Key Points
MTV and ctDNA did not correlate with evidence of disease 1 month after axi-cel in large B-cell lymphoma, suggesting ongoing therapy response.
ctDNA can aid the interpretation of a positive 1-month PET after CAR T-cell therapy for a comprehensive non-invasive response assessment.
Abstract
We examined the meaning of metabolically active lesions on 1-month restaging nuclear imaging of patients with relapsed/refractory large B-cell lymphoma receiving axicabtagene ciloleucel (axi-cel) by assessing the relationship between total metabolic tumor volume (MTV) on positron emission tomography (PET) scans and circulating tumor DNA (ctDNA) in the plasma. In this prospective multicenter sample collection study, MTV was retrospectively calculated via commercial software at baseline, 1, and 3 months after chimeric antigen receptor (CAR) T-cell therapy; ctDNA was available before and after axi-cel administration. Spearman correlation coefficient (rs) was used to study the relationship between the variables, and a mathematical model was constructed to describe tumor dynamics 1 month after CAR T-cell therapy. The median time between baseline scan and axi-cel infusion was 33 days (range, 1-137 days) for all 57 patients. For 41 of the patients with imaging within 33 days of axi-cel or imaging before that time but no bridging therapy, the correlation at baseline became stronger (rs, 0.61; P < .0001) compared with all patients (rs, 0.38; P = .004). Excluding patients in complete remission with no measurable residual disease, ctDNA and MTV at 1 month did not correlate (rs, 0.28; P = .11) but correlated at 3 months (rs, 0.79; P = .0007). Modeling of tumor dynamics, which incorporated ctDNA and inflammation as part of MTV, recapitulated the outcomes of patients with positive radiologic 1-month scans. Our results suggested that nonprogressing hypermetabolic lesions on 1-month PET represent ongoing treatment responses, and their composition may be elucidated by concurrently examining the ctDNA.
Introduction
Treatment with CD19-targeted chimeric antigen receptor (CAR) T-cell therapy has led to unprecedented response rates in patients with relapsed or refractory (R/R) large B-cell lymphomas (LBCL).1-3 Response determination 1 month after CAR T-cell therapy is critical to guide the clinical care decisions. However, interpretation of fluorine-18 fluorodeoxyglucose positron emission tomography/computed tomography (18F-FDG PET/CT) scan imaging can be challenging because of the lack of specificity of FDG-avid lesions, which may represent tumor, infection, and/or inflammation. Although tissue biopsy remains the gold standard for assessment, there is a strong desire to identify tumor noninvasively, efficiently, and accurately to minimize patients’ physical pain and emotional distress and expedite disease management.
We have previously demonstrated that metabolic tumor volume (MTV)4 and circulating tumor DNA (ctDNA),5,6 although based on different principles, can each serve to identify and quantify tumors noninvasively and provide for the prognostication of clinical response after CAR T-cell therapy.
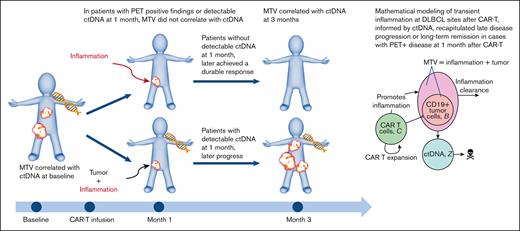
We hypothesized that inflammation generated by continued CAR T-cell antitumor activity contributes to FDG-avidity upon 18F-FDG PET/CT imaging and may confound standard radiological evaluation of response per the Lugano classification,7 particularly within a few months of CAR T-cell therapy administration. The primary objective of this retrospective study was to examine the relationship between MTV and ctDNA before and after axicabtagene ciloleucel (axi-cel) in patients with R/R LBCL in order to elucidate the etiology of nonprogressing lesions with FDG uptake of unclear significance on 18F-FDG PET/CT. Our secondary goal was to mathematically describe the ongoing treatment response to axi-cel after infusion and evaluate if inflammation may factor into the 1-month imaging assessment.
Methods
Patients
Seventy-two patients were originally enrolled in a prospective multiinstitutional study that assessed the role of ctDNA in prognostication before and after standard-of-care axi-cel in patients with R/R LBCL treated from February 2018 to June 2019 at Stanford University, Moffitt Cancer Center, and the University of Maryland Medical Center.6 Based on the baseline 18F-FDG PET/CT scan availability, 57 patients were included in the current retrospective study to form the entire cohort. To account for baseline MTV most closely representing disease at the time of treatment, patients who received bridging therapy with baseline 18F-FDG PET/CT imaging beyond the median time measured from the time of PET-CT imaging to infusion of axi-cel were excluded to create a focused cohort (n = 41). Bridging therapy was defined as any therapy in-between apheresis and axi-cel infusion used to control lymphoma. Clinical data was collected retrospectively. No biopsies were performed of FDG-avid lesions in patients that had not already exhibited frank progression per the Lugano response criteria. Approval for the review of patient records was obtained from each center’s institutional review board.
Tumor burden derivation
MTV was measured in mL, with a threshold >41% maximum standardized uptake value, and was calculated retrospectively in a step-wise process on 18F-FDG PET/CT performed before, and at 1 and 3 months after axi-cel using custom tools on MIM PACS version 6.8.4 (MIM Software Inc., Cleveland, OH), as described in our previous work.4 The processing time per scan was recorded in minutes. Baseline MTV results for 15 of the patients had been derived and reported previously.4 Official radiology responses per the Lugano criteria were available for all patients who had a month 1 and 3 restaging scan.
ctDNA values in lymphoma genomes per mL of plasma (Lg/mL) were previously derived via next-generation sequencing from plasma in a CFD tube (Roche Diagnostics, Indianapolis, IN) before lymphodepletion (baseline) and after axi-cel.6 The clonotype found at the highest concentration was tracked after first being identified via polymerase chain reaction amplification of rearranged immunoglobulin H (IgH)-VDJ, IgH-DJ, and Igκ/λregions using universal consensus primers from archival formalin-fixed, paraffin-embedded samples or from the initial plasma sample collected. Minimal residual disease sensitivity threshold was 10−6.
Mathematical modeling
To parameterize our mechanistic model, we used both previously established parameter values8 as well as patient-level data from the focused cohort for (MTV) and (ctDNA) at all available time points (described later in the article) for 2 representative patients. ctDNA () was assumed to enter the peripheral blood when the tumor died, consistent with empirical observations.9,10 The variable is the rate of degradation of ctDNA, which we assumed to be constant. The tumor B grows autonomously, with a birth rate , dies with rate , and experiences tumor killing at a rate γB, proportional to the number of CAR T cells. The parameter that modulates ctDNA includes with units (Lg/mL)/(mL), which can be thought of as the average amount of ctDNA introduced into the peripheral blood per mL of the tumor. The parameters are dimensionless and reflect the probability of ctDNA being released through cell death. represents the degree of inflammation caused by the CAR T cells killing the tumor, and is the clearance rate at which inflammation is removed. All parameters are assumed to be positive, and .
We integrated the mathematical model and data from the focused cohort in the following way: for model fitting, we used the Julia packages DifferentialEquations, Optim, and BlackBoxOptim, which would find a set of best-fit parameters for equations 4 and 5 assuming previously established cancer cell proliferation rates8 of 0.15 per day. This provided a set of best-fit parameters for individual patients, describing the kinetics of ctDNA and tumor volume/inflammation according to equations 3 to 5. We selected to present and discuss the fits for 2 representative patients based upon day 30 staging results: 1 PET+/ctDNA+ and 1 PET+/ctDNA–.
Statistical analysis
All correlations between MTV and ctDNA were performed by Spearman correlation and the coefficient (rs) was reported.
The median follow-up for survivors was calculated between the date of CAR T-cell administration and the date of last contact, and overall survival (OS) and progression-free survival (PFS) were calculated since the time of CAR T-cell infusion until death, progression, or last contact. Differences in OS and PFS were found via the Kaplan-Meier and log-rank tests, and hazard ratios (HRs) and 95% confidence intervals (95% CIs) were reported. Survival was reported for the focused cohort at baseline per low vs high tumor burden group, defined by our previously derived and validated baseline MTV cutoff value of 147.5 mL4 and prelymphodepletion ctDNA cutoff of 100 Lg/mL.6 Overall response, including partial (PR) and complete response (CR) at 3 and 6 months, respectively, as well as the overall response and CR if achieved by the last follow-up, were reported for the entire cohort. Cytokine release syndrome and CAR T-cell related encephalopathy syndrome rates were reported for the entire cohort.
P < .05 was defined as statistically significant. Analysis was conducted using GraphPad Prism version 9.0.2 (161) for Windows (GraphPad Software, San Diego, CA; www.graphpad.com).
Results
Patient characteristics
Baseline patient characteristics and clinical outcomes for all patients are presented in Table 1. The median time between baseline 18F-FDG PET/CT and administration of CAR T-cell therapy was 33 days (range, 1-137 days; interquartile range [IQR], 44.5 days).
Patient characteristics at the time of axi-cel infusion and clinical outcomes (n = 57)
| Characteristic . | N = 57 (%) . |
|---|---|
| Age (y) | |
| Median, range | 59, 19-76 |
| Sex | |
| Male | 34 (60) |
| ECOG (0-5) | |
| 0-1 | 54 (95) |
| 2 | 3 (5) |
| Histology | |
| DLBCL, NOS | 32 (56) |
| Unknown MYC and BCL2/BCL6 status | 4 (7) |
| High grade B-cell lymphoma, with MYC and BCL2 and/or BCL6 rearrangements | 10 (18) |
| B-cell lymphoma, unclassifiable, with features intermediate between DLBCL and classical Hodgkin lymphoma | 11 (19) |
| Unknown MYC and BCL2/BCL6 status | 3 (5) |
| Primary mediastinal B-cell lymphoma | 4 (7) |
| Stage (I-IV) | |
| I/II | 15 (26) |
| III/IV | 42 (74) |
| LDH level before conditioning > ULN | |
| Yes | 37 (65) |
| CRP level before conditioning > ULN | |
| Yes | 25 (44) |
| Ferritin level before conditioning > ULN | |
| Yes | 31 (54) |
| IPI score (1-5) | |
| 0 | 4 (7) |
| 1-2 | 23 (40) |
| 3-5 | 25 (44) |
| N/A or primary mediastinal B-cell lymphoma | 5 (9) |
| Prior lines of therapy | |
| Median, range | 3, 1-7 |
| Bridging therapy | |
| Yes | 33 (58) |
| Chemotherapy/targeted therapy | 15 (26) |
| Steroids | 3 (5) |
| Radiation therapy | 7 (12) |
| Combination chemotherapy/targeted therapy ± steroids ± radiation therapy | 8 (14) |
| Received before baseline 18F-FDG PET/CT | 10 (18) |
| N/A | 1 (2) |
| Characteristic . | N = 57 (%) . |
|---|---|
| Age (y) | |
| Median, range | 59, 19-76 |
| Sex | |
| Male | 34 (60) |
| ECOG (0-5) | |
| 0-1 | 54 (95) |
| 2 | 3 (5) |
| Histology | |
| DLBCL, NOS | 32 (56) |
| Unknown MYC and BCL2/BCL6 status | 4 (7) |
| High grade B-cell lymphoma, with MYC and BCL2 and/or BCL6 rearrangements | 10 (18) |
| B-cell lymphoma, unclassifiable, with features intermediate between DLBCL and classical Hodgkin lymphoma | 11 (19) |
| Unknown MYC and BCL2/BCL6 status | 3 (5) |
| Primary mediastinal B-cell lymphoma | 4 (7) |
| Stage (I-IV) | |
| I/II | 15 (26) |
| III/IV | 42 (74) |
| LDH level before conditioning > ULN | |
| Yes | 37 (65) |
| CRP level before conditioning > ULN | |
| Yes | 25 (44) |
| Ferritin level before conditioning > ULN | |
| Yes | 31 (54) |
| IPI score (1-5) | |
| 0 | 4 (7) |
| 1-2 | 23 (40) |
| 3-5 | 25 (44) |
| N/A or primary mediastinal B-cell lymphoma | 5 (9) |
| Prior lines of therapy | |
| Median, range | 3, 1-7 |
| Bridging therapy | |
| Yes | 33 (58) |
| Chemotherapy/targeted therapy | 15 (26) |
| Steroids | 3 (5) |
| Radiation therapy | 7 (12) |
| Combination chemotherapy/targeted therapy ± steroids ± radiation therapy | 8 (14) |
| Received before baseline 18F-FDG PET/CT | 10 (18) |
| N/A | 1 (2) |
| Outcome . | n= 57 (%) . |
|---|---|
| Clinical response to ax-cel | |
| CR by last follow-up | 35 (61) |
| ORR by last follow-up | 52 (91) |
| ORR at 3 months | 31 (54) |
| ORR at 6 months | 26 (46) |
| Follow-up and survival | |
| Median follow-up for survivors in months (range) | 20.7 (2.7-32.9) |
| Median OS in months (95% CIs) | Not reached |
| Median PFS in months (95% CIs) | 13.4 (12.2-13.5) |
| Toxicity, grade | |
| CRS 1-3 | 50 (88) |
| 3 | 1 (2) |
| N/A | 3 (5) |
| CRES 1-4 | 30 (53) |
| 3-4 | 15 (26) |
| N/A | 3 (5) |
| Outcome . | n= 57 (%) . |
|---|---|
| Clinical response to ax-cel | |
| CR by last follow-up | 35 (61) |
| ORR by last follow-up | 52 (91) |
| ORR at 3 months | 31 (54) |
| ORR at 6 months | 26 (46) |
| Follow-up and survival | |
| Median follow-up for survivors in months (range) | 20.7 (2.7-32.9) |
| Median OS in months (95% CIs) | Not reached |
| Median PFS in months (95% CIs) | 13.4 (12.2-13.5) |
| Toxicity, grade | |
| CRS 1-3 | 50 (88) |
| 3 | 1 (2) |
| N/A | 3 (5) |
| CRES 1-4 | 30 (53) |
| 3-4 | 15 (26) |
| N/A | 3 (5) |
CRES, CAR-T-cell-related encephalopathy syndrome; CRP, C-reactive protein; CRS, cytokine release syndrome; DLBCL, diffuse large B-cell lymphoma; ECOG, Eastern Cooperative Oncology Group performance status; LDH, lactate dehydrogenase; IPI, International Prognostic Index; N/A, not applicable; NOS, not otherwise specified; ORR, overall response rate; ULN, upper limit or normal.
Biomarker values and correlation
Baseline and available posttreatment MTV and ctDNA values are reported in Tables 2 and 3. The median processing time of a scan at baseline (n = 42) was 30 minutes (range, 8-217 minutes; IQR, 84.5 minutes); at 1 month (n = 47), was 8 minutes (range, 2-180 minutes; IQR, 4 minutes); and at 3 months (n = 35), was 5 min (range, 2-145 minutes; IQR, 11 minutes). Processing time per scan correlated strongly with MTV at baseline (rs, 0.71; P < .0001), 1 month (rs, 0.81; P < .0001), and 3 months (rs, 0.89; P < .0001).
MTV and ctDNA values at the times of PET assessment before and after axi-cel
| Biomarker . | Baseline . | 1 month . | 3 months . | |
|---|---|---|---|---|
| MTV [mL] | n = 57 | N = 41 | N = 56∗ | N = 46† |
| Entire cohort | Focused cohort | |||
| Median | 52.93 | 42.68 | 0.74 | 0 |
| Range | 2.3-2256.24 | 2.3-2106.56 | 0-3369.13 | 0-697.97 |
| IQR | 283.3 | 130.24 | 12.06 | 15.73 mL |
| ctDNA [Lg/mL] | n = 57‡ | n = 41 | n = 54§ | n = 42‖ |
| Entire cohort | Focused cohort | |||
| Median | 95.09 | 58.1 | 0 | 0 |
| Range | 0-17903.02 | 0-6542.75 | 0-2944.85 | 0-10291.34 |
| IQR | 878.84 | 856.89 | 11.98 | 13.67 |
| Biomarker . | Baseline . | 1 month . | 3 months . | |
|---|---|---|---|---|
| MTV [mL] | n = 57 | N = 41 | N = 56∗ | N = 46† |
| Entire cohort | Focused cohort | |||
| Median | 52.93 | 42.68 | 0.74 | 0 |
| Range | 2.3-2256.24 | 2.3-2106.56 | 0-3369.13 | 0-697.97 |
| IQR | 283.3 | 130.24 | 12.06 | 15.73 mL |
| ctDNA [Lg/mL] | n = 57‡ | n = 41 | n = 54§ | n = 42‖ |
| Entire cohort | Focused cohort | |||
| Median | 95.09 | 58.1 | 0 | 0 |
| Range | 0-17903.02 | 0-6542.75 | 0-2944.85 | 0-10291.34 |
| IQR | 878.84 | 856.89 | 11.98 | 13.67 |
Because of unavailable PET, MTV could not be derived for 1 patient not in CR and was marked as 0 mL for 8 patients in CR.
Similarly, MTV could not be derived for 2 patients not in CR and marked as 0 mL for 10 patients in CR.
ctDNA on day 0 was used as baseline in 1 patient without prelymphodepletion ctDNA.
ctDNA was not checked in 3 patients without disease progression.
ctDNA was not checked in 11 patients without disease progression.
ctDNA values at additional time points before and after axi-cel
| . | Day 0 . | Day 7 . | Day 14 . | Day 21 . | Day 56 . | Day 180 . | Day 270 . | Day 300 . | Day 360 . |
|---|---|---|---|---|---|---|---|---|---|
| ctDNA | n = 43 | n = 57 | n = 54 | n = 49 | n = 35 | n = 26 | n = 12 | n = 1 | n = 20 |
| Median | 16.14 | 4.85 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Range | 0-12449 | 0-31664 | 0-14551 | 0-1540 | 0-8131 | 0-4916 | 0-1637 | 0-0 | 0-3180 |
| IQR | 342.2 | 101.8 | 28.06 | 30.6 | 7.850 | 0.1725 | 0 | 0 | 0 |
| . | Day 0 . | Day 7 . | Day 14 . | Day 21 . | Day 56 . | Day 180 . | Day 270 . | Day 300 . | Day 360 . |
|---|---|---|---|---|---|---|---|---|---|
| ctDNA | n = 43 | n = 57 | n = 54 | n = 49 | n = 35 | n = 26 | n = 12 | n = 1 | n = 20 |
| Median | 16.14 | 4.85 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Range | 0-12449 | 0-31664 | 0-14551 | 0-1540 | 0-8131 | 0-4916 | 0-1637 | 0-0 | 0-3180 |
| IQR | 342.2 | 101.8 | 28.06 | 30.6 | 7.850 | 0.1725 | 0 | 0 | 0 |
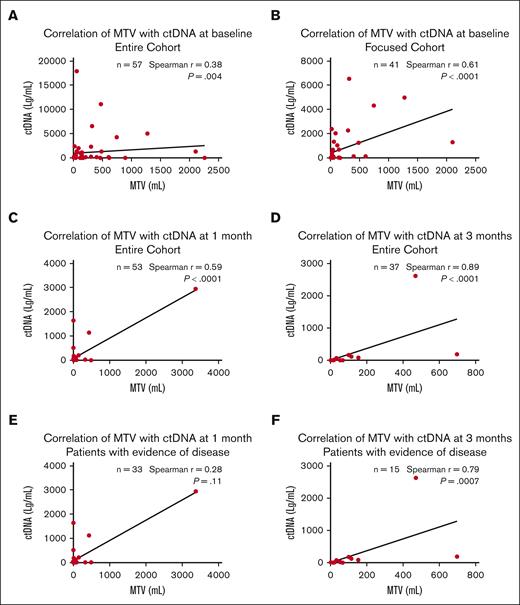
We correlated MTV and ctDNA values at baseline, 1 month, and 3 months after axi-cel. At baseline, in the entire cohort, MTV correlated weakly with ctDNA (rs, 0.38; P = .004); in the focused cohort, the correlation became stronger (rs, 0.61; P < .0001) (Figure 1A,B). We evaluated the impact of bridging therapy on measurements of baseline tumor burden. For 23 patients who received bridging therapy after baseline imaging, there was no correlation of MTV and ctDNA (rs, 0.08; P = .71). However, for 34 patients who did not receive bridging therapy or received it before baseline imaging, there was a significant correlation (rs, 0.57; P = .0004). The timing of the baseline scan in relation to axi-cel administration also affected the relationship between MTV and ctDNA, with significant correlation found when the scans were performed closer to therapy but not otherwise (quartile [Q] 1 rs, 0.78 [P = .0008]; Q2 rs, 0.58 [P = .0296]; Q3 rs, 0.075; [P = .79]; and Q4 rs, −0.066 [P = .82]). At 1 month after CAR T-cell therapy, in the entire cohort (n = 53), MTV had a moderate positive correlation with ctDNA (rs, 0.59; P < .0001; Figure 1C). At 3 months, in the entire cohort (n = 37), the correlation between the 2 biomarkers was the strongest (rs, 0.89; P < .0001; Figure 1D). When patients with undetectable MTV and ctDNA were excluded, the correlation between the biomarkers became nonsignificant at 1 month (n = 33; rs, 0.28; P = .11; Figure 1E) but remained significant at 3 months (n = 15; rs, 0.79; P = .0007; Figure 1F).
Correlation of MTV with ctDNA at baseline, 1 month, and 3 months. The correlation of baseline MTV with ctDNA strengthened in the focused cohort (B) compared with the entire cohort (A). The correlation was moderate at 1 month (C) and the strongest at 3 months (D). For patients with evidence of disease, there was no correlation at 1 month (E), but the correlation persisted at 3 months (F).
Correlation of MTV with ctDNA at baseline, 1 month, and 3 months. The correlation of baseline MTV with ctDNA strengthened in the focused cohort (B) compared with the entire cohort (A). The correlation was moderate at 1 month (C) and the strongest at 3 months (D). For patients with evidence of disease, there was no correlation at 1 month (E), but the correlation persisted at 3 months (F).
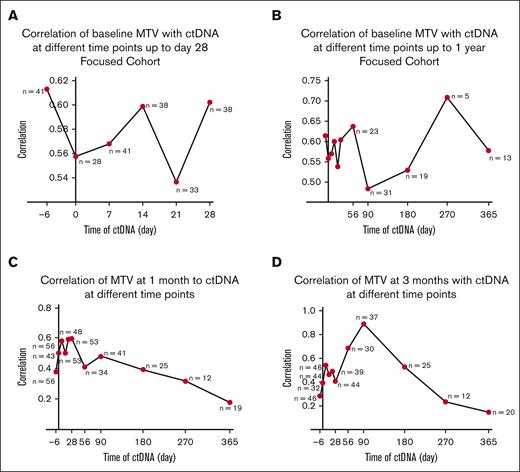
We further evaluated the relationship between MTV obtained at each of the 3 time points to all ctDNA values. In the focused cohort, baseline MTV correlated best with ctDNA at prelymphodepletion (rs, 0.61; P < .0001). The correlation varied more during the first month compared with later time points (Figure 2A). The correlation after day 28 was overall poorer (Figure 2B). In the entire cohort, MTV at 1 month correlated best with ctDNA on day 28 (n = 53; rs, 0.60; P < .0001; Figure 2C), and MTV at 3 months correlated with ctDNA on day 90 (n = 37; rs, 0.89; P < .0001; Figure 2D).
Correlation of baseline, 1 month, and 3 months MTV with ctDNA at different time points. (A-B) Baseline MTV correlated best with ctDNA at prelymphodepletion (n = 41) (rs, 0.61; P < .0001) within the first month and overall: day 0 (n = 28) (rs, 0.56; P = .002), day 7 (n = 41) (rs, 0.57; P = .0001), day 14 (n = 38) (rs, 0.6; P < .0001), day 21 (n = 33) (rs, 0.54; P = .001), day 28 (n = 38) (rs, 0.6; P < .0001), day 56 (n = 23) (rs, 0.64; P = .001), day 90 (n = 31) (rs, 0.48; P = .006), day 180 (n = 19) (rs, 0.53; P = .02), day 270 (n = 5) (rs, 0.71; P = .4), and day 365 (n = 13) (rs, 0.58; P = .04). (C) MTV at 1 month correlated best with ctDNA on day 28 (n = 53) (rs, 0.60; P < .0001): prelymphodepletion (n = 56) (rs, 0.38; P = .004), day 0 (n = 43) (rs, 0.50; P = .0007), day 7 (n = 56) (rs, 0.58; P < .0001), day 14 (n = 53) (rs, 0.50; P = .0001), day 21 (n = 48) (rs, 0.59; P < .0001), day 56 (n = 34) (rs, 0.41; P = .02), day 90 (n = 41) (rs, 0.48; P = .002), day 180 (n = 25) (rs, 0.39; P = .05), day 270 (n = 12) (rs, 0.32; P = .15), and day 365 (n = 19) (rs, 0.18; P = .47). (D) MTV at 3 months correlated best with ctDNA on day 90 (n = 37) (rs, 0.89; P < .0001): prelymphodepletion (n = 46) (rs, 0.28; P = .06), on day 0 (n = 32) (rs, 0.39; P = .03), day 7 (n = 46) (rs, 0.54; P = .0001), day 14 (n = 44) (rs, 0.46; P = .002), day 21 (n = 39) (rs, 0.49; P = .002), day 28 (n = 44) (rs, 0.41; P = .006), day 56 (n = 30) (rs, 0.69; P < .0001), day 180 (n = 25) (rs, 0.53; P = .007), day 270 (n = 12) (rs, 0.23; P = .64), day 365 (n = 20) (rs, 0.15; P = .54).
Correlation of baseline, 1 month, and 3 months MTV with ctDNA at different time points. (A-B) Baseline MTV correlated best with ctDNA at prelymphodepletion (n = 41) (rs, 0.61; P < .0001) within the first month and overall: day 0 (n = 28) (rs, 0.56; P = .002), day 7 (n = 41) (rs, 0.57; P = .0001), day 14 (n = 38) (rs, 0.6; P < .0001), day 21 (n = 33) (rs, 0.54; P = .001), day 28 (n = 38) (rs, 0.6; P < .0001), day 56 (n = 23) (rs, 0.64; P = .001), day 90 (n = 31) (rs, 0.48; P = .006), day 180 (n = 19) (rs, 0.53; P = .02), day 270 (n = 5) (rs, 0.71; P = .4), and day 365 (n = 13) (rs, 0.58; P = .04). (C) MTV at 1 month correlated best with ctDNA on day 28 (n = 53) (rs, 0.60; P < .0001): prelymphodepletion (n = 56) (rs, 0.38; P = .004), day 0 (n = 43) (rs, 0.50; P = .0007), day 7 (n = 56) (rs, 0.58; P < .0001), day 14 (n = 53) (rs, 0.50; P = .0001), day 21 (n = 48) (rs, 0.59; P < .0001), day 56 (n = 34) (rs, 0.41; P = .02), day 90 (n = 41) (rs, 0.48; P = .002), day 180 (n = 25) (rs, 0.39; P = .05), day 270 (n = 12) (rs, 0.32; P = .15), and day 365 (n = 19) (rs, 0.18; P = .47). (D) MTV at 3 months correlated best with ctDNA on day 90 (n = 37) (rs, 0.89; P < .0001): prelymphodepletion (n = 46) (rs, 0.28; P = .06), on day 0 (n = 32) (rs, 0.39; P = .03), day 7 (n = 46) (rs, 0.54; P = .0001), day 14 (n = 44) (rs, 0.46; P = .002), day 21 (n = 39) (rs, 0.49; P = .002), day 28 (n = 44) (rs, 0.41; P = .006), day 56 (n = 30) (rs, 0.69; P < .0001), day 180 (n = 25) (rs, 0.53; P = .007), day 270 (n = 12) (rs, 0.23; P = .64), day 365 (n = 20) (rs, 0.15; P = .54).
Baseline biomarker association with survival
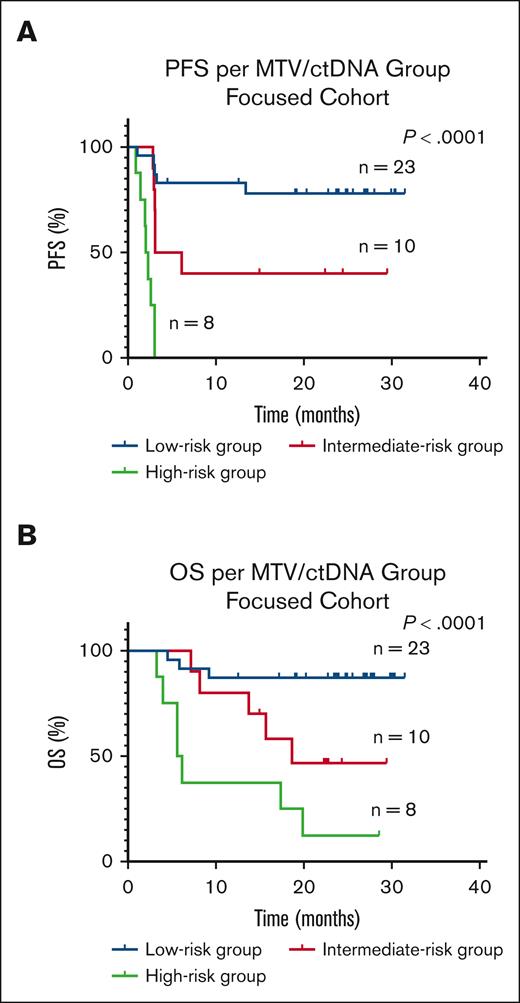
At baseline, in the focused cohort, 32 patients had low (range, 2.3-145.86 mL) and 9 patients had high (range, 166.18-2106.56 mL) MTV; 24 patients had low (range, 0-98.16 Lg/mL) and 17 patients had high (range, 127.92-6542.75 Lg/mL) ctDNA. Low vs high MTV, and low vs high ctDNA, were associated with better PFS (HR, 0.17; 95% CI, 0.03-0.71; P < .0001 vs HR, 0.11; 95% CI, 0.04-0.3; P < .0001) and OS (HR, 0.19; 95% CI, 0.04-0.8; P = .0003 vs HR, 0.13; 95% CI, 0.04-0.39; P = .0002), respectively. Patients with low MTV/low ctDNA (low-risk group) had better survival compared with those with either low MTV/high ctDNA or high MTV/low ctDNA (intermediate-risk group) or high MTV/ high ctDNA (high-risk group) (Figure 3).
Kaplan-Meier survival curves and log-rank P values in the focused cohort at baseline by risk group. (A) PFS (B) OS is worse in the high-risk group (high MTV/high ctDNA) compared with the intermediate-risk (high MTV/low ctDNA or low MTV/high ctDNA) or low-risk (low MTV/low ctDNA) groups. MTV cutoff was >147.5 mL and ctDNA cutoff was >100 Lg/mL.
Kaplan-Meier survival curves and log-rank P values in the focused cohort at baseline by risk group. (A) PFS (B) OS is worse in the high-risk group (high MTV/high ctDNA) compared with the intermediate-risk (high MTV/low ctDNA or low MTV/high ctDNA) or low-risk (low MTV/low ctDNA) groups. MTV cutoff was >147.5 mL and ctDNA cutoff was >100 Lg/mL.
Patterns of lymphoma response and progression
From baseline to 1 month, 51 patients had a confirmed decrease in MTV on imaging (n = 56), with a median reduction of MTV of 47.78 mL (range, 1.91-2251 mL; IQR, 235.63 mL). The day 30 scan of 1 patient with PR was unavailable for review. This correlated to the Lugano response criteria, in which 4 patients progressed and 2 patients had stable disease (SD) (of which 1 had increased MTV, and the other had reduced MTV). The scans of 19 patients were available for review at the time of progression: 15 patients had an increase in MTV >50% compared with that observed in the prior scan, with half of them developing new lesions in addition to an increase in FDG uptake in a lesion.
Responses to axi-cel are shown over time (Figure 4). At 1 month after axi-cel, 28 patients were in CR: 5 patients had positive ctDNA, with 2 patients experiencing progression of disease; 21 patients had negative ctDNA, with 14 remaining in CR; and 2 patients who remained in CR did not have ctDNA values available. At the same assessment time point, 25 patients were in PR: 14 patients had positive ctDNA, with 13 whose disease progressed; 8 had negative ctDNA, with 6 converting to CR; ctDNA was unavailable for 1 patient with progression of disease. There were 2 patients with SD: 1 who had positive ctDNA had progression of the disease by 3 months, and the other who had negative ctDNA achieved CR by 1 year. All 4 patients with progression of disease at 1 month had positive ctDNA.
Responses to axi-cel shown in relation with time. The response of the 57 patients is shown in relation with time. Responses evolved initially with most becoming final by 3 months. The patients with positive ctDNA at 1 month are marked with a dash by the colored boxes and their outcome tracked. At 1 month, 3 patients in CR who remained in durable remission had detectable ctDNA, whereas 5 of 6 patients who progressed by month 6 had undetectable ctDNA. Six of 7 patients initially in PR who achieved CR by 6 months had ctDNA 0 Lg/mL at 1 month. One of 2 patients initially in SD had 0 Lg/mL ctDNA at all available time points before and after axi-cel. Fourteen of 17 PR/ SD who had partial disease had detectable ctDNA at 1 month (1 was unavailable).
Responses to axi-cel shown in relation with time. The response of the 57 patients is shown in relation with time. Responses evolved initially with most becoming final by 3 months. The patients with positive ctDNA at 1 month are marked with a dash by the colored boxes and their outcome tracked. At 1 month, 3 patients in CR who remained in durable remission had detectable ctDNA, whereas 5 of 6 patients who progressed by month 6 had undetectable ctDNA. Six of 7 patients initially in PR who achieved CR by 6 months had ctDNA 0 Lg/mL at 1 month. One of 2 patients initially in SD had 0 Lg/mL ctDNA at all available time points before and after axi-cel. Fourteen of 17 PR/ SD who had partial disease had detectable ctDNA at 1 month (1 was unavailable).
For the patients with evidence of disease at 1 month (n = 33), 19 had both detectable MTV and ctDNA, with 18 experiencing progression of disease (4 at 1 month, 13 at 3 months, and 1 at 6 months) and 1 achieving CR at 3 months. There were 9 patients with detectable MTV but no ctDNA, of which 7 achieved CR (1 marked as CR based on the Lugano criteria at 1 month, 3 at 3 months, 2 at 6 months, and 1 at 1 year), whereas 2 had progression of disease at 3 months. The remaining 5 of the 33 patients with undetectable MTV but detectable ctDNA, 3 remained in CR, whereas 2 progressed (1 at 3 months and 1 at 1 year).
Dynamics of tumor burden, ctDNA, and inflammation
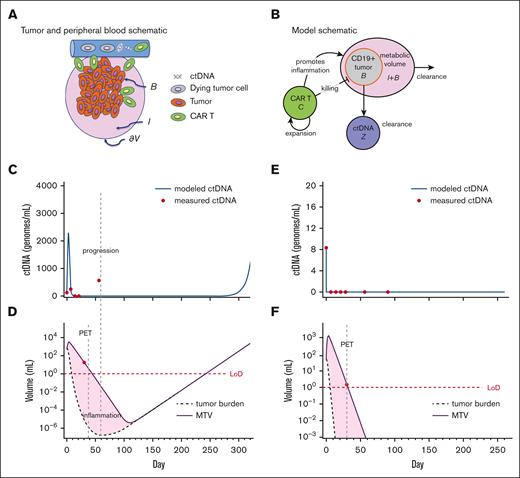
Given the uncertain response or durability of response at 1 month after CAR T-cell therapy based on imaging alone, we used a deterministic version of our previously described mathematical model of CAR T-cell dynamics,8 expanded to include ctDNA and inflammation as part of the MTV measurement (Figure 5A). We assumed that a portion of metabolic activity near or at the tumor site may be related to inflammation associated with the completed or ongoing killing of cancer cells by CAR T cells (Figure 5B). The mathematical model was fitted to 2 patients with PET+ disease on day 30 and adequate longitudinal data to compare with that of the model (Methods): 1 patient who was tested PET+ and ctDNA+ on day 30 and later progressed, and 1 who was tested PET+ but ctDNA– on day 30 and achieved long-term remission. Comparing these cases, the model recapitulated the dynamics of the first 30 or 60 days but showed some discrepancy at later time points (Figure 5C-F) and correctly recapitulated progression (Figure 5C,D) and durable remission (Figure 5E,F).
Dynamical systems model of ctDNA, tumor, and inflammation. (A) Qualitative schematic of the tumor burden (represented as B), surrounded by inflammation (represented as I), and ctDNA in the peripheral blood. (B) Compartment model schematic, including ctDNA (represented as Z). (C) Model kinetics inferred from a patient with MTV+/ctDNA+ undergoing CAR T-cell therapy with ctDNA measurements (dots). The curve represents the modeled (fitted) ctDNA compartment dynamics. (D) Inferred model kinetics of MTV (I + B, solid line) and tumor alone (B, dashed) of the same patient. MTV is measured from the PET scan at day 30 (red dot). This modeling successfully captures disease progression, although delayed compared with the actual clinical progression because of the tumor burden. (E) and (F) Inferred model kinetics for a patient who was tested MTV+/ctDNA– on day 30 and after. Here, the MTV eventually consists entirely of inflammation and is still undetectable at day 365, in line with the cDNA kinetics and the end point of complete response. LoD, estimated level of detection of 1 mL.
Dynamical systems model of ctDNA, tumor, and inflammation. (A) Qualitative schematic of the tumor burden (represented as B), surrounded by inflammation (represented as I), and ctDNA in the peripheral blood. (B) Compartment model schematic, including ctDNA (represented as Z). (C) Model kinetics inferred from a patient with MTV+/ctDNA+ undergoing CAR T-cell therapy with ctDNA measurements (dots). The curve represents the modeled (fitted) ctDNA compartment dynamics. (D) Inferred model kinetics of MTV (I + B, solid line) and tumor alone (B, dashed) of the same patient. MTV is measured from the PET scan at day 30 (red dot). This modeling successfully captures disease progression, although delayed compared with the actual clinical progression because of the tumor burden. (E) and (F) Inferred model kinetics for a patient who was tested MTV+/ctDNA– on day 30 and after. Here, the MTV eventually consists entirely of inflammation and is still undetectable at day 365, in line with the cDNA kinetics and the end point of complete response. LoD, estimated level of detection of 1 mL.
Discussion
The results of our multicenter, prospective sample study in patients with R/R LBCL receiving axi-cel demonstrated an ongoing treatment response in patients with nonprogressing FDG-avid lesions on 1-month posttherapy 18F-FDG PET/CT scans. The lack of correlation between quantifiable MTV and detectable ctDNA implied activity, either continued cancer killing and/or inflammation, at known prior disease sites. Because patients had not undergone tissue biopsies, we used a modified version of an existing mathematical model of the dynamics of CAR T-cell treatment at 1 month that successfully recapitulated clinical responses in patients with PR or SD. Our results indicated that plasma ctDNA served as a physical biologic identifier of a radiologically visualized, metabolically active, residual tumor. Given the test’s low level of detection, its absence suggested that FDG-avid lesions likely represented localized inflammation induced by CAR T-cell therapy. Thus, we showed that concurrent ctDNA added specificity to the 1-month positive restaging 18F-FDG PET/CT.
Our work focused on developing comprehensive noninvasive response assessments of patients undergoing CAR T-cell therapy. We learned about the imaged tumor’s response through the use of consecutively detected ctDNA via next-generation sequencing. LBCL ctDNA monitoring has been shown to be a promising technique with prognostic potential for accurate identification and quantification of disease at diagnosis,11 during and after first-line therapy,12 and before13 and after CAR T-cell therapy.5,6,13 An alternative technique of low-pass whole-genome sequencing of cell-free DNA to find somatic copy number alterations has also been investigated in patients with LBCL treated with CAR T-cell therapy,14 and in one study, results at baseline were combined with surrogates of disease burden, lactate dehydrogenase, and the number of extranodal sites to prognosticate clinical outcomes.15 Liquid biopsies can be particularly advantageous in cases for which tissue biopsies are not feasible or the risks outweigh the benefits, as in patients with PR or SD 1 month after CAR T-cell therapy with small residual FDG-avid lesions on imaging.
By accurately and efficiently deriving MTV, we quantified the total amount of metabolic activity on imaging. This allowed us to go beyond recognizing a scan result as a binary positive or negative for disease. Importantly, baseline MTV and ctDNA are concordant, particularly when removing the impacts of bridging therapy and timing. Both high-baseline ctDNA and MTV were associated with poor clinical outcomes. This is a redemonstration of ctDNA’s value as an emerging biomarker within a proportion (n = 57) of the same cohort of patients we reported on previously (n = 69)6 and another validation of MTV as an imaging biomarker in a largely unique patient cohort (n = 42/57).4 Combining the 2 biomarkers, patients in the low-risk group before axi-cel showed significantly better PFS and OS than those in the intermediate- or high-risk groups. This prognostic model is yet to be validated prospectively but may be possible after the completion of an ongoing clinical study (NCT05255354).
Here, the lack of correlation between ctDNA and MTV at 1 month may be explained by the mechanism of action of CAR T-cell therapy and the ongoing treatment response. Oscillations between the 2 variables during the first month after infusion likely resulted from active tumor killing induced directly or indirectly by axi-cel, which has a median time to radiologic response of 0.9 months.16 The death of the tumor was confirmed by the reduction of median MTV and ctDNA in our nonprogressing patients at 1 month. Although LBCL tumor typically consists of up to 90% tumor cells causing FDG uptake on scans,17 we suspected localized inflammation caused by the CAR T-cell therapy clearing tumor contributed to residual FDG-avid lesions on 1-month restaging 18F-FDG PET/CT. Loss of correlation at 1 month, but not 3 months, after therapy for those patients with either persistent MTV, ctDNA, or both was likely due to ongoing treatment response.
To reproduce tumor response at 1 month after axi-cel, we proposed a mathematical model that is an extension of previous work.8 This modeling is a novel approach to conceptualizing immunotherapy effects.18 We used a deterministic version of a hidden Markov model, a type of stochastic method representing an autonomous system whose future state depends only on its current state at any given time.19 The hidden nature of the model comes from the fact that although MTV is observable, it not only comprises tumor but also inflammation. The difference between these 2 quantitates must be inferred. This inference can be accomplished by using detectable ctDNA, which provides a proxy for tumor growth activity (vs inflammation). Incorporating ctDNA in our study distinguishes it from other published works, in which FDG-avidity obtained from 1-month imaging alone was used to predict the risk of lymphoma progression.20,21 By depicting tumor dynamics, our model was able to predict clinical outcomes in patients with measurable MTV 1 month after axi-cel. The model supports the hypothesis that localized tumor inflammation plays a role in ongoing tumor PET avidity, thus implying that in the absence of ctDNA, FDG-avid lesions on a scan may represent inflammation. The current data does not allow complete parametrization and validation of our mathematical model. Rather, the model effort is a starting point for novel hypotheses regarding the joint kinetics of ctDNA, MTV, and inflammation.
Serial plasma ctDNA measurements assisted with capturing the patients’ ongoing response to therapy. The response is otherwise clearly visible only on spaced-out imaging studies. The clearance rate of ctDNA may explain the remaining detectable ctDNA at 1 month via PET in patients in CR who eventually achieved durable CR.22 Although ctDNA may be undetectable in patients with CR at 1 month, upon relapse of disease, ctDNA can rise sharply. In our original prospective study of the test,6 the dynamics of ctDNA were described in detail, and we reported sensitivity of 94% and specificity of 82% for the test in subjects with PR or SD. The results in this follow-up study confirm that a one-time value of ctDNA reflects instantaneous tumor burden, as evidenced by the mismatched timing of measurement with MTV, leading to a poorer correlation. Future work should further elucidate ctDNA dynamics and account for the timing of the 2 tests.
Strengths of the study include the multicenter approach, the use of ctDNA, which relies on clonotype identification of tumors with high specificity and sensitivity, the ability to quantify tumor burden accurately via MTV, the incorporation of data from multiple time points, and the modification of an existing mathematical model as evidence for inflammation leading to PET positivity after CAR T-cell therapy. Limitations include its retrospective nature, unavailable imaging or ctDNA results in all patients at all time points, different timing of MTV and ctDNA at baseline, preventing capturing tumor burden contemporaneously, limitations of the ctDNA test itself (in particular, nonsecretion by certain tumors and inability to detect disease below the current limit of detection), lack of biopsies in patients with FDG-avid PET lesions after CAR T-cell therapy in the absence of frank progression, and the hypothesis-generating nature of the modified mathematical model, which is not yet validated as a tool for clinical response prediction.
In conclusion, we showed that plasma ctDNA might serve as a valuable supplementary test to standard 18F-FDG PET/CT scan imaging at 1 month by enhancing the accuracy of noninvasive response assessment of LBCL in patients receiving CAR T-cell therapy.
Acknowledgments
M.J.F. is supported by the National Institutes of Health, National Cancer Institute grant K08 CA248968. N.M.H. is funded by an American Society of Transplantation and Cellular Therapy New Investigator grant. D.B.M. is supported by National Cancer Institute grant PO1 CA049605 and California Institute for Regenerative Medicine grant CLIN2-10846. F.L.L. is supported by the National Institutes of Health, National Cancer Institute grants P30 CA076292 and R01CA244328-01, and as a Clinical Scholar of the Leukemia and Lymphoma Society. Funding for this work was provided in part by the National Institutes of Health, National Cancer Institute grants P30 CA076292 and 1U54CA193489-01A1, and generous donations from the Hyer Family Foundation. Adaptive Biotechnologies provided ClonoSEQ assay free of charge as part of a previous prospective effort. No additional ClonoSEQ data was generated specifically for this analyses.
Authorship
Contribution: F.L.L., D.B.M., M.J.F., and N.M.H. conceived the study; M.J.F., M.D.J., S.D., D.B.M., and F.L.L. contributed to clinical patient management, enrollment of patients onto the prospective trial, and sample collection; E.A.D., M.J.F., and A.B. performed data collection; E.A.D. performed statistical analysis; G.J.K., F.L.L., and P.M.A. performed mathematical modeling analysis; E.A.D., G.J.K., P.M.A., and F.L.L. wrote the manuscript; and all authors reviewed and approved the manuscript.
Conflict-of-interest disclosure: E.A.D. reports unlicensed patents (no royalties) held by Moffitt Cancer Center in her name. M.J.F. served as a scientific advisory for Adaptive Biotechnology, Kite Pharma, and Cargo Inc; has received research funding from Kite Pharma, Allogene, and Adaptive Biotechnology; and has served as a consultant for EcoR1 and BRV Capital Management. M.D.J. reports consultancy or advisory for Kite/Gilead and Myeloid Therapeutics, and research funding from Kite/Gilead, Incyte, and Loxo@Lilly. S.D. reports research funding from Miltenyi Biotec, Jazz Pharmaceuticals, and Kite Pharma/Gilead, and advisory board or consulting from Kite Pharma/Gilead, Bristol Myers Squibb (BMS), and Incyte. D.B.M. is supported by the National Cancer Institute, Kite sponsored scientific research agreement, and multiple clinical trials with sponsors including Kite-Gilead, BMS, Novartis, Miltenyi, Allogene, 2Seventy, Adicet, and Fate Therapeutics. P.M.A. received funding from Kite Pharma and consultancy fees from CRISPR Therapeutics. F.L.L. received compensation for scientific advisory roles from A2, Allogene, Amgen, bluebird bio, BMS/Celgene, Calibr, Caribou, Cellular Biomedicine Group, Daiichi Sankyo, GammaDelta Therapeutics, Iovance, Kite Pharma, Janssen, Legend Biotech, Novartis, Sana, Takeda, Wugen, and Umoja; his institution has received research funding from Kite Pharma (Institutional), Allogene (Institutional), Novartis (Institutional), bluebird bio (Institutional), BMS (Institutional), National Cancer Institute, and Leukemia and Lymphoma Society; his institution holds patents in his name, but none are licensed and he receives no royalties; he has served as a consultant for Cowen, EcoR1, Emerging Therapy Solutions, Gerson Lehrman Group; and has received compensation for education or Editorial Activity from American Society of Transplant and Cellular Therapy, Aptitude Health, ASH, BioPharma Communications CARE Education, Clinical Care Options Oncology, Imedex, and Society for Immunotherapy of Cancer. The remaining authors declare no completing financial interests.
Correspondence: Frederick L. Locke, Moffitt Cancer Center, 12902 USF Magnolia Dr, Tampa, FL 33612; e-mail: frederick.locke@moffitt.org.
References
Author notes
Data are available upon request from the corresponding author, Frederick L. Locke (frederick.locke@moffitt.org).