assessed via 2 analysis approaches. Firstly, we attempted to identify a threshold for the recipient chimerism proportion, above which relapse risk became statistically significant. Cox regression HRs and their associated P values were computed over a range of chimerism thresholds incrementally increasing between 1.0e–5 and 0.5 for the BMAs, CD33+, and CD3+ specimens (outside this range, the number of events on either side of the threshold would become too skewed for the statistical model to work properly). The CD19+ subset was not analyzed because it only counted 10 patients and 1 relapse event. In BMAs, the first statistically significant association (unadjusted and adjusted models) was observed at a chimerism proportion threshold of 0.06 (Figure 3B-D). This association was similar in the CD33+ subset (myeloid lineage and relevant in AML and MDS), with statistical significance reached, at a threshold of 0.018 (Figure 3B-D). In T cells, recipient chimerism proportions were trending toward protection against relapse, because thresholds between 0.033 and 0.046 correlated with protection in the multivariate model (P = .04) before continuing to trend in the opposite direction toward neutral association (Figure 3B).
For these analyses, statistically significant thresholds all start at levels >0.01, well within the STR regimen of detection. We then questioned whether similar to ultrasensitive chimerism, an STR-based chimerism threshold could be found in association with relapse risk. No such threshold was found in BMAs, and only within a limited threshold range (between 0.01-0.02) in CD33+ (unadjusted model only; supplemental Figure 8). This hints at the superiority of our technology, even in regimens within the LOD of alternative chimerism techniques.
Overall, there was an increased relapse risk as ultrasensitive chimerism threshold increased, noticeable in whole BMAs and in the CD33+ subset.
Association of chimerism dynamics with relapse risk
The second approach was to identify a threshold for the rate of change of the recipient chimerism affecting the risk of leukemia relapse. The idea was to test whether using a technique with high quantitative precision could meaningfully track chimerism changes over time after transplant. For patients with detectable chimerism in ≥2 time points, recipient chimerism proportion fold change per day was calculated for each 2 consecutive time points (Figure 4A; supplemental Figure 9). Cox regression HRs and their associated P values were computed over a range of thresholds incrementally increasing between 0.9-fold per day (ie, 10% daily decrease) and 1.1-fold per day (ie, 10% daily increase) for BMA, CD33+, and CD3+ specimens. Relapse risk was significantly increased when the rate of chimerism change was >1.042-fold per day in the univariate model in BMA samples (Figure 4B-D). In the CD33+ subsets, relapse risk trended toward increase with an increasing rate of chimerism change, with the effect peaking at 1.025-fold change per day (Figure 4D). No clear trends were observed for the CD3+ subsets. These analyses were valid in the unadjusted models but not in the adjusted models, in which Cox regressions were difficult to compute, giving rise to HRs with infinite intervals on most of the analysis range (Figure 4C).
Evaluating recipient chimerism dynamics to identify a threshold of chimerism rate of change affecting the risk of leukemia relapse after allo-HCT. (A) Day-to-day fold change of chimerism proportions, calculated between 2 consecutive highly sensitive chimerism measurements after transplant in BMAs, PB-derived CD33+ myeloid cells, and PB-derived CD3+ T cells in the first 540 days after allo-HCT, excluding time points at which relapsed disease was already present and/or time points that self-fulfilled the relapse prophecy (see “Methods”). P values (B) and HRs (C) from the Cox proportional hazard regression (unadjusted and adjusted for confounding covariates) with chimerism fold change per day as a time-dependent covariate, computed over a range of thresholds incrementally increasing between 0.9-fold per day and 1.1-fold per day. (D) Probability (cumulative incidence) of relapse with follow-up in a 5-year window (1825 days) after allo-HCT for a representative chimerism fold change. Ticks represent individuals who were censored using the interval censoring approach. A generalized additive model was used to smooth fit the P values (blue line), which were truncated when HRs gave infinite CIs; the HR gray ribbon represents the 95% CI; n.max, the maximum number of patients at risk; adj, adjusted.
Evaluating recipient chimerism dynamics to identify a threshold of chimerism rate of change affecting the risk of leukemia relapse after allo-HCT. (A) Day-to-day fold change of chimerism proportions, calculated between 2 consecutive highly sensitive chimerism measurements after transplant in BMAs, PB-derived CD33+ myeloid cells, and PB-derived CD3+ T cells in the first 540 days after allo-HCT, excluding time points at which relapsed disease was already present and/or time points that self-fulfilled the relapse prophecy (see “Methods”). P values (B) and HRs (C) from the Cox proportional hazard regression (unadjusted and adjusted for confounding covariates) with chimerism fold change per day as a time-dependent covariate, computed over a range of thresholds incrementally increasing between 0.9-fold per day and 1.1-fold per day. (D) Probability (cumulative incidence) of relapse with follow-up in a 5-year window (1825 days) after allo-HCT for a representative chimerism fold change. Ticks represent individuals who were censored using the interval censoring approach. A generalized additive model was used to smooth fit the P values (blue line), which were truncated when HRs gave infinite CIs; the HR gray ribbon represents the 95% CI; n.max, the maximum number of patients at risk; adj, adjusted.
Overall, certain rates of recipient chimerism proportions that increased in the BM over time could be identified in association with an increased risk of leukemia relapse after allo-HCT (unclear after accounting for confounders).
Posttransplant MRD and the risk of relapse
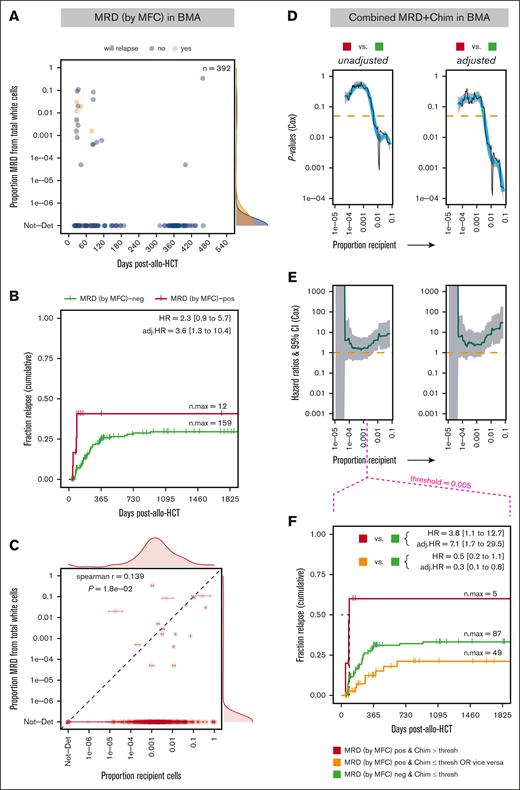
The association of posttransplant MRD with posttransplant relapse was assessed via MFC as an orthogonal approach to our technique. Because MFC is performed on BMA samples, MFC measurements at or within 2 days of chimerism measurements corresponding to each available BMA time point were used in the analyses. Of the 298 patients with leukemia originally included in the study, 292 had ≥1 BMA samples left over. Of these patients, 230 had their MRD measured at or within a maximum of 2 days of a chimerism measurement (STR and/or ultrasensitive). After removing MRD data points from beyond day 540 after transplant and those of patients with relapsed disease, within <1 week of relapse or at time points after relapse, 213 patients (with 392 data points) remained included. As for chimerism, posttransplant MRD via MFC was modeled as a time-dependent covariate in the Cox regression, with any detectable level of residual disease considered MRD positive.25
After transplant, 93.4% of MFC measurements returned no identifiable MRD (Figure 5A). Although a trend toward increased relapse risk was observed with MRD positivity, the correlation was statistically significant only in the multivariate model, suggesting that ≥1 of the covariates may be acting as negative confounder(s), allowing the association between relapse and MRD to emerge when tested independently of those confounder(s) (Figure 5B). In a 1-on-1 comparison, recipient cell proportions, via ultrasensitive chimerism, correlated significantly with MRD levels (P = .018), but the effect was modest (r = 0.139; Figure 5C). We then questioned whether analyzing posttransplant MRD in combination with recipient chimerism data could improve the ability of MRD via MFC to predict posttransplant relapse. Of the 213 patients in the MRD analysis, 179 also had ultrasensitive chimerism data (286 data points). We classified the patients as follows: MRD positive (any level) and above a certain chimerism threshold in the BM; MRD negative and below a certain chimerism threshold; and either MRD positive with decreased chimerism or MRD negative with increased chimerism. Starting at thresholds of recipient chimerism proportions in BMAs as low as 0.005, MRD positivity after transplant correlated significantly with relapse vs MRD negativity and chimerism below the threshold (in both univariate and multivariate models; Figure 5D-F). This was not the case if only MRD was positive or only chimerism was above the threshold (Figure 5F). MRD via MFC dynamics after transplant (analogous to the chimerism dynamics analysis) did not show statistically significant correlation with relapse in time-dependent Cox models, although becoming or remaining positive for MRD trended toward increased relapse (supplemental Figure 10).
MRD measured via MFC in BMAs is marginally associated with relapse risk after allo-HCT and augmented when adding recipient chimerism data. (A) The proportion of MRD cells identified via MFC in BMA-derived total white cells within the first 540 days after allo-HCT, excluding time points at which relapsed disease was already present and/or time points that self-fulfilled the relapse prophecy (see “Methods”). (B) Probability (cumulative incidence) of relapse with follow-up in a 5-year window (1825 days) post-allo-HCT for MRD status (via MFC). (C) Comparison of MRD levels (via MFC) vs ultrasensitive recipient chimerism measurements in BMAs. P values (D) and HRs (E) from the Cox proportional hazard regression (unadjusted and adjusted for confounding covariates) with the combination of MRD and chimerism levels as a time-dependent covariate, computed over a range of thresholds of chimerism proportions incrementally increasing between 1.0e–5 and 0.5. The comparison of the categories MRD positive and chimerism above threshold vs MRD negative and chimerism below threshold is represented. (F) Probability (cumulative incidence) of relapse with follow-up in a 5-year window (1825 days) after allo-HCT for combination of MRD and chimerism status. Ticks represent individuals who were censored using the interval censoring approach. A generalized additive model was used to smooth fit the P values (blue line), which were truncated when HRs gave infinite CIs; the HR gray ribbon represents the 95% CI; thresh, threshold; Chim, chimerism proportion (recipient); n.max, the maximum number of patients at risk; Not-Det, not detected; adj, adjusted; pos, positive; neg, negative.
MRD measured via MFC in BMAs is marginally associated with relapse risk after allo-HCT and augmented when adding recipient chimerism data. (A) The proportion of MRD cells identified via MFC in BMA-derived total white cells within the first 540 days after allo-HCT, excluding time points at which relapsed disease was already present and/or time points that self-fulfilled the relapse prophecy (see “Methods”). (B) Probability (cumulative incidence) of relapse with follow-up in a 5-year window (1825 days) post-allo-HCT for MRD status (via MFC). (C) Comparison of MRD levels (via MFC) vs ultrasensitive recipient chimerism measurements in BMAs. P values (D) and HRs (E) from the Cox proportional hazard regression (unadjusted and adjusted for confounding covariates) with the combination of MRD and chimerism levels as a time-dependent covariate, computed over a range of thresholds of chimerism proportions incrementally increasing between 1.0e–5 and 0.5. The comparison of the categories MRD positive and chimerism above threshold vs MRD negative and chimerism below threshold is represented. (F) Probability (cumulative incidence) of relapse with follow-up in a 5-year window (1825 days) after allo-HCT for combination of MRD and chimerism status. Ticks represent individuals who were censored using the interval censoring approach. A generalized additive model was used to smooth fit the P values (blue line), which were truncated when HRs gave infinite CIs; the HR gray ribbon represents the 95% CI; thresh, threshold; Chim, chimerism proportion (recipient); n.max, the maximum number of patients at risk; Not-Det, not detected; adj, adjusted; pos, positive; neg, negative.
Overall, MRD measured via MFC after allo-HCT appeared as a weak predictor of relapse after transplant. However, complementing MRD data with ultrasensitive chimerism significantly augmented the ability of the MRD assay to predict relapse.
Discussion
We designed and developed a panel of highly sensitive and specific qPCR-based assays to quantify recipient cells, distinguishing them from donor-derived cells, in blood and BM specimens of patients who underwent allo-HCT. Testing these assays in a cohort of patients with leukemia who underwent a transplant showed that recipient chimerism was detected in almost all BMA samples after transplant, compared with in less than one-third of the patients when using the current standard-of-care STR-based technique. Detectability was lower in FAC-sorted cellular subsets, ranging between 45% and 75% but still higher than that with the STR-based technique (between 0% and 37%). Our assay showed an association of risk of relapse after transplant with recipient chimerism levels in the BM and in purified cells expressing CD33, which are frequently displayed on AML and MDS blasts.45,46 The association of relapse risk with MRD after transplant measured via MFC was weak, statistically significant only in the multivariate model. Remarkably, detectable MRD combined with increased proportions of recipient chimerism in BMA became strongly associated with relapse, demonstrating the ability of our ultrasensitive chimerism assay to augment the MRD assay and bringing forth a previously underappreciated usefulness of clinical chimerism. Additionally, chimerism dynamics in BMA, that is, the rate at which recipient chimerism increased, correlated with relapse risk.
From a technical aspect, our panel has high analytic specificity, detecting only target gDNA, and high analytic sensitivity, detecting a single target copy in a background of up to a million copies (when enough gEq was available). The technique was designed to be scalable (possibility to use more replicate wells), with a LOD depending only on the amount of total genomic material available. The panel was extended and optimized, with the future goal of creating an ultrasensitive chimerism test for clinical use. The output measurements were in high agreement with STR-based measurements when chimerism proportions were ≥0.01, the limit below which only our technology could detect chimerism. In a separate study, our panel was benchmarked against a next-generation sequencing (NGS)-based chimerism technique, with an LOD comparable with ours, and showed significant agreement with the results.32
Our study had a number of limitations. It was a retrospective study that used leftover samples from patients with leukemia who underwent transplants with known outcomes, collected at specific time points per clinical protocols, over which we had no control. Moreover, leftover genomic material at all time points were not always available (∼50% of patients had samples available from only 1 time point). The cohort was heterogeneous, with 3 diagnostic groups, including AML and MDS, both with their own extra levels of heterogeneity. Molecular features were not routinely collected (especially for early patients), and our data set missed variables such as leukemia-driving mutations/gene fusions. Another limitation was that not all the relevant cellular subsets were available for testing in acute leukemia. Prognostic value of chimerism testing on clinical outcome has been shown for CD34+ cells in ALL, AML, and MDS,47-50 and for CD19+51 and CD8+52 cells in ALL. The number of CD19+ samples were insufficient for risk association analyses, and even fewer CD34+ samples were available (not included in this study). A technical limitation was the quantity of available genomic material per sample. Although we developed the technique to deliver highly accurate measurements, the resolution remained constrained by the availability of gDNA per test. FAC-sorted subsets yielded cell counts ≤30 000, which was lower than those of unsorted BMAs. This resulted in more frequent 0 values in recipient chimerism proportions, which could have affected our ability to detect associations with risk of relapse, specifically in the chimerism dynamics analysis approach.
Measuring disease burden after therapy is at the core of modern leukemia management. Standardized and emerging technologies are used to make this assessment, including gene rearrangement/mutation-specific qPCR and NGS,21,53,54 or MFC-based55 approaches, each with their respective merits and limitations. Those limitations can be technical (qPCR and NGS techniques not universally applicable, MFC subjective and requiring high-level expertise, etc) but also more fundamental, addressing the complexity of disease biology, because no single feature or biomarker accounts for all relapse mechanisms. One technology may separate a group of patients with significantly higher relapse risk than those at baseline, whereas another technology may identify another group. This is best exemplified in a recent study in which the usage of molecular MRD (via NGS) together with the established MFC had independent and additive prognostic value for predicting risk of leukemia relapse.53 Using our combined posttransplant MFC and chimerism threshold model, we were able to identify a small group of patients with significantly high relapse risk (5-year relapse rate of 60.0% with MRD positivity and chimerism above threshold vs 29.1% otherwise; Figure 5F; supplemental Figure 11A). The diseases of these patients tended to relapse rapidly (all events at, or before, day 90 after allo-HCT). Conceivably, patients with similar features could benefit from rapid therapy adjustments, such as introducing donor lymphocyte infusions or hypomethylating agents such as azacytidine that has shown promise in preventing or delaying overt relapse after allo-HCT.56,57 In patients who are already at high risk of relapse (pretransplant MRD positive or active disease at transplant), we observed trends toward our technique performing better in predicting relapse when pretransplant MRD was known but not necessarily so when disease status at transplant was known (supplemental Figure 11). However, we could not conclusively interpret these trends because the numbers in the risk strata were small. This, together with the other data presented herein, provide the impetus to pursue larger studies and clinical trials, which will determine the extent to which our highly sensitive technique, alone or in combination with emerging MRD technologies, could improve patient risk stratification and relapse prediction.
Acknowledgments
The authors are grateful to Emily Stevens, Oyku Sensoy, and Elena Kahn for initial analytic assistance and helpful discussions and to Nathalie C. Lambert, in France, for generously providing the HLA-A∗30 assay to test in our panel. The authors thank the entire team at the Fred Hutchinson Cancer Center’s Clinical Immunogenetics Laboratory for help in locating and retrieving archived patient samples, Ted Gooley and his clinical biostatistics team at the Fred Hutchinson Cancer Center for statistical guidance, and Louise Sudour for her artistic contributions to the figures. The authors extend their gratitude to the patients who consented to participate in the study.
This research was supported by the National Institutes of Health through the SBIR/STTR programs, by grant R41CA232805 received by Chimerocyte Inc.
Authorship
Contribution: S.B.K. conceived the study, designed the experiments, performed data analyses, and wrote the paper; F.U. performed the experiments and contributed to data analyses; and J.P.R. and J.L.N. provided resources for the study including patient data and laboratory space/equipment, and contributed helpful guidance and critical review.
Conflict-of-interest disclosure: S.B.K. and J.L.N. are cofounders of Chimerocyte Inc, which develops highly sensitive chimerism analysis technologies. The remaining authors declare no competing financial interests.
Correspondence: Sami B. Kanaan, Translational Science and Therapeutics Division, Fred Hutchinson Cancer Center, 1100 Fairview Ave N, Seattle, WA 98109; e-mail: skanaan@fredhutch.org.
References
Author notes
Molecular assay oligonucleotide sequences for highly sensitive chimerism, following the establishment of data/material transfer agreements, are available upon reasonable request from corresponding author, Sami B. Kanaan (skanaan@fredhutch.org). Additional user guidelines, templates for planning, running, and analyzing experiments as well as the computational code used in this study can be found on GitHub at https://github.com/sbkanaan/UltraSensChim.
The full-text version of this article contains a data supplement.