Key Points
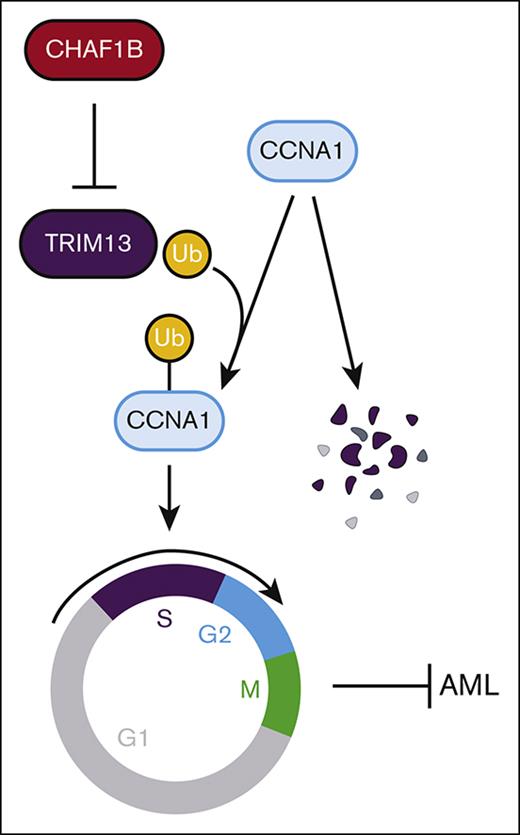
The chromatin assembly machinery protects leukemic cell self-renewal by directly regulating transcription of the E3 ubiquitin ligase TRIM13.
TRIM13 is nuclear localized and represses self-renewal by driving cell cycle entry through the stabilization of cyclin A1.
Abstract
Acute myeloid leukemia (AML) is an aggressive blood cancer that stems from the rapid expansion of immature leukemic blasts in the bone marrow. Mutations in epigenetic factors represent the largest category of genetic drivers of AML. The chromatin assembly factor CHAF1B is a master epigenetic regulator of transcription associated with self-renewal and the undifferentiated state of AML blasts. Upregulation of CHAF1B, as observed in almost all AML samples, promotes leukemic progression by repressing the transcription of differentiation factors and tumor suppressors. However, the specific factors regulated by CHAF1B and their contributions to leukemogenesis are unstudied. We analyzed RNA sequencing data from mouse MLL-AF9 leukemic cells and bone marrow aspirates, representing a diverse collection of pediatric AML samples and identified the E3 ubiquitin ligase TRIM13 as a target of CHAF1B-mediated transcriptional repression associated with leukemogenesis. We found that CHAF1B binds the promoter of TRIM13, resulting in its transcriptional repression. In turn, TRIM13 suppresses self-renewal of leukemic cells by promoting pernicious entry into the cell cycle through its nuclear localization and catalytic ubiquitination of cell cycle–promoting protein, CCNA1. Overexpression of TRIM13 initially prompted a proliferative burst in AML cells, which was followed by exhaustion, whereas loss of total TRIM13 or deletion of its catalytic domain enhanced leukemogenesis in AML cell lines and patient-derived xenografts. These data suggest that CHAF1B promotes leukemic development, in part, by repressing TRIM13 expression and that this relationship is necessary for leukemic progression.
Introduction
Acute myeloid leukemia (AML) is an aggressive blood cancer characterized by blasts in the bone marrow (BM). Though AML tumors generally harbor a low mutational load, the mutations that they possess frequently occur in genes that encode epigenetic regulatory proteins.1,2 This has led to the categorization of AML a disease of epigenetic dysregulation, and therapeutic compounds that target epigenetic regulators are some of the most exciting new treatment strategies for AML.3,4 The chromatin assembly factor (CAF) CHAF1B is a master epigenetic regulator of transcription associated with cell state and self-renewal.5-7 CHAF1B is the p60 subunit of the heterotrimeric CAF1 complex also containing p150 CHAF1A and p48 RBBP4, responsible for facilitating histone H3/H4 heterodimer assembly at the replication fork during the S-phase of cell cycle.8 In addition to its role in nucleosome assembly, CHAF1B has a regulatory role in transcription and is linked to cell state–directed transcriptional regulation in many metazoan organisms.6,9-11 CHAF1B is upregulated in a wide variety of cancers, and enhanced expression correlates with poor prognosis in almost every cancer studied.12-17 We recently reported that CHAF1B is upregulated in AML, in which it binds to chromatin at promoters and enhancers of differentiation genes, resulting in the repression of the transcription of differentiation genes through the displacement of transcription factors (such as C/EBPα).5 Reduction of CHAF1B to expression levels in healthy tissues is sufficient to release the transcriptional repression on differentiation genes and resolve the tumors in vitro and in vivo.5 This led us to hypothesize that CHAF1B promotes tumorigenicity by repressing tumor suppressors and differentiation genes. Because there are no therapeutic interventions that can directly target CHAF1B or the CAF1 complex, we are interested in understanding the transcriptional targets of CHAF1B so that we can leverage those as potential therapies.
TRIM13 is an E3 ubiquitin ligase (tripartite motif protein), characterized by a RING motif located near the N-terminus, zinc-binding B-box motif, and coiled-coil B-box motif.18 TRIM13 also has a transmembrane domain that is responsible for anchoring it to the endoplasmic reticulum (ER) in some cell types, such as those in samples from patients with multiple myeloma.19 TRIM13 is located on human chromosome 13q14, an area frequently deleted in B-cell chronic lymphocytic leukemia (CLL).20-23 Because of its frequent deletion in CLL, TRIM13 was originally referred to as ret finger protein deleted in leukemia “LEU5/RFP2.”24,25 Although not every patient with CLL has an accompanying loss of TRIM13,26 TRIM13 expression is reduced in CLL tumors upon progression when compared with initial diagnosis.27 Because of this relationship, TRIM13 is considered a tumor suppressor in CLL.25,28 Chromosome 13 deletions are also present in myeloma, mantle cell lymphoma, diffuse large B-cell lymphoma, prostate cancer, and head and neck cancers,29-34 and low expression of TRIM13 is a marker of poor prognosis in breast cancer.35 Furthermore, elevated expression of TRIM13 is considered a marker of good prognosis in non–small cell lung carcinoma through the initiation of apoptosis in the tumor tissue in vivo.36 In contrast, 1 study so far suggested that TRIM13 may act as an oncogene in the MMS1 multiple myeloma cell line by activating NF-κB through targeted the degradation of the NF-κB inhibitor IκBα.19 We demonstrate that TRIM13 represses leukemogenesis by localizing at the nucleus and ubiquitinating the cell cycle gene cyclin A1 (CCNA1), prompting premature entry into the cell cycle resulting in loss of self-renewal. CHAF1B, in turn, promotes leukemic transformation by repressing TRIM13, enforcing self-renewal of leukemic blasts, and promoting progression.
Materials and methods
Cell culture
AML cell lines (U937, THP1, K562, NB4, and MOLM13) were maintained in RPMI-1640 supplemented with 10% heat-inactivated fetal bovine serum (HI-FBS), Pen-Strep, and L-glutamine. CMK cells were maintained in RPMI-1640 supplemented with 20% HI-FBS, Pen-Strep, and L-glutamine. The AML cell line, HL60, was maintained in Iscove modified Dulbecco medium supplemented with 10% HI-FBS, Pen-Strep, and L-glutamine. Mouse MLL-AF9 leukemic cells were maintained in RPMI-1640 supplemented with 10% HI-FBS, Pen-Strep, L-glutamine, and cytokines (100 ng/mL of stem cell factor, 50 ng/mL of interleukin-6 [IL-6], and 20 ng/mL of IL-3). Human CD34+ peripheral blood stem cells (PBSCs) were maintained in Stemspan II serum-free expansion medium, with CD34 supplement and primocin. Patient-derived xenograft (PDX) samples were maintained for a short-term in Iscove modified Dulbecco medium supplemented with 20% FBS, Pen-Strep, and 10 ng/mL each of IL-3, IL-6, stem cell factor, FLT3L, and thrombopoietin. 293T cells were maintained in Dulbecco’s modified Eagle medium supplemented with 10% HI-FBS, Pen-Strep, and L-glutamine.
Cell cycle assay
Cells were pulsed with 10 μM 5-ethynyl-2'-deoxyuridine (EdU) for 1 hour at 37°C. After EdU pulse, cells were stained with fixable viability dye–red at 4 °C for 30 minutes. After washing, cells were fixed in 4% paraformaldehyde for 15 minutes and permeabilized in saponin buffer. Allophycocyanin-azide was clicked onto EdU for 30 minutes at room temperature, and DNA content was stained using FxCycle Violet for 30 minutes at 4°C. Cells were read on either a CantoII or Fortessa flow cytometer. Data were analyzed using FlowJo.
CRISPR knockout of TRIM13 RING domain
Complete single guide RNA targeting early in the coding sequence of TRIM13 were ordered from Synthego. These single guide RNAs were complexed with HiFi Cas9 V3 (IDT) for 20 minutes at room temperature before being resuspended in Neon Buffer R (Thermo Fisher) and Cas9 Electroporation Enhancer (IDT). A total of 1 × 105 cells were electroporated in a 10 μL Neon Tip (human AML cells = 1350 V, 35 ms, 1 pulse; mouse AML cells = 1700 V, 20 ms, 1 pulse) and plated in complete media without antibiotics for 24 hours. After 48 hours of electroporation, CRISPR efficiency was measured via Sanger sequencing and inference of CRISPR edits analysis. Cells were then plated in methylcellulose, and single colonies were selected for expansion. TRIM13 knockout was confirmed in candidate colonies, first via Sanger sequencing, followed by western blot. Knockout clones were expanded. In-frame deletions (RINGdel) or out-of-frame knockout (TRIM13 knock out) were confirmed via Sanger and western blot, using expanded clones.
Oligonucleotides, antibodies, and expanded methods are presented in the supplemental figures and methods.
Results
CHAF1B regulates TRIM13 promoter preference in mouse MLL-AF9 leukemic cells
To identify mechanisms by which CHAF1B regulates transcription associated with leukemogenesis, we knocked out Chaf1b (Chaf1bΔ/Δ) in mouse MLL-AF9 leukemic cells (Figure 1A-B), followed by RNA sequencing (supplemental Figure 1A). As previously shown,5 CHAF1B loss leads to transcriptional upregulation of many genes, including differentiation genes (supplemental Figure 1A; supplemental Table 1). We found that many genes were expressed from alternative promoters in the absence of CHAF1B (supplemental Figure 1B). Because our previous work demonstrated that CHAF1B occupies promoters of differentiation genes resulting in their transcriptional repression,5 we focused on this subset of genes for our analysis. We ranked these genes in the order of most to least exclusive use of alternative promoters using AltAnalyze37 and found that the top ranked gene transcribed by an alternative promoter, after CHAF1B loss, was TRIM13. In Chaf1b+/+ leukemic cells, TRIM13 is transcribed exclusively from the proximal exon (Trim13-201 isoform). Upon Chaf1b deletion, TRIM13 can be transcribed from either the proximal or a distal (Trim13-202) exon (supplemental Figure 1C). This matched with POLR2A chromatin occupancy at the proximal promoter associated with Trim13-201 but redistributed to the distal exon after Chaf1b loss (supplemental Figure 1D).5 This was also associated with an increase in H3K4me3 and assay for transposase-accessible chromatin signal at the distal exon, as measured via chromatin immunoprecipitation sequencing (supplemental Figure 1E).5 We confirmed the increased expression and use of the alternative exon from our RNA sequencing data via Taqman quantitative polymerase chain reaction, with probes that exclusively recognize the 201 or 202 isoforms of TRIM13 (supplemental Figure 1F-G).
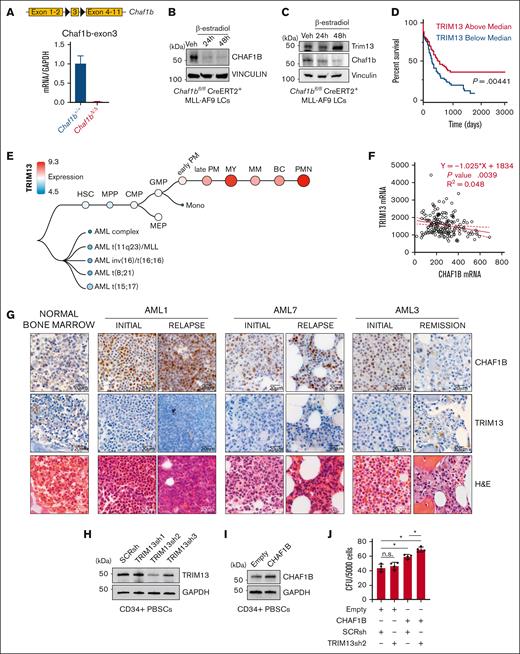
CHAF1B upregulation and TRIM13 downregulation are associated with AML progression. (A-B) Schematic representation of Chaf1b-floxed allele in mouse MLL-AF9 leukemic cells and confirmation of Chaf1b deletion after Cre activation via quantitative polymerase chain reaction (qPCR) (A) and western blot (B). Value shown is mean ± standard deviation (SD) of n = 3 independent experiments. (C) Western blot at 24 and 48 hours after Cre induction in MLL-AF9 leukemic cells. (D) Survival of patients with AML with high or low TRIM13 expression in the Cancer Genome Atlas (TCGA). P value shown. (E) Expression of TRIM13 throughout human hematopoiesis and leukemia from BloodSpot. (F) Scatterplot of expression comparing CHAF1B and TRIM13 in TCGA. The solid red line represents best fit with ± SD (dashed red lines). Line equation, P value, and R2 value listed. (G) Immunohistochemistry (IHC) in BM biopsies for CHAF1B (top), TRIM13 (middle), and hematoxylin and eosin (H&E) staining of biopsies of healthy BM and from 3 matched patients with initial/relapse or initial/remission AML. Scale bar, 20 μm; IHC and H&E imaged from the same section, if possible. (H) Western blot confirming TRIM13 short hairpin RNA (shRNA) efficacy in mobilized CD34+ PBSCs. Results shown are representative of 3 independent assays. (I) Western blot confirming the CHAF1B overexpression efficacy. Results shown are representative of 3 independent assays. (J) Colony assay in PBSCs expressing indicated viral constructs. Results shown are mean ± SD, dots indicate individual replicates. ∗P < .05, as determined using 1-way analysis of variance (ANOVA) (J). SCRsh, scrambled control short hairpin; Veh, vehicle.
CHAF1B upregulation and TRIM13 downregulation are associated with AML progression. (A-B) Schematic representation of Chaf1b-floxed allele in mouse MLL-AF9 leukemic cells and confirmation of Chaf1b deletion after Cre activation via quantitative polymerase chain reaction (qPCR) (A) and western blot (B). Value shown is mean ± standard deviation (SD) of n = 3 independent experiments. (C) Western blot at 24 and 48 hours after Cre induction in MLL-AF9 leukemic cells. (D) Survival of patients with AML with high or low TRIM13 expression in the Cancer Genome Atlas (TCGA). P value shown. (E) Expression of TRIM13 throughout human hematopoiesis and leukemia from BloodSpot. (F) Scatterplot of expression comparing CHAF1B and TRIM13 in TCGA. The solid red line represents best fit with ± SD (dashed red lines). Line equation, P value, and R2 value listed. (G) Immunohistochemistry (IHC) in BM biopsies for CHAF1B (top), TRIM13 (middle), and hematoxylin and eosin (H&E) staining of biopsies of healthy BM and from 3 matched patients with initial/relapse or initial/remission AML. Scale bar, 20 μm; IHC and H&E imaged from the same section, if possible. (H) Western blot confirming TRIM13 short hairpin RNA (shRNA) efficacy in mobilized CD34+ PBSCs. Results shown are representative of 3 independent assays. (I) Western blot confirming the CHAF1B overexpression efficacy. Results shown are representative of 3 independent assays. (J) Colony assay in PBSCs expressing indicated viral constructs. Results shown are mean ± SD, dots indicate individual replicates. ∗P < .05, as determined using 1-way analysis of variance (ANOVA) (J). SCRsh, scrambled control short hairpin; Veh, vehicle.
To interrogate the function of this alternative promoter, we used CRISPR to delete the proximal (Trim13Δ201) or the distal exon (Trim13Δ202) (supplemental Figure 1H). Using isoform-specific Taqman probes, we found that in Chaf1b+/+ MLL-AF9 leukemic cells, the proximal exon appeared necessary for TRIM13 expression. However, in Chaf1bΔ/Δ MLL-AF9 leukemic cells, TRIM13 expression could be restored entirely from the distal promoter (supplemental Figure 1I). Although the proximal promoter was necessary for both messenger RNA (mRNA) transcription and protein production of TRIM13 in Chaf1b+/+ MLL-AF9 leukemic cells (supplemental Figure 1J), this alternative promoter appeared to be associated with a significant increase in TRIM13 protein production after CHAF1B loss (Figure 1C). Reduction of TRIM13 expression, either by CRISPR-mediated Trim13 deletion or loss of the upstream exons leads to a slight but significant increase in the colony forming capacity of MLL-AF9 leukemic cells (supplemental Figure 1K).
CHAF1B upregulation and TRIM13 repression are associated with AML progression and relapse
Elevated levels of TRIM13 are associated with enhanced survival (Figure 1D). Analysis of the Bloodspot38 database helped confirm our finding that TRIM13 was repressed in AML but upregulated in mature myeloid cells (Figure 1E). To determine whether CHAF1B/TRIM13 inverse expression also occurs in human AML, we first analyzed the Cancer Genome Atlas data and confirmed our observation that increased CHAF1B transcription correlates with decreased TRIM13 transcription (Figure 1F). Next, we compared BM aspirates from healthy donors to matched samples taken from patients either at diagnosis and relapse (initial/relapse) or diagnosis and remission (initial/remission) and observed the upregulation of CHAF1B in the leukemic marrows that coincided with the repression of TRIM13 (Figure 1G). In 1 patient with a matched initial diagnosis and remission sample AML3, CHAF1B returned to lower levels, whereas TRIM13 was re-expressed upon remission (Figure 1G). We also observed a similar pattern of high CHAF1B and low TRIM13 expression in unmatched AML samples (supplemental Figure 2). These findings suggest that concurrent upregulation of CHAF1B and repression of TRIM13 may be proleukemic. To test this, we knocked down TRIM13 (Figure 1H) and caused the overexpression of CHAF1B (Figure 1I) together in CD34+ PBSCs from healthy donors and measured the effects on colony formation in vitro. We found that colony formation capacity was significantly enhanced in PBSCs when CHAF1B was overexpressed and TRIM13 was repressed (Figure 1J).
CHAF1B negatively regulates TRIM13 expression through promoter binding
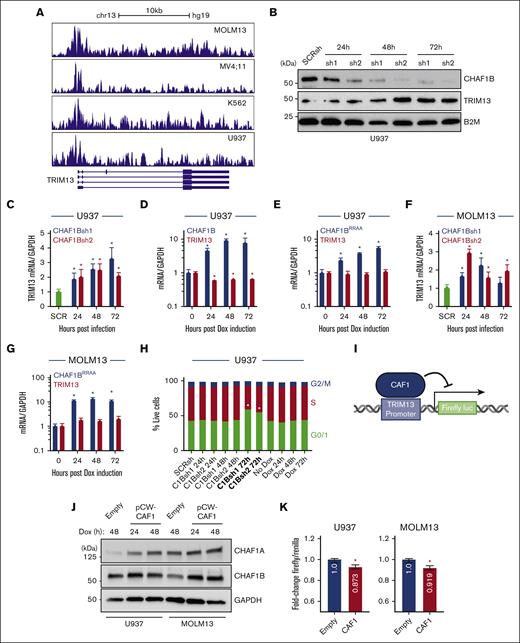
We confirmed, via chromatin immunoprecipitation sequencing analysis, that CHAF1B occupies the human TRIM13 promoter in multiple AML cell lines (Figure 2A).5 We noted that the exon distribution around the promoter of the Homo sapiens TRIM13 was different compared with that of M. musculus Trim13 (supplemental Figures 1C and 2A), so we continued the examination of TRIM13 function in human models. U937 cells showed increases in TRIM13 expression at both the protein level (Figure 2B) and the mRNA level (Figure 2C) after knockdown of CHAF1B. CHAF1B overexpression in U937 cells led to the repression of TRIM13 mRNA production (Figure 2D), which was dependent on the binding ability of CHAF1B chromatin because overexpression of the CHAF1BRR482/483AA (CHAF1BRRAA) chromatin–binding-deficient mutant39 was unable to repress TRIM13 transcription (Figure 2E). We confirmed this relationship in the MOLM13 AML cell line (Figure 2F-G). We noted that all changes in TRIM13 expression occurred before differentiation-driven changes in the cell cycle after CHAF1B overexpression or knockdown at 72 hours (Figure 2H). Because CHAF1B (working through the CAF1 complex) exerts its transcriptional repressive function at the chromatin, we hypothesized that CHAF1B represses TRIM13 by occupying its promoter in the chromatin. To test this, we cloned the promoter sequence bound by CHAF1B (Figure 2A) into a promoterless lentivirus to drive firefly luciferase (Figure 2I), with which we cotransduced cells, lentiviruses encoding a doxycycline-induced CHAF1B-T2A-CHAF1A (Figure 2J) lentivirus, and a Renilla luciferase control. We observed a slight but significant reduction in firefly luciferase activity upon CAF1 overexpression (Figure 2K), suggesting that promoter binding is likely responsible for some of the transcriptional repression of TRIM13.
CHAF1B represses TRIM13 expression by binding to its promoter. (A) CHAF1B chromatin immunoprecipitation sequencing comparing the occupancies in MOLM13, MV4;11, K562, and U937 AML cell lines. (B) Western blot at indicated timepoints after CHAF1B knockdown in U937 cells. (C,F) Taqman QPCR measuring TRIM13 expression after CHAF1B knockdown. Values shown are mean ± SD relative to SCRsh. (D) Taqman QPCR of TRIM13 expression after CHAF1B overexpression. Values shown are mean ± SD relative to those of nondoxycycline (Dox) control. (E,G) Taqman QPCR measuring TRIM13 expression after CHAF1BRRAA overexpression. Values shown are mean ± SD relative to those of non-Dox control. (H) Cell cycle assay in U937 cells expressing indicated CHAF1Bsh or CHAF1B complementary DNA for the indicated times. (± SD is omitted for simplicity) Bolded conditions are significantly different from that of control. (I) Schematic design of luciferase assay. (J) Western blot of dox-inducible CAF1 or empty vector in indicated cell lines. (K) Luciferase assay normalized for Renilla expression. Values shown are mean values from 3 biological replicates experiments. (H). ∗P < .05, as determined using one-way ANOVA with Bonferroni post hoc test (C-H) or t test with Welch correction (K).
CHAF1B represses TRIM13 expression by binding to its promoter. (A) CHAF1B chromatin immunoprecipitation sequencing comparing the occupancies in MOLM13, MV4;11, K562, and U937 AML cell lines. (B) Western blot at indicated timepoints after CHAF1B knockdown in U937 cells. (C,F) Taqman QPCR measuring TRIM13 expression after CHAF1B knockdown. Values shown are mean ± SD relative to SCRsh. (D) Taqman QPCR of TRIM13 expression after CHAF1B overexpression. Values shown are mean ± SD relative to those of nondoxycycline (Dox) control. (E,G) Taqman QPCR measuring TRIM13 expression after CHAF1BRRAA overexpression. Values shown are mean ± SD relative to those of non-Dox control. (H) Cell cycle assay in U937 cells expressing indicated CHAF1Bsh or CHAF1B complementary DNA for the indicated times. (± SD is omitted for simplicity) Bolded conditions are significantly different from that of control. (I) Schematic design of luciferase assay. (J) Western blot of dox-inducible CAF1 or empty vector in indicated cell lines. (K) Luciferase assay normalized for Renilla expression. Values shown are mean values from 3 biological replicates experiments. (H). ∗P < .05, as determined using one-way ANOVA with Bonferroni post hoc test (C-H) or t test with Welch correction (K).
TRIM13 overexpression represses leukemogenesis in AML
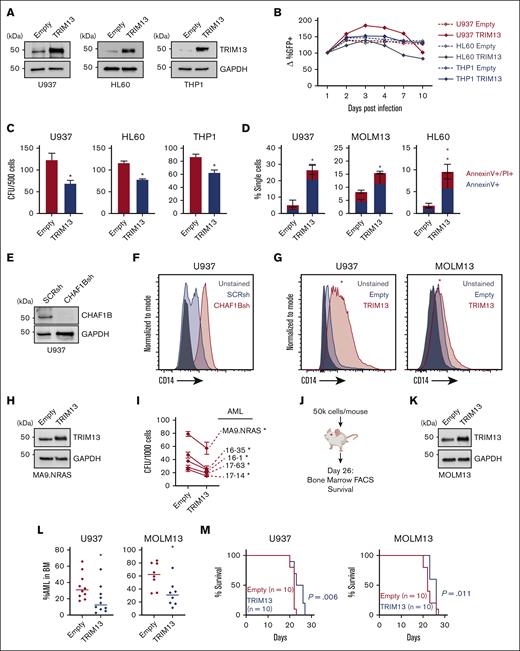
TRIM13 expression is a marker of good prognosis in the the Cancer Genome Atlas AML data set (Figure 1D-E). Conversely, CHAF1B is a marker of poor prognosis in AML.5 To test the negative effects of enhanced TRIM13 expression on AML leukemogenicity, we caused the overexpression of TRIM13 in several AML cell lines (Figure 3A). This proved detrimental for long-term productivity (Figure 3B), as demonstrated by the reduced colony forming capacity (Figure 3C) and a small increase in apoptosis (Figure 3D; supplemental Figure 3A). To test whether this was because of differentiation, we used a panel of potential differentiation markers (Figure 3E-F; supplemental Figure 3B-E) and found that the overexpression of TRIM13 was able to induce CD14 expression in both U937 and MOLM13 AML cell lines (Figure 3G). We then demonstrated that the overexpression of TRIM13 repressed the colony formation capacity in 5 different primary samples of patients with AML (Figure 3H-I). We also caused TRIM13 to be overexpressed in U937 and MOLM13 cell lines and engrafted them into recipient NSG mice (Figure 3J-K). BM aspirates 26 days after the transplant revealed a significant reduction in TRIM13-overexpressing AML cells in the BM (Figure 3L), which coincided with a slight but significant extension of survival (Figure 3M). Together, these results suggest that upregulation of TRIM13 is detrimental to leukemic progression.
TRIM13 overexpression represses leukemogenesis. (A) Western blot confirmation of TRIM13 overexpression or knockdown in AML cell lines. (B) In vitro suspension culture competitive growth assay. Values shown are mean of 3 independent replicates. (C) Colony formation assay in indicated AML cell lines expressing empty vector or TRIM13 overexpression. (D) Cell death assay measuring annexin V and propidium iodide staining in AML cell lines expressing empty vector of TRIM13 overexpression, taken at day 5. (E) Western blot confirmation of CHAF1B knockdown by shRNA. (F) CD14 surface marker expression on U937 cells 72-hours after CHAF1B knockdown. (G) CD14 surface marker expression on U937 and MOLM13 AML cell lines 5-days after TRIM13 overexpression. Red star indicates significant increase in CD14 expression when compared with empty vector. (H) Western blot confirmation of TRIM13 overexpression in sample of patients with AML. (I) Colony formation assay in indicated sample of patients with AML expressing empty vector or TRIM13 overexpression. (J) Schematic for xenograft transplantation of AML cell lines into NSG mice. (K) Western blot confirmation of TRIM13 overexpression in MOLM13 AML cell line. (L) Percent human CD45+ mononuclear cells in BM aspirates 26 days after transplantation. Each dot represents an individual mouse (n = 10), and line indicates mean. (M) Survival curve of recipient mice receiving TRIM13 overexpressing or control AML cell lines (n = 10 mice). ∗P < .05, as determined using nonparametric t test with Welch correction (C,D,L), one-way ANOVA (I), and log-rank test (M). Data shown are representative of 3 independent assays (A,E-H,K) and the mean of 3 biological replicates (B) with SD (C-D). FACS, fluorescence-activated cell sorting.
TRIM13 overexpression represses leukemogenesis. (A) Western blot confirmation of TRIM13 overexpression or knockdown in AML cell lines. (B) In vitro suspension culture competitive growth assay. Values shown are mean of 3 independent replicates. (C) Colony formation assay in indicated AML cell lines expressing empty vector or TRIM13 overexpression. (D) Cell death assay measuring annexin V and propidium iodide staining in AML cell lines expressing empty vector of TRIM13 overexpression, taken at day 5. (E) Western blot confirmation of CHAF1B knockdown by shRNA. (F) CD14 surface marker expression on U937 cells 72-hours after CHAF1B knockdown. (G) CD14 surface marker expression on U937 and MOLM13 AML cell lines 5-days after TRIM13 overexpression. Red star indicates significant increase in CD14 expression when compared with empty vector. (H) Western blot confirmation of TRIM13 overexpression in sample of patients with AML. (I) Colony formation assay in indicated sample of patients with AML expressing empty vector or TRIM13 overexpression. (J) Schematic for xenograft transplantation of AML cell lines into NSG mice. (K) Western blot confirmation of TRIM13 overexpression in MOLM13 AML cell line. (L) Percent human CD45+ mononuclear cells in BM aspirates 26 days after transplantation. Each dot represents an individual mouse (n = 10), and line indicates mean. (M) Survival curve of recipient mice receiving TRIM13 overexpressing or control AML cell lines (n = 10 mice). ∗P < .05, as determined using nonparametric t test with Welch correction (C,D,L), one-way ANOVA (I), and log-rank test (M). Data shown are representative of 3 independent assays (A,E-H,K) and the mean of 3 biological replicates (B) with SD (C-D). FACS, fluorescence-activated cell sorting.
Repression of TRIM13 activity promotes AML leukemogenicity
Because TRIM13 overexpression represses leukemogenesis, and low levels of TRIM13 are associated with poor outcome in AML, we hypothesized that TRIM13 may act as a putative tumor suppressor in AML. To test this, we knocked down TRIM13 in a pediatric PDX of AML (CBFA2T3-GLIS2 driven AML).40 We were able to expand TRIM13 knockdown cells (Figure 4A) and found that TRIM13 knockdown drove the enhanced expansion of leukemic cells within the BM of recipient mice (Figure 4B) and contributed to a more aggressive disease (Figure 4C). We then confirmed the role of TRIM13 in 7 different pediatric AML samples (supplemental Table 2) by knocking down TRIM13 and assaying the colony forming capacity in vitro (Figure 4D), which led to an increase in colony formation rates in all samples (Figure 4E).
TRIM13 repression promotes leukemogenesis. (A) Western blot confirmation of TRIM13 knockdown in PDX 16 to 35 before transplant. (B) Percent human CD45+ mononuclear cells in BM aspirates on indicated days after transplantation. Each dot represents an individual mouse (n = 10), middle line represents mean, box represents SD, and whiskers represent minimum-maximum range. Individual points represent individual mice (n = 10). (C) Survival of recipient mice after transplantation of 1 × 106 live AML16 to AML35 cells. (D) Western blot confirmation of TRIM13 knockdown in AML17 to AML129. (E) Colony forming assay in indicated PDXs expressing TRIM13sh2 (T13sh2). (F) Schematic diagram of RING domain deletion in TRIM13 by CRISPR. (G) Western blot confirmation of deletion clones in AML cell lines. ∗ indicates full-length TRIM13. (H) Colony assay in indicated AML cell lines comparing CTRLsg with T13RINGdel cells. (I) Cell death assay at steady state in T13RINGdel clones compared with CTRLsg. Red star indicates significant difference in AnnexinV/propidium iodide between conditions, black start indicates significant differences in AnnexinV between conditions. (J) Cell surface CD14 expression at steady state in AML cell lines. Unless otherwise noted, ∗P < .05, as determined using t test with Welch correction (B), one-way ANOVA (E,H-I), or log-rank (C). All experiments shown are representative of 3 biological replicates (A,D,G,J), mean + SD of 3 biological replicates (E,H,I), or from 10 mice (B-C). CTRLsg, cells electroporated with a nontargeting control single guide RNA.
TRIM13 repression promotes leukemogenesis. (A) Western blot confirmation of TRIM13 knockdown in PDX 16 to 35 before transplant. (B) Percent human CD45+ mononuclear cells in BM aspirates on indicated days after transplantation. Each dot represents an individual mouse (n = 10), middle line represents mean, box represents SD, and whiskers represent minimum-maximum range. Individual points represent individual mice (n = 10). (C) Survival of recipient mice after transplantation of 1 × 106 live AML16 to AML35 cells. (D) Western blot confirmation of TRIM13 knockdown in AML17 to AML129. (E) Colony forming assay in indicated PDXs expressing TRIM13sh2 (T13sh2). (F) Schematic diagram of RING domain deletion in TRIM13 by CRISPR. (G) Western blot confirmation of deletion clones in AML cell lines. ∗ indicates full-length TRIM13. (H) Colony assay in indicated AML cell lines comparing CTRLsg with T13RINGdel cells. (I) Cell death assay at steady state in T13RINGdel clones compared with CTRLsg. Red star indicates significant difference in AnnexinV/propidium iodide between conditions, black start indicates significant differences in AnnexinV between conditions. (J) Cell surface CD14 expression at steady state in AML cell lines. Unless otherwise noted, ∗P < .05, as determined using t test with Welch correction (B), one-way ANOVA (E,H-I), or log-rank (C). All experiments shown are representative of 3 biological replicates (A,D,G,J), mean + SD of 3 biological replicates (E,H,I), or from 10 mice (B-C). CTRLsg, cells electroporated with a nontargeting control single guide RNA.
We used CRISPR editing to generate clonally derived populations of in-frame RING domain deletion mutants (T13RINGdel) to inactivate the catalytic activity of the endogenous TRIM13 loci (Figure 4F). We generated 2 independent clones of U937, HL60, and THP1 cell lines each with in-frame RING domain deletions (Figure 4G). T13RINGdel did not appear to have a negative effect on the truncated protein stability (Figure 4G). Mutant clones had increased colony forming capacity (Figure 4H) and reduced cell death (Figure 4I). Loss of TRIM13 catalytic activity was also sufficient to reduce the surface CD14 expression levels in the AML cells lines, suggesting a slightly less differentiated cell state (Figure 4J).
CHAF1B loss is associated with both enhanced TRIM13 expression (Figures 1C and 2C,F) and loss of colony formation capacity (supplemental Figure 4B-C). To test whether TRIM13 upregulation contributed to the colony formation loss phenotype of CHAF1B-depleted AML cells (supplemental Figure 4A), we performed a pathway suppressor experiment to determine whether ablation of TRIM13 function could synthetically rescue the loss of the colony formation capacity from CHAF1B-depleted AML cell lines. We found that inactivation of TRIM13 led to a partial rescue of the colony formation capacity in CHAF1B-depleted cell lines (supplemental Figure 4B). Despite the different promoter organization between mouse and human TRIM13, we were able to confirm thefunctional conservation of TRIM13 between the 2 species using Chaf1b deletion in our MLL-AF9 AML cell lines with either TRIM13 knocked out or upstream exons deleted (supplemental Figure 4C). These data, when taken together with our finding that TRIM13 upregulation is detrimental to leukemogenesis (Figure 3), suggest that TRIM13 plays a putative tumor suppressor role in AML.
TRIM13 localizes at the nucleus of immature hematopoietic cells
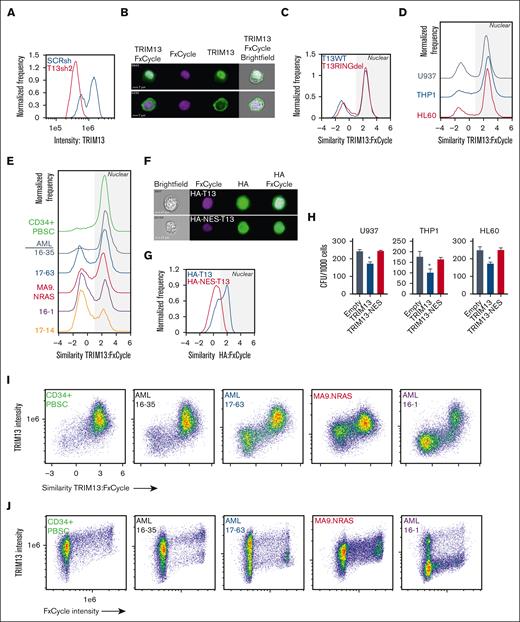
TRIM13 has previously been reported to localize to the ER through a transmembrane domain on TRIM13 C-terminus.41 To determine its localization in AML, we used imaging cytometry to assay TRIM13 localization in wild-type T13 (T13WT), T13RINGdel U937, HL60, and THP1 cells. We first confirmed the specificity of our antibody for endogenous TRIM13 (Figure 5A). We found that TRIM13 was predominantly localized at the nucleus in the U937 cells, a localization pattern that was not dependent on presence of the RING domain (Figure 5B-C). We did note some perinuclear staining that may correspond to previously described ER localizations,42,43 but this was found in a minority of the cells. This localization pattern was almost identical in THP1 and HL60 cell lines (Figure 5D), PDX models, and healthy mobilized CD34+ PBSCs (Figure 5E), suggesting that this may be a feature of immature hematopoietic cells. To determine whether nuclear localization of TRIM13 was necessary for its role as an antileukemic repressor, we caused a TRIM13 allele tagged with a nuclear exchange signal to be overexpressed to promote cytoplasmic expression. We confirmed the activity of the nuclear exchange signal tag via imaging cytometry (Figure 5F-G). Cytoplasmic localization of TRIM13 did not lead to a loss in clonogenic capacity of the AML cell lines, which led us to conclude that it is indeed the nuclear localization of TRIM13 that is responsible for its repressive phenotype in AML (Figure 5H). We then found that the most intense TRIM13 staining occurred in the nucleus (Figure 5I) and that TRIM13 staining intensity increased with progression through the cell cycle (Figure 5J), suggesting that it may play a role in cell cycle regulation.
TRIM13 nuclear localization is necessary for its antileukemic function. (A) Confirmation of TRIM13 antibody specificity (histogram = TRIM13 staining intensity vs frequency). Result shown is quantified from 10 000 U937 cells expressing either SCRsh or TRIM13sh2. (B) Representative images of TRIM13 localization (top image, nuclear localized; bottom image, nonnuclear localized) in U937 AML cell line. Scale bar, 7 μm; cell number (top left). (C-E) Pixel analysis of 10 000 cells: (C) U937 T13RINGdel vs T13WT, (D) AML cell lines, (E) CD34+ PBSCs and PDXs; TRIM13 localization compared with DNA localization (FxCycle Violet). Higher similarity score means TRIM13 and DNA images are more similar. Nuclear localization indicated by gray shading. (F) Representative images of U937 cells expressing either HA-TRIM13 (top) or HA-NES-TRIM13 (bottom). Scale bar, 7 μm; cell number (top left). (G) Pixel analysis of 10 000 U937 cells expressing HA-TRIM13 or HA-NES-TRIM13. Gray shading indicates nuclear localization. (H) Colony forming unit assay in AML cell lines expressing empty vector, HA-TRIM13, or HA-NES-TRIM13. (I) Pixel analysis of 10 000 CD34+ PBSCs or PDXs cells comparing TRIM13 staining intensity to similarity to DNA stain (FxCycle). (J) Pixel analysis of 10 000 CD34+ PBSCs or PDXs cells showing the similarity between TRIM13 staining intensity and DNA staining (FxCycle) intensity. ∗P < .05, determined using one-way ANOVA, compared with empty vector (H). Results shown are mean ± SD from 3 biological replicates (H), histogram from 10 000 cells (A,C,D,E,G), or representative of 3 biological replicates (I,J).
TRIM13 nuclear localization is necessary for its antileukemic function. (A) Confirmation of TRIM13 antibody specificity (histogram = TRIM13 staining intensity vs frequency). Result shown is quantified from 10 000 U937 cells expressing either SCRsh or TRIM13sh2. (B) Representative images of TRIM13 localization (top image, nuclear localized; bottom image, nonnuclear localized) in U937 AML cell line. Scale bar, 7 μm; cell number (top left). (C-E) Pixel analysis of 10 000 cells: (C) U937 T13RINGdel vs T13WT, (D) AML cell lines, (E) CD34+ PBSCs and PDXs; TRIM13 localization compared with DNA localization (FxCycle Violet). Higher similarity score means TRIM13 and DNA images are more similar. Nuclear localization indicated by gray shading. (F) Representative images of U937 cells expressing either HA-TRIM13 (top) or HA-NES-TRIM13 (bottom). Scale bar, 7 μm; cell number (top left). (G) Pixel analysis of 10 000 U937 cells expressing HA-TRIM13 or HA-NES-TRIM13. Gray shading indicates nuclear localization. (H) Colony forming unit assay in AML cell lines expressing empty vector, HA-TRIM13, or HA-NES-TRIM13. (I) Pixel analysis of 10 000 CD34+ PBSCs or PDXs cells comparing TRIM13 staining intensity to similarity to DNA stain (FxCycle). (J) Pixel analysis of 10 000 CD34+ PBSCs or PDXs cells showing the similarity between TRIM13 staining intensity and DNA staining (FxCycle) intensity. ∗P < .05, determined using one-way ANOVA, compared with empty vector (H). Results shown are mean ± SD from 3 biological replicates (H), histogram from 10 000 cells (A,C,D,E,G), or representative of 3 biological replicates (I,J).
TRIM13 ubiquitinates cell cycle regulatory proteins in AML cells
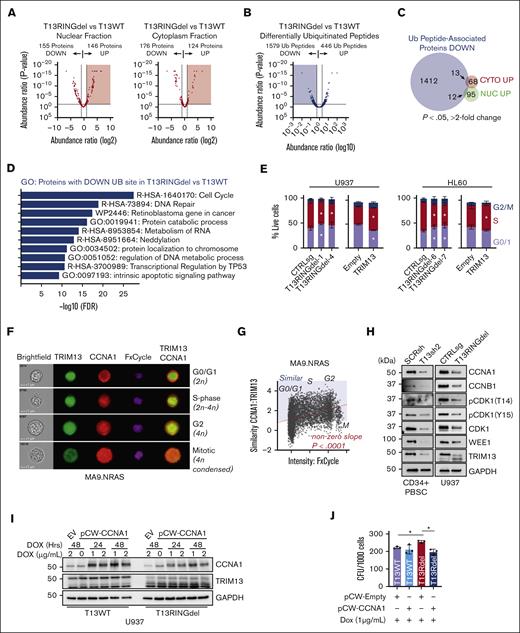
Previous studies have linked TRIM13 to driving the degradation of target proteins via ubiquitin-linked proteasomal degradation.19,36,44,45 We chose to focus on U937 cells for the mechanistic study of TRIM13 in AML because these cells responded most similarly to the PDX lines and displayed the greatest regulatory effect of TRIM13 by CHAF1B of the cell lines we tested. To identify potential degradation targets of TRIM13, we assayed the global proteome of T13WT and T13RINGdel U937 cells (supplemental Figure 5A; supplemental Table 3). To increase our depth of analysis, we also measured the nuclear/cytoplasmic fractions (Figure 6A; supplemental Figure 5B-C). To identify the specific degradation targets of TRIM13 because of ubiquitination, we also compared the differentially ubiquitinated peptides in T13RINGdel U937 cells vs T13WT (Figure 6B; supplemental Table 4). This yielded a substantial number of peptides with a loss of ubiquitination in T13RINGdel cells, suggesting that TRIM13 may have a wide variety of catalytic targets. To find potential degradation targets of TRIM13, we searched the overlap of proteins with both increased expression and loss of a ubiquitinated peptide in T13RINGdel U937 cells (25 total proteins; Figure 6C; supplemental Figure 5D). To attempt the confirmation of a target most likely to be proleukemic based on expression in adverse outcomes in AML, we selected the premRNA splicing factor WTAP. WTAP expression is increased in adverse outcome AML (supplemental Figure 5E), but we were not able to detect a significant change in expression in our nuclear or cytoplasmic fractions of the U937 AML cell line (supplemental Figure 5A).
TRIM13 stabilizes proteins associated with cell cycle. (A) Volcano plot comparing identified protein expression from T13WT with T13RINGdel in nuclear or cytoplasmic extracts. Results are calculated from 3 independent submissions. Red shaded region indicates proteins significantly upregulated (P < .05) in T13RINGdel compared with T13WT. (B) Volcano plot comparing differentially ubiquitinated peptides in T13RINGdel vs T13WT U937 cells. Blue shading indicates ubiquitinated peptides significantly lost in T13RINGdel (P < .05). (C) Venn diagram comparing differentially ubiquitinated peptides lost in T13RINGdel with proteins significantly upregulated (P < .05; more than twofold change in peptide spectrum match, or PSMs). (D) Gene ontology (GO) analysis of proteins with a ubiquitination site lost in T13RINGdel U937 cells. (E) Cell cycle (EdU vs FxCycle) analysis in multiple T13RINGdel clones and TRIM13 overexpression on day 5 after infection in each AML cell line. Bars indicate mean ± SD, and ∗P < .05 for each specific cell cycle phase within each cell type compared with CTRLsg, as determined using one-way ANOVA on 3 independent biological replicates. (F) Representative imaging cytometry of CCNA1 and TRIM13 localization through cell cycle in MA9.NRAS. (G) Similarity of CCNA1/TRIM13 through cell cycle. Red dashed line is line of best fit by linear regression. Nonzero slope calculated by linear regression modeling and P value listed. Blue shading indicates more similar TRIM13/CCNA1 than dissimilar, and cell cycle stages listed (G0/G1, S, G2, and M) as determined by DNA content and nuclear morphology. (H) Western blot analysis of PBSC expressing TRIM13sh 72-hours after infection and U937 T13RINGdel AML cell line at steady state. (I) Western blot analysis of T13WT vs T13RINGdel U937 cells expressing a dox-inducible CCNA1 at indicated time points after dox induction. Results shown are representative of 2 biological replicates. (J) Colony assay of indicated genotypes of U937 cells expressing either empty vector or CCNA1 in the presence of dox. Results shown are mean ± SD from 4 biological replicates, dots indicate individual replicates. ∗P < .05 between noted groups (E,H), as determined using one-way ANOVA with Bonferroni post hoc test (H).
TRIM13 stabilizes proteins associated with cell cycle. (A) Volcano plot comparing identified protein expression from T13WT with T13RINGdel in nuclear or cytoplasmic extracts. Results are calculated from 3 independent submissions. Red shaded region indicates proteins significantly upregulated (P < .05) in T13RINGdel compared with T13WT. (B) Volcano plot comparing differentially ubiquitinated peptides in T13RINGdel vs T13WT U937 cells. Blue shading indicates ubiquitinated peptides significantly lost in T13RINGdel (P < .05). (C) Venn diagram comparing differentially ubiquitinated peptides lost in T13RINGdel with proteins significantly upregulated (P < .05; more than twofold change in peptide spectrum match, or PSMs). (D) Gene ontology (GO) analysis of proteins with a ubiquitination site lost in T13RINGdel U937 cells. (E) Cell cycle (EdU vs FxCycle) analysis in multiple T13RINGdel clones and TRIM13 overexpression on day 5 after infection in each AML cell line. Bars indicate mean ± SD, and ∗P < .05 for each specific cell cycle phase within each cell type compared with CTRLsg, as determined using one-way ANOVA on 3 independent biological replicates. (F) Representative imaging cytometry of CCNA1 and TRIM13 localization through cell cycle in MA9.NRAS. (G) Similarity of CCNA1/TRIM13 through cell cycle. Red dashed line is line of best fit by linear regression. Nonzero slope calculated by linear regression modeling and P value listed. Blue shading indicates more similar TRIM13/CCNA1 than dissimilar, and cell cycle stages listed (G0/G1, S, G2, and M) as determined by DNA content and nuclear morphology. (H) Western blot analysis of PBSC expressing TRIM13sh 72-hours after infection and U937 T13RINGdel AML cell line at steady state. (I) Western blot analysis of T13WT vs T13RINGdel U937 cells expressing a dox-inducible CCNA1 at indicated time points after dox induction. Results shown are representative of 2 biological replicates. (J) Colony assay of indicated genotypes of U937 cells expressing either empty vector or CCNA1 in the presence of dox. Results shown are mean ± SD from 4 biological replicates, dots indicate individual replicates. ∗P < .05 between noted groups (E,H), as determined using one-way ANOVA with Bonferroni post hoc test (H).
Because of the small number of proteins that fit our original criteria, this led us to consider that TRIM13 may have a novel function in AML. Gene ontology analysis of our ubiquitination screen revealed a strong enrichment in proteins associated with cell cycle regulation (Figure 6D). We confirmed that T13RINGdel repressed cell cycle entry and TRIM13 overexpression enhanced cell cycle entry in U937 and HL60 cell lines (Figure 6E; supplemental Figure 5F). This led us to hypothesize that TRIM13 may be stabilizing cell cycle proteins through ubiquitination, and we found several cell cycle genes (CCNA1, CCNB1, and CDK1) containing peptides with high levels of ubiquitination in T13WT and undetectable ubiquitination in T13RINGdel (supplemental Table 4). We focused on CCNA1 because of its high degree of differential ubiquitination in T13RINGdel cells and its role in driving cell cycle progression and found that colocalization of CCNA1 and TRIM13 (Figure 6F) increased in a cell cycle stage–dependent manner in AML samples and cell lines (Figure 6G; supplemental Figure 5G).
We also found that the protein products of these key cell cycle regulatory genes are repressed in T13RINGdel U937 cells and TRIM13 knockdown PBSCs (and U937), compared with the mRNA products (Figure 6H; supplemental Figure 5H-I). These data suggest that TRIM13 may stabilize these cell cycle targets through its catalytic capacity as a ubiquitin ligase. To test this hypothesis, we caused the expression of CCNA1 using a doxycycline inducible lentivirus and found that T13RINGdel U937 cells were unable to sustain the levels of CCNA1 expression in T13WT (Figure 6I). In addition, we were able to functionally reverse the increase in clonogenic capacity of T13RINGdel U937 cells by re-expressing CCNA1 (Figure 6J).
TRIM13 stabilizes CCNA1 through ubiquitination
To test whether TRIM13 was involved in stabilizing CCNA1 through ubiqutination, we caused the epitope-tagged TRIM13 and CCNA1 to be overexpressed in 293T cells and demonstrated that these 2 proteins could form a precipitable complex (Figure 7A-B). We next made domain-deletion mutants of TRIM13 and found that the canonical protein-protein interaction domains (coiled-coil and B-box) of TRIM13 were predominantly responsible for mediating the interaction with CCNA1 (Figure 7C). We then found that CCNA1 is capable of being ubiquitinated by TRIM13 through most K-linkage conditions with 2 main exceptions, K11 and K33 (Figure 7D; supplemental Figure 6A). In addition, recombinant TRIM13 was able to directly ubiquitinate recombinant CCNA1 in cell-free ubiquitination assay using an E1 and an E2 panel. These experiments revealed that TRIM13 can use multiple E2s to ubiquitinate CCNA1, except for UbcH5a (Figure 7E; supplemental Figure 6B), which may explain the many different K-linked ubiquitin chains that we found built via CCNA1 by TRIM13 in Figure 7D. Because several of these noncanonical ubiquitination chains (including K27-linked) have been previously associated with protein stability and signaling,46 and TRIM13 catalytic inactivation was causing reduction in CCNA1 protein levels, we hypothesized that TRIM13-mediated ubiquitination was stabilizing CCNA1. In support of this hypothesis, it was found that the half-life of CCNA1 was reduced in T13RINGdel U937 cells exposed to a cycloheximide chase (Figure 7F-G). To test which degradation pathway of CCNA1 was being prevented by TRIM13, we inhibited the activity of either the proteosome (using MG-132, CTNNB1 as positive control for proteosome inhibitor function) or the lysosome (using Bafilomycin, LC3 as positive control for lysosome inhibitor function) and found that the TRIM13 catalytic function was to protect CCNA1 from lysosomal degradation (Figure 7G-H). We further confirmed lysosomal degradation of CCNA1 in the absence of TRIM13 by rescuing protein levels with chloroquine treatment (supplemental Figure 6C). Finally, we significantly rescued CCNA1 degradation in T13RINGdel U937 cells with lysosomal inhibition (Figure 7J-K). Together, these data demonstrate that TRIM13 stabilizes CCNA1 through ubiquitination, preventing its degradation by the lysosome.
TRIM13 stabilizes CCNA1 through ubiquitination. (A-B) Coimmunoprecipitation (Co-IP) of indicated factors 48 hours after transfection in 293T cells. (C) Co-IP of indicated TRIM13 domain mutants 48 hours after transfection in 293T cells. (D) Denaturing IP for FLAG-CCNA1 in 293T cells expressing TRIM13-V5 and indicated HA-tagged R-K ubiquitin. (E) Cell-free ubiquitination assay with a library of E2s. Bottom panel is ponceau stain, E1/GST-CCNA1/TRIM13-E2s are indicated. (Top) Blot for ubiquitin. (F) Western blot analysis of indicated genotypes U937 cells after cycloheximide chase. (G) Statistical analysis of CCNA1 half-life replicates. (H-J) Western blot analysis of T13WT and T13RINGdel U937 cells treated with indicated inhibitors: MG132 (H), Bafilomycin (BAF) (I-J), and cyclohexamide (CHX) (J). (K) Statistical analysis of CCNA1 half-life replicates in the presence of BAF. Results shown are representative of 3 biological replicates (A-D,F,H-J), or are mean + SD from 3 biological replicates (G,K). (G,K) Normalized to T13WT CCNA1 Veh level.
TRIM13 stabilizes CCNA1 through ubiquitination. (A-B) Coimmunoprecipitation (Co-IP) of indicated factors 48 hours after transfection in 293T cells. (C) Co-IP of indicated TRIM13 domain mutants 48 hours after transfection in 293T cells. (D) Denaturing IP for FLAG-CCNA1 in 293T cells expressing TRIM13-V5 and indicated HA-tagged R-K ubiquitin. (E) Cell-free ubiquitination assay with a library of E2s. Bottom panel is ponceau stain, E1/GST-CCNA1/TRIM13-E2s are indicated. (Top) Blot for ubiquitin. (F) Western blot analysis of indicated genotypes U937 cells after cycloheximide chase. (G) Statistical analysis of CCNA1 half-life replicates. (H-J) Western blot analysis of T13WT and T13RINGdel U937 cells treated with indicated inhibitors: MG132 (H), Bafilomycin (BAF) (I-J), and cyclohexamide (CHX) (J). (K) Statistical analysis of CCNA1 half-life replicates in the presence of BAF. Results shown are representative of 3 biological replicates (A-D,F,H-J), or are mean + SD from 3 biological replicates (G,K). (G,K) Normalized to T13WT CCNA1 Veh level.
Discussion
In this study, we found that CHAF1B exerts its proleukemic function by suppressing the transcription of TRIM13 by binding to the promoter (Figures 1 and 2). We then found that TRIM13 is a putative tumor suppressor because overexpression is detrimental to leukemogenesis (Figure 3), and the loss of TRIM13 catalytic activity enhances AML tumorigenicity concurrent with an immature phenotype (Figure 4). We found that TRIM13 has a novel nuclear localization in AML that is necessary for its tumor suppressor function (Figure 5) and that TRIM13 catalyzes the ubiquitination of many different cell cycle regulators (Figure 6). The catalytic ubiquitination activity of TRIM13 on these cell cycle regulators promotes their stability. We then found that TRIM13 can directly interact with CCNA1 and promote a variety of different K-linked ubiquitination events associated with protein stability by preventing lysosomal degradation (Figure 7).
In our AML models, we found that K27-linked ubiquitination of CCNA1 by TRIM13 led to a reduced degradation of the CCNA1 protein product by the lysosome (Figure 7). What was most striking, though, was that (1) CCNA1 could be ubiquitinated by essentially every known K-linkage of ubiquitin and (2) TRIM13 was promiscuous in the E2 required to catalyze CCNA1 ubiquitination in a cell-free system (Figure 7). In other models, when K63 ubiquitinated, TRIM13 has been shown to drive ER-phagy to degrade aggregates of proteins resistant to the normal autophagic degradation by the lysosomal degradation pathway.47 In addition, other studies have shown that clonogenicity can be negatively affected by TRIM13 because of ER stress and induction of autophagy via p62 in the adherent cell lines HEK293 and HeLa, though it should be noted that these are quite different from leukemic cells.43,47 Our proteomics data showed significant changes in the ubiquitination of lysine residues in these proteins that are not canonically associated with degradation, therefore we propose that this is a result of stabilizing ubiquitination by TRIM13 on the other residues (supplemental Table 4). Another possibility is that the ubiquitination of these factors by TRIM13 in the nucleus (instead of in the ER42) is contributing to their stability. In conclusion, we mechanistically demonstrated how the chromatin assembly machinery regulates the transcription of genes associated with cell cycle in a manner that promotes the immature state of leukemic cells.
CHAF1B (and the overall CAF1 complex) are functionally associated with the cell cycle, facilitating the assembly of histone H3/H4 heterodimers at the replication fork during S-phase. However, neither the overexpression nor depletion of CHAF1B alone is sufficient to drive changes in cell cycle in AML before differentiation (Figure 2H).5 In addition, these data suggest that CHAF1B loss may not lead to an immediate reduction in cell cycling in AML because the associated upregulation of TRIM13 promotes cell cycle entry and proliferation. This is supported by our observation that TRIM13 loss partially rescues the colony formation capacity of CHAF1B-depleted cells (supplemental Figure 4). When CHAF1B is depleted in cells lacking functional TRIM13, we propose that the progression into cell cycle is slowed, thereby allowing cells to potentially activate parallel antidifferentiation pathways and maintain their state throughout the process of cell division.
Some mutations or modulations observed in AMLs that cause precocious entry into cell cycle (FOXM1 deletion, a recent example) also cause loss of self-renewal and eventual exhaustion of the malignant stem cell pool.48,49 In our AML models, overexpression of TRIM13 leads to an increase in cell proliferation coupled with a loss of colony formation and in vivo leukemogenesis. Our observation that loss of TRIM13, in contrast, leads to both increased colony formation and a reduction in the number of cells entering cell cycle, suggests that it is beneficial for leukemic cells to control cell cycle entry by downregulating TRIM13. However, our experiments are unable to conclude if this effect is predominating in leukemia stem or progenitor cells. Based on our data, we propose that TRIM13 is detrimental to leukemogenesis in general by promoting premature entry into or progression through cell cycle.
A patient with Trisomy 21 developed acute megakaryoblastic leukemia in the absence of GATA1s, and it was later uncovered that there was a biallelic deletion of 13q14.2 and 13q14.3, the region that includes TRIM13. Though this study attributed acute megakaryoblastic leukemia progression to RB1 loss, which also resides in that region, our studies suggest that the TRIM13 loss could also contribute to the proleukemic state of those cells.50 Several studies suggest that TRIM13 promoter methylation is predominantly responsible for its transcriptional repression in some models.51-53 Although a reduction in TRIM13 expression is seen in the tumor of patients with CLL, the loss of transcriptional activity does not appear to be caused by promoter methylation.54 In contrast, a study focusing on pediatric patients with AML FAB M5 who showed specific patterns of hypermethylation around TRIM13, there was no significant change in the TRIM13 (or RFP2/DLEU5 by their notation) expression, although the DLEU1/2 genes were affected.53 We interpret our data considering this study by suggesting that TRIM13 is being repressed by CHAF1B binding to its promoter.
Acknowledgments
The authors acknowledge Nicolas Nassar for the thoughtful discussions and Charlie Nims and Levi Fox for their participation in the laboratory activities.
This work was supported by a seed grant from Ohio Cancer Research as well as National Institutes of Health grants (National Cancer Institute R00CA230314 and National Institute of General Medical Sciences R35GM142452) (A.G.V.). A.G.V. is supported by a Special Fellow Award from Leukemia and Lymphoma Society as well as a Junior Faculty Scholar Award from American Society of Hematology. All flow cytometric data were acquired using equipment maintained by the Research Flow Cytometry Core in the Division of Rheumatology at Cincinnati Children’s Hospital Medical Center, and the authors would like to acknowledge the support of the Coordinating Center of Excellence in Hematology grant U54DK126108 to Cincinnati Children’s Hospital Medical Center. The Proteomics data were collected on a mass spectrometer that was supported in part by a National Institutes of Health S10 Shared Instrumentation grant (S10OD026717) (K.D.G.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We dedicate this work to Mattie Louise Martin (1921-2020).
Authorship
Contribution: S.T.D., C.I., X.Z., S.W., J.K.J., and T.N. contributed to formal analysis, investigation, methodology, and manuscript review and editing; S.H. contributed to the investigation, methodology, and manuscript review; M. Wyder contributed to the investigation, mass spectrometry methodology, and manuscript review; N.S. contributed to the methodology, conceptualization, and RNA sequencing splicing analysis; M. Wunderlich contributed to resource acquisition, methodology, conceptualization, in vivo and PDX investigation, and manuscript review and editing; K.D.G. contributed to the methodology, conceptualization, mass spectrometry investigation, and manuscript review and editing; D.T.S. contributed to the formal analysis, methodology, and manuscript review and editing; and A.G.V. contributed to the funding and resource acquisition, formal analysis, investigation, methodology, writing of the original draft, and project administration.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andrew G. Volk, Division of Experimental Hematology and Cancer Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH 45229; e-mail: andrew.volk@cchmc.org.
References
Author notes
Next-generation sequencing data for this study have been deposited in the Gene Expression Omnibus database (accession number GSE231512).
Requests for resources or reagents should be directed to the corresponding author, Andrew G. Volk (andrew.volk@cchmc.org).
The full-text version of this article contains a data supplement.