Key Points
A considerable subset of classic Hodgkin lymphoma recurrences represents clonally unrelated second primary lymphoma.
Assessment of T-cell clonality combined with pathogenic mutation analysis can identify TCL mimicking cHL.
Abstract
Despite high cure rates in classic Hodgkin lymphoma (cHL), relapses are observed. Whether relapsed cHL represents second primary lymphoma or an underlying T-cell lymphoma (TCL) mimicking cHL is underinvestigated. To analyze the nature of cHL recurrences, in-depth clonality testing of immunoglobulin (Ig) and T-cell receptor (TCR) rearrangements was performed in paired cHL diagnoses and recurrences among 60 patients, supported by targeted mutation analysis of lymphoma-associated genes. Clonal Ig rearrangements were detected by next-generation sequencing (NGS) in 69 of 120 (58%) diagnoses and recurrence samples. The clonal relationship could be established in 34 cases, identifying clonally related relapsed cHL in 24 of 34 patients (71%). Clonally unrelated cHL was observed in 10 of 34 patients (29%) as determined by IG-NGS clonality assessment and confirmed by the identification of predominantly mutually exclusive gene mutations in the paired cHL samples. In recurrences of >2 years, ∼60% of patients with cHL for whom the clonal relationship could be established showed a second primary cHL. Clonal TCR gene rearrangements were identified in 14 of 125 samples (11%), and TCL-associated gene mutations were detected in 7 of 14 samples. Retrospective pathology review with integration of the molecular findings were consistent with an underlying TCL in 5 patients aged >50 years. This study shows that cHL recurrences, especially after 2 years, sometimes represent a new primary cHL or TCL mimicking cHL, as uncovered by NGS-based Ig/TCR clonality testing and gene mutation analysis. Given the significant therapeutic consequences, molecular testing of a presumed relapse in cHL is crucial for subsequent appropriate treatment strategies adapted to the specific lymphoma presentation.
Introduction
Hodgkin lymphoma (HL) affects both young adults and older patients and has an age-adjusted incidence of ∼3 in 100 000 people in the Western world.1 HL can be divided into 2 distinct entities, nodular lymphocyte–predominant HL and classic HL (cHL), of which cHL is the most common form, representing a spectrum of 4 different morphological subtypes.2 Current multiagent chemotherapy with or without radiotherapy results in high cure rates of patients with cHL, with a 5-year overall survival exceeding 90%.3-5 However, up to 25% of patients with cHL show refractory or relapsed disease, often involving patients with advanced-stage disease.6-8 In most patients, relapsed disease develops within 2 years after the primary diagnosis, but recurrences may even occur after 5 years.9-11
The lymphoid neoplasm cHL originates from transformed germinal center B cells. The malignant CD30+ Hodgkin and Reed-Sternberg (HRS) cells have lost most phenotypic B-cell characteristics, including the expression of the B-cell receptor, but clonal immunoglobulin (IG) gene rearrangements are still detectable.12 This molecular fingerprint has confirmed the clonal outgrowth of malignant B cells and can serve as a diagnostic marker to establish the clonal relationship between primary cHL and its recurrences.12,13 Because of the limited number of HRS cells (usually <1%-5%) in an inflammatory background, Ig clonality assessment in whole tissue specimens has been challenging in a diagnostic setting. The recently developed EuroClonality IG–next-generation sequencing (NGS) assay,14,15 which has shown high sensitivity, allows for improved detection of malignant HRS clones in cHL tissues.16 Besides detection of Ig clonality, clonal T-cell receptor (TR) gene rearrangements have been observed in cHL,17,18 even in isolated CD30+ HRS cells,19-23 suggesting a T-cell origin in very rare cases of cHL. More common are T-cell lymphomas (TCLs), with atypical B cells mimicking cHL, such as nodal T-follicular helper cell lymphomas (nTFHLs), including both the angioimmunoblastic24-26 and the follicular types.25,27,28
In cases in which it remains challenging to distinguish cHL from TCL with HRS-like cells, gene mutation analysis on tissue samples can be helpful to complement clinicopathological evaluation. Recurrently mutated genes driving cHL pathogenesis include SOCS1, TNFAIP3, and B2M,29-31 whereas genetic alterations associated with the angioimmunoblastic type of nTFHL, here referred to as angioimmunoblastic TCL (AITL), commonly involve RHOA, TET2, DNMT3A, and IDH2.32,33 Notably, some of these affected genes, such as TET2 and DNMT3A mutations, are drivers of clonal hematopoiesis of indeterminate potential (CHIP), which predisposes to hematologic malignancies,34,35 and can also be detected in HRS cells themselves.36
It is commonly assumed that recurrence of cHL after treatment is a relapse of the original disease. However, we hypothesize, in line with a previous study,37 that some patients may actually present with a new primary cHL, unrelated to the original tumor. Therefore, we analyzed paired diagnoses and recurrence tissue samples in a cohort of 60 patients with cHL to determine the clonal relationship between these cHL presentations by performing NGS-based Ig and TCR clonality assessment and targeted mutation analysis. Our in-depth molecular analysis demonstrated that recurrences of cHL represent clonally unrelated second primary cHL in a considerable proportion of cases. Moreover, several cHL and its recurrences appeared upon retrospective pathology review TCL with HRS-like cells mimicking cHL.
Methods
Patient cohort and tissue samples
The study cohort consisted of 60 patients whose tissue samples were available from both cHL primary diagnosis and secondary disease. Archival material from 1995 to 2019 was obtained from pathology departments within the Netherlands (Radboud University Medical Center, Canisius Wilhelmina Hospital, Rijnstate Hospital, Jeroen Bosch Hospital, and University Medical Center Groningen), Germany (University Hospital Tübingen), Poland (Medical University of Lodz), and Belgium (University Hospital Brussels). Each tissue sample was reviewed by 2 experienced hematopathologists (H.v.K. and M.v.d.B.) according to the 2017 World Health Organization classification.2 Clinical and pathological information is summarized in supplemental Table 1. Tissue biopsy specimen from recurrences in patients with an interval of ≤1 year after primary diagnosis and without confirmed complete response (CR; n = 4; cases 20, 30, 31, and 52) were still considered as relapses in our study descriptions. An age-matched cHL control cohort of patients without a relapse (median follow-up of 16 years) was included to establish whether TCR clonality was associated with relapsed cHL (supplemental Table 2). All samples and clinical information were collected in accordance with the Declaration of Helsinki and Declaration of Taipei and received approval of the local medical ethical review board (approval number #2020-6390).
NGS-based clonality assessment
NGS-based clonality assays were performed as previously described by the EuroClonality-NGS Working Group14,38,39 to detect Ig heavy chain (IGH) and Ig kappa light chain (IGK) gene rearrangements or TRB and TRG gene rearrangements. The 5 standard targets of the EuroClonality Ig-NGS assay (framework 3 [FR3] IGHV-IGHD-IGHJ, IGHD-IGHJ, IGKV-IGKJ, and IGKV/intron recombination signal sequence [RSS]- kappa deleting element [KDE]) were analyzed for all samples. Samples without detectable IG gene rearrangements for these 5 targets and sufficient DNA quality were subsequently tested for FR2 IGHV-IGHD-IGHJ gene rearrangements (supplemental Materials and Methods; supplemental Tables 3 and 4). IG-NGS clonality assessment was performed in duplicates with 40- to 80ng DNA input. TR-NGS clonality assay involved detection of TRBV-TRBD-TRBJ, TRBD-TRBJ, and TRGV-TRGJ gene rearrangements and was performed with 40ng DNA input for each polymerase chain reaction. Library preparations were made compatible for sequencing either on an IonTorrent or Illumina platform. NGS data sets (Bioproject accession number PRJNA1003679) were analyzed using the bioinformatics analysis tool ARResT/Interrogate (http://arrest.tools/interrogate).40 Guidelines for Ig clonality assessment in cHL have been described previously,16 and for TCR clonality detection, it is provided in the supplemental Materials and Methods.
Mutation analysis
Mutation analysis of clonally unrelated cHL was performed with the Trusight Oncology 500 (TSO500; Illumina, San Diego, CA) NGS panel (supplemental Table 5A) on total genomic DNA and sequenced on an Illumina NextSeq500 platform, as described previously.41 Lymphomas with clonal TR gene rearrangements were analyzed with AmpliSeq Custom panels (AITL32 and BLYMF20042; supplemental Table 5B-C, respectively) and sequenced on an IonTorrent platform. Further details are described in the supplemental Information.
Results
Clinicopathological characteristics of paired diagnosis and recurrence in the cHL patient cohort
The study cohort consisted of archived paired diagnosis and secondary cHL tissue samples from 60 patients diagnosed with cHL based on clinical and histomorphological criteria. A total of 130 tissue samples were included, which involved 99 formalin-fixed, paraffin-embedded (FFPE) and 31 fresh frozen (FF) tissue specimens. Within this cohort, the median age at the time of primary diagnosis was 26 years (range, 4-76 years), and 18 patients were diagnosed with cHL in childhood (age, ≤18 years; Table 1). At primary cHL diagnosis, 55% of the patients presented with advanced-stage disease (Ann Arbor stage III-IV), which is higher than that of the general cHL population (30%).43 The majority of primary cHL tissue samples showed nodular sclerosis (NSHL) morphology (n = 34, 57%), followed by mixed cellularity (MCHL; n = 22, 37%). Epstein-Barr virus (EBV)–positive HRS cells were detected in 13 of 60 (22%) primary cHL diagnosis samples and in 17 of 68 (25%) of the recurrences (supplemental Table 6). Considering all diagnosis and recurrence samples with known EBV status (n = 114), EBV positivity was more frequently present in MCHL (39%) than in NSHL (16%; P = .007). The median time to the first recurrence was 1.5 years (range, 0.4-13.6 years), and 25 patients (42%) developed a first recurrence >2 years after primary diagnosis. From 12 patients, 2 independent recurrences were included for molecular analysis.
Clinicopathological data of paired diagnosis and recurrence cHL cohort at the time of primary diagnosis
| Total cohort (N = 60) . | n (%) . |
|---|---|
| Sex (N = 60) | |
| Male | 37 (62) |
| Female | 23 (38) |
| Median age (N = 60) | 26 y (4-76 y) |
| Pediatric (≤18 y) | 18 (30) |
| Adults (>18 y) | 42 (70) |
| Disease stage Ann Arbor (n = 52) | |
| I | 6 (12) |
| II | 19 (37) |
| III | 17 (33) |
| IV | 10 (19) |
| Risk groups (n = 40) | |
| Stages I to II (n = 24) | |
| EORTC favorable | 9 (23) |
| EORTC unfavorable | 15 (38) |
| Stages III to IV (n = 16) | |
| IPS 0 to 2 | 8 (20) |
| IPS ≥3 | 8 (20) |
| First line treatment (n = 52)∗ | |
| Chemotherapy | 49 (94) |
| Radiotherapy | 20 (38) |
| Immunotherapy | 1 (2) |
| Time to first recurrence (N=60) | |
| Median interval (range) (y) | 1.5 (0.4-13.6) |
| cHL subtype (N = 60) | |
| NSHL | 34 (57) |
| MCHL | 22 (37) |
| LRHL | 1 (2) |
| cHL-NOS | 3 (5) |
| EBV status (N = 60) | |
| Negative | 47 (78) |
| Positive | 13 (22) |
| Total cohort (N = 60) . | n (%) . |
|---|---|
| Sex (N = 60) | |
| Male | 37 (62) |
| Female | 23 (38) |
| Median age (N = 60) | 26 y (4-76 y) |
| Pediatric (≤18 y) | 18 (30) |
| Adults (>18 y) | 42 (70) |
| Disease stage Ann Arbor (n = 52) | |
| I | 6 (12) |
| II | 19 (37) |
| III | 17 (33) |
| IV | 10 (19) |
| Risk groups (n = 40) | |
| Stages I to II (n = 24) | |
| EORTC favorable | 9 (23) |
| EORTC unfavorable | 15 (38) |
| Stages III to IV (n = 16) | |
| IPS 0 to 2 | 8 (20) |
| IPS ≥3 | 8 (20) |
| First line treatment (n = 52)∗ | |
| Chemotherapy | 49 (94) |
| Radiotherapy | 20 (38) |
| Immunotherapy | 1 (2) |
| Time to first recurrence (N=60) | |
| Median interval (range) (y) | 1.5 (0.4-13.6) |
| cHL subtype (N = 60) | |
| NSHL | 34 (57) |
| MCHL | 22 (37) |
| LRHL | 1 (2) |
| cHL-NOS | 3 (5) |
| EBV status (N = 60) | |
| Negative | 47 (78) |
| Positive | 13 (22) |
EORTC, European Organization of Research and Treatment of Cancer; IPS, international prognostic score; LRHL, lymphocyte rich Hodgkin lymphoma; NOS, not otherwise specified.
If patients received combination therapy, multiple treatment categories were applicable, making the sum of all categories >100%. For detailed information, see supplemental Table 1.
Identification of clonally related relapsed cHL by NGS-based detection of IG gene rearrangements
Clonality analysis was performed with the EuroClonality IG-NGS assay, and in 120 of 130 tissue samples, good quality NGS clonality data sets were obtained with interpretable results. Clonal IG gene rearrangements were detected in 69 of 120 samples (58%; 46 of 89 FFPE = 52%; 23 of 31 FF = 74%), whereas 51 samples (43%; 43 of 89 FFPE = 48%; 8 of 31 FF = 26%) displayed a polyclonal pattern (supplemental Table 7). In the remaining 10 samples (7 primary diagnoses and 3 recurrences), IG-NGS generated poor-quality data sets that were noninterpretable, mainly because of inferior genomic DNA quality. Therefore, cHL tissue samples of 7 of the 60 patients had to be excluded from subsequent analysis. In the other 53 patients, Ig clonotype comparison between primary diagnosis and recurrences revealed identical Ig-related clonotypes in 16 patients, confirming their clonal relationship and thus an actual relapse (supplemental Table 7). In 15 of the remaining 37 patients, a dominant Ig clonotype could be detected in only 1 of the cHL tissue samples, whereas the paired sample(s) lacked detection of a clonal Ig gene rearrangement. However, backtracking of the specific cHL-associated clonotypes in the 15 reciprocal samples resulted in the identification of identical clones below the defined clonality thresholds in 5 additional cases (see supplemental Information for clonality threshold definitions). Therefore, these 5 samples were also classified as clonally related cHL cases (supplemental Figure 1). In addition, NGS-based detection of FR2 IGHV-IGHD-IGHJ gene rearrangements was performed for a selected set of unresolved cases with sufficient DNA quality (n = 14 cases; n = 29 samples). This revealed detectable clonal IG gene rearrangements in 10 of 29 samples (supplemental Table 7), thereby identifying 3 additional clonally related cases. Collectively, NGS-based clonality assessment by detection of IG gene rearrangements demonstrated clonally related relapsed cHL in 24 patients (Figure 1).
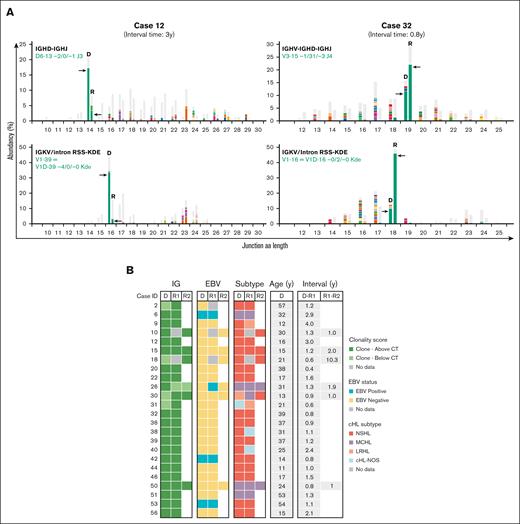
NGS-based clonality analysis identifies clonally related cHL recurrences. (A) Representative data sets indicating NGS-based detection of IG gene rearrangements in paired diagnosis and recurrence tissue samples of 2 patients with clonally related relapsed cHL (cases 12 and 32). For each patient, the data for 2 clonal IG gene rearrangements are shown; IGKV-IGKJ and IGKV/intron RSS-KDE for case 12 and IGHV-IGHD-IGHJ and IGKV/intron RSS-KDE for case 32. The specific clonotype for the dominant IG gene rearrangement is indicated in green. On the x-axis, the junction length in amino acids (junction aa length) is shown, and the abundancy of clonotypes is shown in percentages on the y-axis. (B) Summary of clonality assessment detecting relapsed cHL in 24 patients, together with EBV status, cHL subtype, and time interval between the paired cHL samples. CT, clonality threshold; D, primary diagnosis sample; NOS, not otherwise specified; R, recurrence sample; y, years.
NGS-based clonality analysis identifies clonally related cHL recurrences. (A) Representative data sets indicating NGS-based detection of IG gene rearrangements in paired diagnosis and recurrence tissue samples of 2 patients with clonally related relapsed cHL (cases 12 and 32). For each patient, the data for 2 clonal IG gene rearrangements are shown; IGKV-IGKJ and IGKV/intron RSS-KDE for case 12 and IGHV-IGHD-IGHJ and IGKV/intron RSS-KDE for case 32. The specific clonotype for the dominant IG gene rearrangement is indicated in green. On the x-axis, the junction length in amino acids (junction aa length) is shown, and the abundancy of clonotypes is shown in percentages on the y-axis. (B) Summary of clonality assessment detecting relapsed cHL in 24 patients, together with EBV status, cHL subtype, and time interval between the paired cHL samples. CT, clonality threshold; D, primary diagnosis sample; NOS, not otherwise specified; R, recurrence sample; y, years.
The majority of patients with clonally related cHL developed a relapse within 2 years after their primary diagnosis (19 of 24 patients [79%]). Confirmation of clonal relationship was based on identical clonotypes for at least 2 distinct IG rearrangement targets in 13 of 24 patients (54%), whereas for the remaining patients, this involved a single IG target (supplemental Table 7). In 9 of 24 patients with relapsed cHL, the clonal relationship was based on identical FR2/FR3 IGHV-IGHD-IGHJ gene rearrangements. Their complementarity-determining region 3 (CDR3) displayed nucleotide sequence differences with the germ line sequence, indicative of somatic hypermutation (SHM). However, comparison between primary diagnosis and relapsed disease revealed identical sequences without any additional nucleotide substitutions, even after ∼11 years (supplemental Table 8). This demonstrates a lack of ongoing SHM between primary diagnosis and relapsed cHL. Furthermore, 4 patients (cases 6, 9, 40, and 56) showed additional unique clonal IG gene rearrangement(s) besides the shared clonotype(s) in either diagnosis or recurrence (supplemental Table 7). These additional clonotypes, which were present at multiple IG targets for 3 patients, displayed a distinct dominance compared with the shared clonotype(s), indicative for the presence of a second B-cell clone within these tissue samples.
IG-NGS clonality analysis demonstrates clonally unrelated lymphomas in cHL recurrences
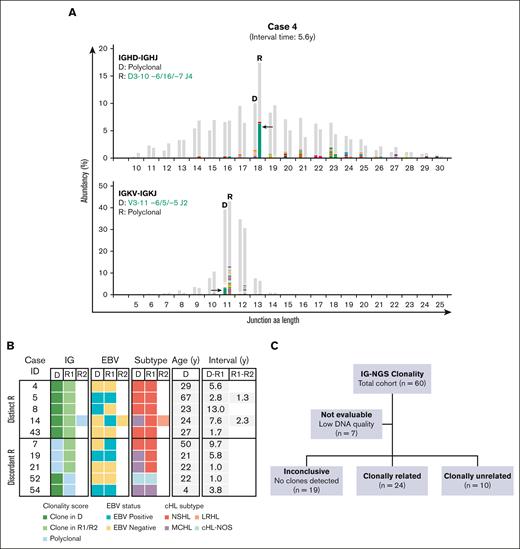
Besides the 24 clonally related cHL recurrences, 5 of 53 patients (cases 4, 5, 8, 14, and 43) showed distinct clonal IG gene rearrangements (termed as distinct rearrangements) in the paired samples, indicating the presence of different B-cell clones and clonally unrelated cHL (Figure 2; supplemental Table 9A). In another 5 patients (cases 7, 19, 21, 52, and 54), specific clonal IG gene rearrangements were detected in either diagnosis or recurrence, whereas these dominant clonotypes were nondetectable in the reciprocal samples (termed as discordant rearrangements; supplemental Table 9B). Genomic DNA quality, HRS cell percentage (as a fraction of total B cells), and the lack of ongoing SHM between diagnosis and recurrence were all parameters that could not explain the inability of detecting these clonotypes in the complete data set for IGHV-IGHD-IGHJ (range clonotypes, 683-3168), IGKV-IGKJ (range clonotypes, 720-1036), and IGKV/Intron-KDE (range clonotypes, 166-678) rearrangements of the reciprocal samples (supplemental Table 10; supplemental Figure 2). This indicated that the paired sample lacked the malignant B-cell clone detected in the reciprocal sample and, instead, harbored a clone that could not be identified with the standard IG-NGS assay, as was the case for the 37 cHL samples of the 19 patients that remained inconclusive for clonal comparison. Therefore, 10 cases were classified as clonally unrelated (Figure 2; supplemental Table 9B).
NGS-based clonality analysis identifies clonally unrelated second primary cHL recurrences. (A) Representative data sets indicating NGS-based detection of IG gene rearrangements in paired diagnosis (D) and recurrence (R) tissue samples of case 4. Data of 2 IG targets (IGHD-IGHJ and IGKV-IGKJ) are shown. The specific clonotypes for the dominant clonal IG gene rearrangement are indicated in green. On the x-axis, the junction aa length is shown, and the abundancy of clonotypes is shown in percentages on the y-axis. (B) Summary of clonality assessment detecting second primary cHL in 10 patients. Two groups of clonally unrelated cases were defined, based on the detection of distinct clonal IG gene rearrangement(s) (distinct R) or clonal IG gene rearrangement(s) in 1 sample absent in the paired sample (discordant R). EBV status, cHL subtype, and time interval between the paired cHL samples are indicated. (C) Schematic overview of the clonal relationship of paired diagnosis and recurrence samples of the complete diagnosis and recurrence cHL cohort, based on IG-NGS clonality assessment.
NGS-based clonality analysis identifies clonally unrelated second primary cHL recurrences. (A) Representative data sets indicating NGS-based detection of IG gene rearrangements in paired diagnosis (D) and recurrence (R) tissue samples of case 4. Data of 2 IG targets (IGHD-IGHJ and IGKV-IGKJ) are shown. The specific clonotypes for the dominant clonal IG gene rearrangement are indicated in green. On the x-axis, the junction aa length is shown, and the abundancy of clonotypes is shown in percentages on the y-axis. (B) Summary of clonality assessment detecting second primary cHL in 10 patients. Two groups of clonally unrelated cases were defined, based on the detection of distinct clonal IG gene rearrangement(s) (distinct R) or clonal IG gene rearrangement(s) in 1 sample absent in the paired sample (discordant R). EBV status, cHL subtype, and time interval between the paired cHL samples are indicated. (C) Schematic overview of the clonal relationship of paired diagnosis and recurrence samples of the complete diagnosis and recurrence cHL cohort, based on IG-NGS clonality assessment.
In the total cohort of clonally unrelated cHL, EBV positivity was present in 4 of 10 diagnosis samples (40%), including 2 MCHL (cases 19 and 54) and 2 NSHL subtypes (cases 5 and 8). This rate of EBV positivity was higher than cHL diagnosis in the clonally confirmed relapse cohort (n = 3 of 24, 13%; P = .071). Notably, the EBV status switched between primary diagnosis and second cHL in 3 of 10 clonally unrelated patients (cases 7, 8, and 14). The clonally unrelated cHL recurrences occurred in all age groups, and 7 of 10 patients (70%) showed a time to recurrence of >2 years (range, 2.8-13.0 years), compared with 5 of 24 patients (21%) with a confirmed relapse (range, 2.1-4.0 years; P = .006).
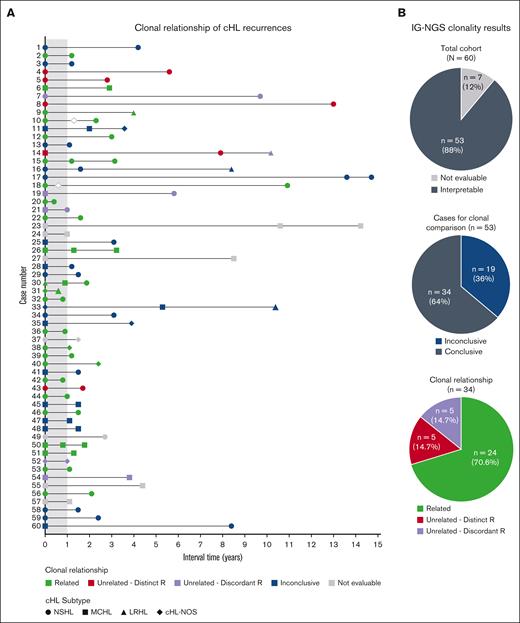
Altogether, the IG-NGS clonality data demonstrate that, in this study cohort, the clonal relationship could be established in 34 of 53 patients (64%) by NGS-based detection of IG gene rearrangements; of which 24 of 34 patients (71%) developed relapsed disease and 10 of 34 patients (29%) displayed a clonally unrelated de novo cHL. In the remaining 19 patients, the absence of detectable clonal IG gene rearrangements in whole tissue specimens of diagnosis and/or recurrence resulted in inconclusive results (Figure 3).
Summary clonal relationship of paired diagnosis and recurrence cHL samples. (A) Clonal comparison revealed the clonal relationship of paired diagnosis and recurrence samples by NGS-based detection of immunoglobulin rearrangements (IG-NGS). Time interval between the diagnoses and the cHL subtype of each biopsy is indicated. The open diamonds represent recurrences for which no tissue biopsy was available; therefore, molecular analysis could not be performed. (B) The top pie chart shows the proportion of cases with interpretable and nonevaluable data sets within the total cohort, the middle pie chart the proportion of cases with conclusive and inconclusive data sets, and the bottom pie chart the clonal relationship of all cases with conclusive results. Clonally unrelated cHL is subdivided in cases with distinct R or discordant R.
Summary clonal relationship of paired diagnosis and recurrence cHL samples. (A) Clonal comparison revealed the clonal relationship of paired diagnosis and recurrence samples by NGS-based detection of immunoglobulin rearrangements (IG-NGS). Time interval between the diagnoses and the cHL subtype of each biopsy is indicated. The open diamonds represent recurrences for which no tissue biopsy was available; therefore, molecular analysis could not be performed. (B) The top pie chart shows the proportion of cases with interpretable and nonevaluable data sets within the total cohort, the middle pie chart the proportion of cases with conclusive and inconclusive data sets, and the bottom pie chart the clonal relationship of all cases with conclusive results. Clonally unrelated cHL is subdivided in cases with distinct R or discordant R.
Mutation analysis confirms the occurrence of second primary cHL
As demonstrated by NGS-based Ig clonality assessment, 10 patients were suspected of a second primary cHL. To further substantiate these results, targeted mutation analysis on whole tissue specimen of the paired diagnosis and recurrence samples (n = 10) was performed with TSO500 assay (600 × average median exon coverage) (supplemental Table 11). This hybrid capture panel harbors different cancer-related genes, including 100 lymphoma-associated, but not cHL-specific, target genes (supplemental Table 5A). The detection of single nucleotide polymorphisms (SNPs) reminiscent of germ line variants confirmed that all sample pairs were from the same patient (average, 5 SNPs; range, 2-9 SNPs; data not shown).
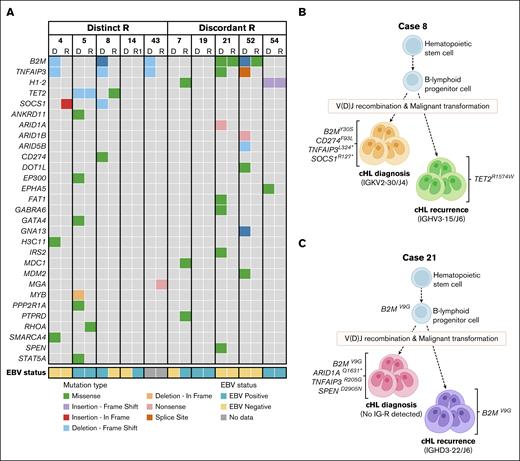
Next, (potential) pathogenic mutations (PA3-PA5) with a variant allele frequency (VAF) from 1% to 30%, were selected to identify candidate HRS-specific gene mutations and compared between paired diagnosis and recurrence sample. In 4 of 5 patients with distinct rearrangements in diagnosis and recurrence, mutually exclusive gene mutations were identified in both samples, except for 1 overlapping TET2 mutation in case 5 (p.Q731Tfs∗22; VAF diagnosis, 3%; VAF recurrence, 14%; Figure 4A; supplemental Table 11A). The mutually exclusive mutations represented known cHL target genes, including ARID1A, B2M, CD274, EP300, GNA13, SMARCA4, SOCS1, SPEN, STAT5A, and TNFAIP3 as well as other cancer-related genes (Figure 4A-B; supplemental Table 11A). Similarly, 4 of 5 patients with discordant rearrangements harbored mutually exclusive mutations in diagnosis and recurrence (eg, ARID1A, ARID1B, DOT1L, GNA13, SPEN, and TNFAIP3). Here, 3 patients showed single overlapping mutations between diagnosis and recurrence, which included 2 patients harboring a B2M mutation (case 21, p.V9G and case 52, p.M1R) with a VAF ranging from 2% to 7% (Figure 4C) and 1 patient (case 54) showing a H1-2 mutation (p.I80Afs∗11; VAF diagnosis and recurrence, 2%; supplemental Table 11B). No pathogenic mutations were detected with TSO500 assay in both diagnosis and recurrence for cases 14 and 19. Together, these targeted mutation analyses supported the identification of clonally unrelated cHL in this study cohort and the occurrence of second primary cHL.
Mutation analysis supports the identification of second primary lymphoma in cHL recurrences. Tissues of paired diagnosis and recurrences representing second primary cHL were subjected to targeted mutation analysis (TSO500 assay). HRS-associated gene mutations were selected based on a VAF of 1% to 30% and a minimal coverage of 350 reads. (A) Left panel shows data of clonally unrelated cHL samples based on distinct R, and right panel shows data of cases with discordant R. Paired data show mutually exclusive mutations and single overlapping mutations for 8 cases, whereas tissue samples of 2 cases (14 and 19) lacked pathogenic gene mutations with TSO500 assay. Mutation types and EBV status are shown. All shown mutations are scored as (potentially) pathogenic (PA3-PA5). For detailed information, see supplemental Table 9. (B-C) Schematic representations of the development of 2 independent lymphomas of case 8 (panel B) and case 21 (panel C). Gene mutations associated with cHL and clonal Ig rearrangement targets are indicated.
Mutation analysis supports the identification of second primary lymphoma in cHL recurrences. Tissues of paired diagnosis and recurrences representing second primary cHL were subjected to targeted mutation analysis (TSO500 assay). HRS-associated gene mutations were selected based on a VAF of 1% to 30% and a minimal coverage of 350 reads. (A) Left panel shows data of clonally unrelated cHL samples based on distinct R, and right panel shows data of cases with discordant R. Paired data show mutually exclusive mutations and single overlapping mutations for 8 cases, whereas tissue samples of 2 cases (14 and 19) lacked pathogenic gene mutations with TSO500 assay. Mutation types and EBV status are shown. All shown mutations are scored as (potentially) pathogenic (PA3-PA5). For detailed information, see supplemental Table 9. (B-C) Schematic representations of the development of 2 independent lymphomas of case 8 (panel B) and case 21 (panel C). Gene mutations associated with cHL and clonal Ig rearrangement targets are indicated.
The clinical data for most of these patients revealed no underlying disease explaining the occurrence of such second primary cHL. However, 1 patient (case 19) showed a complex clinical history, with chronic lymphocytic inflammation with pontine perivascular enhancement responsive to steroids (CLIPPERS) syndrome 6 years after the cHL recurrence and autoimmune hemolytic anemia. This suggested an underlying history of an immune dysregulating disorder that might have contributed to the development of 2 independent lymphomas. Another patient (case 54) with a positive family history of cHL developed cHL twice before the age of 9 years together with serological and molecular evidence for chronic active EBV infection, also suggesting an underlying genetic defect.
Detection of clonal TR gene rearrangements and TCL-associated mutations in cHL
In 43% of the cHL samples, no clonal IG gene rearrangements were detected. Because clonal TR gene rearrangements have been described in cHL, TR-NGS clonality assessment was performed in 57 paired cHL diagnosis and recurrences for which sufficient genomic DNA was available (n = 125 samples). After defining thresholds for T-cell clonality in cHL (supplemental Information), 14 of 125 samples (11%) displayed clonal TRB/TRG gene rearrangements, whereas the remaining samples showed a polyclonal pattern (supplemental Table 12). In 5 of 14 samples, both clonal IG and TR gene rearrangements were present (Figure 5). Overall, in 12 of 57 patients (21%), clonal TR gene rearrangements were detected in diagnosis (n = 5) and/or recurrence (n = 9). Four patients (cases 1, 5, 11, and 23) showed preserved dominant TCR clonotypes in paired diagnosis and relapse (supplemental Table 13). To assess whether detection of TCR clonality was associated with cHL relapse, TR-NGS was performed in a control cohort of patients with cHL without relapse (n = 48). Here, 3 of 48 samples (6%) displayed clonal TR gene rearrangements (supplemental Table 12), indicating that TCR clonality as such is not unique for relapse-prone cHL.
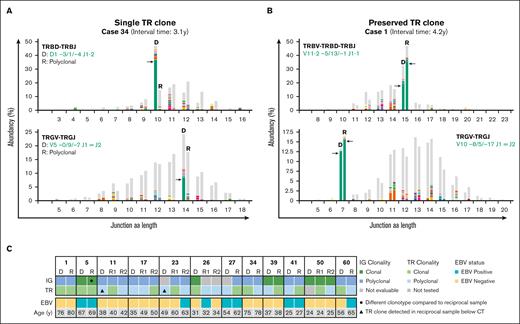
NGS-based T-cell clonality analysis identifies clonal TR gene rearrangements in cHL samples. (A) Representative data set indicating NGS-based detection of TR gene rearrangements in paired diagnosis (D) and recurrence (R) tissue samples of a case with a dominant T-cell clone in only the primary diagnosis but not recurrence (case 34). The results of 2 TCR targets (TRBV-TRBD-TRBJ and TRGV-TRGJ) are shown, and the specific clonotypes for the dominant TR gene rearrangements are indicated. On the x-axis, the junction aa length is shown, and the abundancy of clonotypes is shown in percentages on the y-axis. (B) Representative data set indicating NGS-based detection of TR gene rearrangements in paired samples with a preserved identical dominant T-cell clone (case 1). (C) Summary of IG- and TR-NGS clonality results of all patients with at least 1 sample with a dominant TCR clone. EBV status of all tumor samples and age of the patients are shown.
NGS-based T-cell clonality analysis identifies clonal TR gene rearrangements in cHL samples. (A) Representative data set indicating NGS-based detection of TR gene rearrangements in paired diagnosis (D) and recurrence (R) tissue samples of a case with a dominant T-cell clone in only the primary diagnosis but not recurrence (case 34). The results of 2 TCR targets (TRBV-TRBD-TRBJ and TRGV-TRGJ) are shown, and the specific clonotypes for the dominant TR gene rearrangements are indicated. On the x-axis, the junction aa length is shown, and the abundancy of clonotypes is shown in percentages on the y-axis. (B) Representative data set indicating NGS-based detection of TR gene rearrangements in paired samples with a preserved identical dominant T-cell clone (case 1). (C) Summary of IG- and TR-NGS clonality results of all patients with at least 1 sample with a dominant TCR clone. EBV status of all tumor samples and age of the patients are shown.
To determine whether cHL samples with clonal TR gene rearrangement were instead TCLs mimicking cHL, mutation analysis was performed for TCL-related genes. First, potential AITL-associated mutations in DNMT3A, IDH2, RHOA, and TET2 were investigated in 13 samples with a clonal TR gene rearrangement by a targeted NGS approach, as previously described.32 Here, 7 samples showed TET2 mutations with a VAF ranging from 3% to 50%, which potentially also included CHIP-related mutations (supplemental Table 14). Furthermore, in 3 of 7 TET2-mutated samples, the RHOA hotspot mutation p.G17V was detected. Second, a selected set of 6 samples with a dominant TCR clone were analyzed with a lymphoma-specific 200 gene panel (BLYMF200)42 to identify other TCL-associated mutations. Here, all 6 samples from 5 patients showed TCL-associated gene mutations, including DNMT3A (case 60, p.E856G), STAT3 (case 1, p.S614R and case 11, p.D661Y), VAV1 (case 27, p.M501V), and JAK3 (case 1, p.I239T and case 27, p.T435A; supplemental Table 14).
These molecular findings were indicative of TCL mimicking cHL, and candidate tissue samples were reassessed by pathology review. One patient (case 1), who showed preserved TCR clonotypes in paired diagnosis and relapse samples, exhibited STAT3 and JAK3 mutations, along with large lymphoid cells strongly positive for CD30 but negative for PAX5 and anaplastic lymphoma kinase (ALK). This case was reclassified as ALK–negative anaplastic large cell lymphoma (ALCL). Recurrence samples of 3 other patients (cases 5, 23, and 27) showed RHOA and TET2 mutations and immunohistology in line with AITL mimicking cHL (Figure 6; supplemental Figure 3). Two primary diagnosis samples displayed composite cHL and AITL (cases 5 and 60), based on molecular results (IG/TR clonality and gene mutations) and histomorphology. The recurrence of case 11 harboring a dominant T-cell clone and STAT3 mutation was still considered a cHL. Altogether, in 5 patients (n = 7 samples), retrospective pathology review with integration of the molecular findings indicated the presence of a TCL, of which the HRS-like cells in 3 AITL samples of 2 patients showed EBV-positive results.
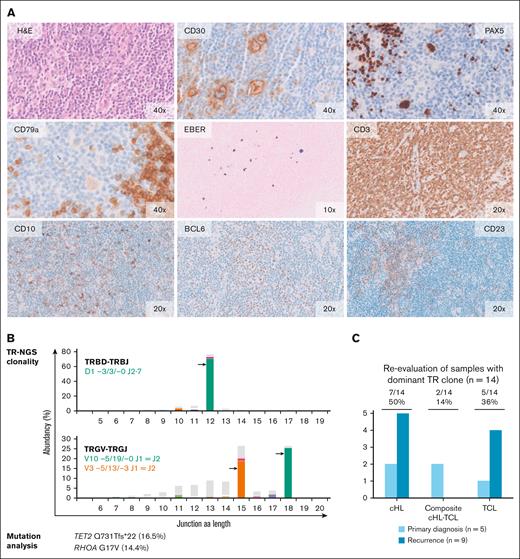
Molecular analysis and histomorphological reevaluation reveal AITL diagnosis in cHL recurrence. Histomorphology and molecular data of the recurrence sample of case 5. (A) The normal lymph node architecture has been replaced by a polymorphous infiltrate consisting of small- and medium-sized lymphocytes with scattered cells having large nuclei and prominent nucleoli, some of which are consistent with HRS-like cells. High-endothelial venules are prominent. The large cells show strong expressions of CD30 and PAX5 and a weak expression of CD79a. Epstein-Barr encoding region (EBER) in situ hybridization shows scattered positive cells of different size. Anti-CD3 staining reveals numerous small- to medium-sized T cells. Many of these cells are positive for programmed cell death protein 1 (PD1) (data not shown) and a small proportion shows expression of CD10 and BCL6. Anti-CD23 staining demonstrates slight expansion of irregular dendritic meshworks. The original magnification (×10, ×20 and ×40) is indicated. (B) TR-NGS clonality analysis shows the presence of clonal TRBD- TRBJ and TRGV- TRGJ rearrangements. Targeted mutation analysis reveals mutations in AITL-associated genes TET2 and RHOA; VAF is indicated. (C) Summary of morphological reevaluation of the samples with a dominant TCR clone with integration of molecular findings. In total, 7 samples of 5 patients were indicative for TCL.
Molecular analysis and histomorphological reevaluation reveal AITL diagnosis in cHL recurrence. Histomorphology and molecular data of the recurrence sample of case 5. (A) The normal lymph node architecture has been replaced by a polymorphous infiltrate consisting of small- and medium-sized lymphocytes with scattered cells having large nuclei and prominent nucleoli, some of which are consistent with HRS-like cells. High-endothelial venules are prominent. The large cells show strong expressions of CD30 and PAX5 and a weak expression of CD79a. Epstein-Barr encoding region (EBER) in situ hybridization shows scattered positive cells of different size. Anti-CD3 staining reveals numerous small- to medium-sized T cells. Many of these cells are positive for programmed cell death protein 1 (PD1) (data not shown) and a small proportion shows expression of CD10 and BCL6. Anti-CD23 staining demonstrates slight expansion of irregular dendritic meshworks. The original magnification (×10, ×20 and ×40) is indicated. (B) TR-NGS clonality analysis shows the presence of clonal TRBD- TRBJ and TRGV- TRGJ rearrangements. Targeted mutation analysis reveals mutations in AITL-associated genes TET2 and RHOA; VAF is indicated. (C) Summary of morphological reevaluation of the samples with a dominant TCR clone with integration of molecular findings. In total, 7 samples of 5 patients were indicative for TCL.
Discussion
Investigating the clonal relationship of lymphoma recurrences reveals the occurrence of a second primary lymphoma. In our study, NGS-based detection of IG gene rearrangements showed clonally related relapsed cHL in approximately two-third of patients with cHL, whereas clonally unrelated second primary cHL was detected in almost one-third of patients with cHL. T-cell clonality analysis demonstrated clonal TR gene rearrangements in ∼10% of the assumed cHL tissue samples. Combined with the identification of TCL-related gene mutations, this led retrospectively to TCL with HRS-like cells mimicking cHL diagnosis in ∼50% of the samples harboring a clonal TR gene rearrangement.
Considering all archival tissues analyzed in this study, IG-NGS clonality assessment detected B-cell clonality in 58% of cHL samples, which is similar to our previous findings in primary cHL whole tissue specimens.16 This allowed for the clonal comparison of primary diagnosis and recurrence in 34 patients with cHL, revealing clonally related relapsed cHL disease in 24 patients, with a median time to relapse of 1.2 years (range, 0.4-4 years). Notably, in 4 clonally related samples, a second B-cell clone was identified with a different abundancy compared with the preserved neoplastic clonotypes. We considered this suggestive of either biclonal cHL, as demonstrated previously,16 or the presence of a reactive B-cell clone.
The occurrence of clonally unrelated second primary cHL was based on the detection of distinct rearrangements in 5 paired primary diagnosis and recurrences. For another 5 patients with cHL, the identified clonal IG gene rearrangements were completely absent in the reciprocal polyclonal sample, representing discordant rearrangements. Because these samples harbored good quality genomic DNA with deep coverage NGS data sets and were considered negative for ongoing SHM between diagnosis and recurrence, the latter 5 cases were also considered clonally unrelated cHL. Thus, IG clonality assessment indicated the occurrence of a second primary cHL in a total of 10 patients. Targeted mutation analysis confirmed these findings in 8 patients (4 patients in each group) by the presence of mutually exclusive mutations in the paired primary diagnosis and recurrence samples, whereas 2 patients showed no detectable somatic mutations with the targeted gene panel. The pathogenic cHL-associated gene alterations included B2M, SOCS1, and TNFAIP3 loss-of-function mutations but also other cancer-associated genes, such as MGA.
Besides the presence of these mutually exclusive mutations, single mutations were identified that were shared between diagnosis and recurrence in 4 patients with clonally unrelated cHL. In 1 patient (case 5), this involved a TET2 mutation combined with a RHOA hotspot mutation (only detected above threshold in recurrence) related to a minor AITL component in diagnosis and AITL in recurrence. In the other 3 patients (age, 4-23 years), these represented mutations in B2M (2 patients) and H1-2 (one patient), with a VAF <7%. We propose that these latter shared mutations might have occurred in an ancestor cell at an early stage of B-cell development (ie, before IG gene rearrangement), and via branching, clonal evolution resulted into 2 independent consecutive cHL tumors. Our findings by IG-NGS of clonally unrelated second primary cHL in approximately one-third of the patients is in line with a previous report detecting clonally unrelated cHL in 8 of 20 patients (40%) with cHL.37 Similar to their observation, we observed a switch in EBV-positivity status between primary diagnosis and recurrence in 3 of 10 patients, which seldomly occurs in clonally related relapsed cHL.
The occurrence of second de novo lymphomas might be linked to an immune dysregulation disorder or lymphoma predisposition. Monozygotic twins of patients with cHL display a highly increased risk (∼100-fold) of developing cHL and relatives of patients with cHL also show increased risk of cHL.44,45 Based on genome wide association studies, certain HLA alleles result in an increased susceptibility for cHL,46 whereas other HLA alleles act as a protective factor for cHL development.47-49 SNPs identified as cHL risk factors are often present in genes related to immunity, such as BMF, GATA3, IRF4, PAX5, STAT3, TCF3, TLR4, and TP63.50-55 Furthermore, specific cHL predisposing genes have been reported, including CTLA4, DICER1, MKL1, ACAN, and KDR.54,56-60 Because at least 2 patients in this study cohort with second primary cHL were linked to immune dysregulation and/or genetic predisposition, future studies using whole genome sequencing might reveal whether specific germ line variants are affected in these patients.
By using IG/TR-NGS clonality assessment combined with targeted mutation analysis, we were able to reevaluate the initial diagnosis of several patients. Tissue biopsy specimen of 5 patients with a dominant TCR clone and TCL-associated mutations were retrospectively diagnosed as TCL instead of cHL. The primary diagnosis of samples of 2 of 5 patients were likely composite lymphomas consisting of cHL and a (minor) TCL component. Dominant TR gene rearrangements in the other patients were considered to represent reactive T-cell clones, because there was no evidence for TCL-associated mutations or histomorphology. In fact, minor T-cell clones (below the clonality threshold of 5%) were also detected in other cHL samples (data not shown). Such T-cell clones probably represent clonal expansions of reactive T cells, for instance, against EBV+ HRS cells with detectable major histocompatibility complex class I and/or class II expression.61 Our findings of misdiagnosed cHL is in line with a recent study in which 18 of 54 selected patients with cHL were reclassified as TCL.62 Indeed, TCL such as angioimmunoblastic type nTFHL/AITL are increasingly recognized as a diagnostic pitfall in the context of cHL. Therefore, TCR clonality assessment and/or targeted TCL-associated gene mutation analysis should be considered in routine diagnostics to avoid misdiagnosis in cases with morphological or clinical doubt and in cases with disease recurrence.
Finally, our mutation analyses in patient samples with a dominant TCR clone also revealed TET2 mutations with VAFs of >20% in 4 of 11 patients (36%), which did not correlate with the percentage of neoplastic HRS cells within the cHL tissue specimens. This possibly reflected CHIP-associated mutations that may also reside within HRS cells themselves, as demonstrated recently.36 The relative high frequency of such variants in this subset of predominantly older patients with cHL (>50 years) with disease recurrence in combination with an underlying TCL suggests a role for these CHIP-associated mutations in lymphoma development, such as in other hematopoietic neoplasms.63-65
In conclusion, our in-depth molecular analysis demonstrates the occurrence of second de novo cHL in approximately one-third of patients with cHL and ∼60% of patients with cHL, with a time to recurrence of >2 years after primary cHL diagnosis. Furthermore, our data show that in retrospect 5 other patients were diagnosed with TCL. These findings obviously have strong clinical implications, because these patients should receive different treatment strategies. The outcome of our study indicates that NGS-based clonality assessment combined with targeted mutation analysis should be considered in routine diagnostics for patients with cHL with recurrences after 2 years.
Acknowledgments
The authors thank Karin Beunen and Harald Verheij (Rijnstate Hospital) and Lieneke Homans-ter Keurs (Gelderse Vallei Hospital) for their assistance in gathering clinical data, supporting staff for collecting tumor samples from pathology archives, Maria Pafiti for DNA isolation and clonality assessment of the nonrelapse control cohort, and the technical staff of the departments of pathology and human genetics for preparing samples for TSO500 assay and loading NGS runs. Panels B and C of Figure 4 were created with BioRender.com.
This work was funded by the Dutch Cancer Society (KWF-11137) and the Dutch Health Insurers’ Innovation Fund (project number 17-179).
Authorship
Contribution: J.H.J.M.v.K. and B.S. designed the research and conceived the project; D.A.G.v.B., J.L.M.v.d.L.-K., J.R., J.A.C.W.L., I.B., R.A.L.d.G., and F.A.d.G. performed the experimental research; D.A.G.v.B., L.I.K., M.R.B., P.J.T.A.G., K.M.H., M.v.d.B., I.B., J.S.P.V., R.A.L.d.G., and B.S. were involved in data analysis and interpretation; D.A.G.v.B., W.B.C.S., E.v.d.S., W.J.P., J.F.M.P., S.D.P.W.M.d.J.-P., G.A.V., C.L., E.R.v.B., B.F., B.M.H., A.P., J.v.d.W.t.B., P.T.G.A.N., K.M.H., F.F., A.D., and M.v.d.B. collected the histological material and/or clinical data; D.A.G.v.B. and B.S. wrote the manuscript; M.v.d.B., J.H.J.M.v.K., K.M.H., and A.D. examined histopathology; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Blanca Scheijen, Pathology, Radboud University Medical Center, Geert Grooteplein Zuid 10, 6525 GA Nijmegen, The Netherlands; e-mail: blanca.scheijen@radboudumc.nl.
References
Author notes
Data are available on request from the corresponding author, Blanca Scheijen (blanca.scheijen@radboudumc.nl).
The full-text version of this article contains a data supplement.