Key Points
Survey results from 18 academic laboratories reveal differences in molecular testing approaches and practices for hematologic neoplasms.
Shared understanding of the capabilities and limitations of current and future molecular testing will help optimize patient care.
Abstract
While molecular testing of hematologic malignancies is now standard of care, there is variability in practice and testing capabilities between different academic laboratories, with common questions arising on how to best meet clinical expectations. A survey was sent to hematopathology subgroup members of the Genomics Organization for Academic Laboratories consortium to assess current and future practice and potentially establish a reference for peer institutions. Responses were received from 18 academic tertiary-care laboratories regarding next-generation sequencing (NGS) panel design, sequencing protocols and metrics, assay characteristics, laboratory operations, case reimbursement, and development plans. Differences in NGS panel size, use, and gene content were reported. Gene content for myeloid processes was reported to be generally excellent, while genes for lymphoid processes were less well covered. The turnaround time (TAT) for acute cases, including acute myeloid leukemia, was reported to range from 2 to 7 calendar days to 15 to 21 calendar days, with different approaches to achieving rapid TAT described. To help guide NGS panel design and standardize gene content, consensus gene lists based on current and future NGS panels in development were generated. Most survey respondents expected molecular testing at academic laboratories to continue to be viable in the future, with rapid TAT for acute cases likely to remain an important factor. Molecular testing reimbursement was reported to be a major concern. The results of this survey and subsequent discussions improve the shared understanding of differences in testing practices for hematologic malignancies between institutions and will help provide a more consistent level of patient care.
Introduction
Next-generation sequencing (NGS)-based testing of hematologic malignancies is currently the standard of care. Improved understanding of the genetics of these neoplasms has resulted in a continually growing list of genes with clinical relevance.1-3 As with solid tumors, hematologic malignancies increasingly require the identification of molecular alterations for initial therapy selection.4-6 In contrast to most solid tumors, acute leukemia requires an extremely rapid (≤1 week) turnaround time (TAT) for the identification of actionable genomic alterations.7,8 This clinical urgency presents difficulties for most clinical laboratory workflows that batch NGS testing to minimize reagent costs and personnel needs.
Non-NGS assays with reduced complexity and time requirements can help provide rapid TAT for recurrent targetable hotspot mutations, particularly IDH1/IDH2,9,10FLT3-internal tandem duplication (ITD),11,12 and, potentially, NPM113,14 mutations. However, certain biomarkers affecting diagnosis, prognosis, and therapy require full gene sequencing (eg, TP53, CEBPA, RUNX1, and others7), which is efficiently achieved only by NGS. Posttherapy/residual disease evaluation largely requires NGS, given the range of variants associated with neoplastic signatures.
The choice of alterations to target and assay specifications needed for various clinical settings present common laboratory challenges.15 At this time, individual laboratories, in consultation with their clinical teams, have developed a balance between TAT and cost-effective testing to meet institutional needs. However, we suggest that collaboration between laboratories to develop consensus gene content and optimized workflows may further promote clinical care. Such common standards represent an important mission of the Genomics Organization for Academic Laboratories (GOAL), a multi-institution consortium focused on the development of molecular testing, currently with 28 academic laboratory members (D.L. Aisner, C.D. Gocke, D. Jones, M. Limson, J. Morrissette, and J.P. Segal, unpublished data, February 2023). GOAL supports member discussions on optimal strategies for genomic profiling through regular meetings and group listservs.
Here, we present the results of a structured survey of current clinical practices and development efforts of laboratories participating in the GOAL Hematopathology Working Group. Details of current and future planned molecular NGS-based testing were assessed, including panel size, gene content, and laboratory operations. This article builds on previous work addressing the current state of molecular testing in hematologic malignancies16 by describing plans for future testing and providing information about operational practices and workflows at academic laboratories. We believe this information will help establish a shared understanding of the capabilities and limitations of molecular testing being developed at these laboratories, with particular insight for cases requiring rapid TAT.
Methods
A 67-question survey was sent out to the hematopathology subgroup members of the GOAL consortium to gather information about hematologic malignancy NGS panels offered clinically and in development as well as laboratory operations and testing algorithms. Unspecified responses and answers that were not applicable or not known were excluded from the calculated statistics. The GOAL consortium developed shared gene content through a common set of NGS hybridization probes, purchased in bulk from Integrated DNA Technologies Inc. (Coralville, IA) and made available to member institutions. Panels using these consortium-purchased hybridization probes were termed as GOAL panels. However, the gene content (ie, subset of purchased gene probes included in a panel), library preparation, and sequencing methods for GOAL panels were individually chosen by institutions. Non-GOAL panels were panels from those laboratories using probes or primers purchased from other commercial vendors, although those institutions are members of GOAL by virtue of their purchase of GOAL probes for either a separate panel or for future use. Respondents provided details, including gene lists, on such GOAL panels or other NGS panels in use for hematologic neoplasms. Consensus gene list content was not preagreed upon but constructed based on the intersection of genes found in common among the lists provided by individual laboratories. The survey was sent out on 25 August 2021, with a final due date of 1 October 2021. Three additional survey responses were received until 6 June 2022. The subset of 31 survey questions and responses used for this publication are given in the supplement. Questions allowing for multiple responses were indicated with “check all that apply.” All participants agreed to the publication of the survey results.
Results
Participant description and hematologic malignancy NGS panel status
Of the 28 academic laboratory GOAL members, survey responses were received from 18 laboratories (see supplement for institution list). Most respondents (17 of 18 [94.4%]) were laboratory directors and lead development MDs, PhDs, and MD/PhDs involved in laboratory direction and operations. Most laboratories have been running in-house hematologic malignancy NGS panels (17 of 18 [94.4%]). Most (13 of 18 [72.2%]) were either running (8 of 18 [44.4%]) or developing (5 of 18 [27.8%]) GOAL panels. The others either had a GOAL panel as a potential future project (“wish list”; 3 of 18 [16.7%]) or no GOAL panel plans (2 of 18 [11.1%]). Survey results were separately analyzed for (1) all panels (GOAL panels in use as primary tests or in development or other panels for institutions without a GOAL panel) and (2) specifically for GOAL panels in use or development.
Hematologic malignancy NGS panel design
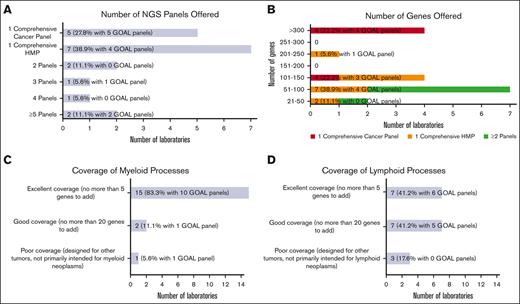
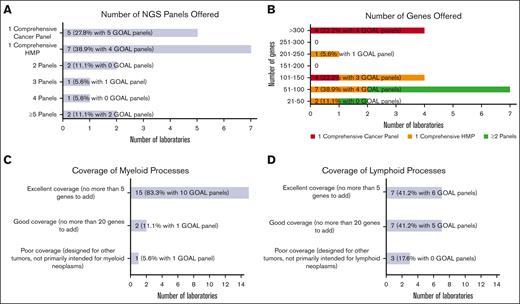
Most laboratories were or will be testing hematologic malignancies with either a single hematologic malignancy panel (HMP) focused on combined myeloid/lymphoid malignancies (7 of 18 [38.9%]) or a single comprehensive cancer panel testing all cancer types (5 of 18 [27.8%]; Figure 1A). The remaining (6 of 18 [33.3%]) were or will be tested using 2 or more NGS panels (eg, separate myeloid and lymphoid panels, or separate myeloid and comprehensive [including lymphoid] panels). Laboratories with GOAL panels (ie, in use or development) favored a single comprehensive cancer panel (5 of 12 [41.7%]) vs a single comprehensive HMP (4 of 12 [33.3%]). Most laboratories (15 of 18 [83.3%]) reported tumor-only sequencing.
Hematologic malignancy NGS panel design. (A) Bar chart showing the number of NGS panels offered for hematologic malignancies by responding laboratories, including a single comprehensive cancer panel, a single comprehensive HMP, and 2 or more panels. (B) Bar chart showing the number of genes offered ranging from small panels with 21 to 50 genes to large panels with more than 300 genes. The number of genes offered are further stratified into 3 categories based on the number of NGS panels offered: respondents with single comprehensive cancer panel, respondents with single comprehensive HMP, and combined group of respondents with >1 panel. (C) Bar chart generally showing excellent coverage of myeloid processes with no more than 5 genes needed to be added for most NGS panels. (D) Bar chart showing more moderate coverage of lymphoid processes with wider range of number of genes needed.
Hematologic malignancy NGS panel design. (A) Bar chart showing the number of NGS panels offered for hematologic malignancies by responding laboratories, including a single comprehensive cancer panel, a single comprehensive HMP, and 2 or more panels. (B) Bar chart showing the number of genes offered ranging from small panels with 21 to 50 genes to large panels with more than 300 genes. The number of genes offered are further stratified into 3 categories based on the number of NGS panels offered: respondents with single comprehensive cancer panel, respondents with single comprehensive HMP, and combined group of respondents with >1 panel. (C) Bar chart generally showing excellent coverage of myeloid processes with no more than 5 genes needed to be added for most NGS panels. (D) Bar chart showing more moderate coverage of lymphoid processes with wider range of number of genes needed.
The number of genes included in the panels varied widely from >300 genes (4 of 18 [22.2%]) down to 21 to 50 genes (2 of 18 [11.1%]; Figure 1B). The 4 laboratories covering >300 genes were or will be offering testing through comprehensive cancer panels. Of the 7 laboratories testing through a comprehensive HMP, panel sizes ranged from 21 to 50 genes to 201 to 250 genes. Gene lists for panels used to test hematologic malignancies were received from 13 laboratories (see supplement for institution list). Two gene lists were used for comprehensive cancer panels, each covering >300 genes, and 11 for comprehensive HMPs, ranging in size from 21 to 50 genes to 201 to 250 genes. Consensus gene lists are shown in Table 1, in which each row shows the genes found in a given number of the 13 gene lists (most to least common), with the resultant cumulative panel size including more frequently covered genes. All laboratories that submitted the lists (100%) included core gene content of 29 genes, and a cumulative total of 58 genes was covered by 10 of 13 laboratories (76.9%).
Consensus gene lists
| Genes . | Number of panels with listed genes (N = 13) . | Number of genes . | Cumulative panel size . |
|---|---|---|---|
| ASXL1, BCOR, BRAF, CBL, CSF3R, DNMT3A, ETV6, EZH2, FLT3, GATA2, IDH1, IDH2, JAK2, KIT, KRAS, MPL, NPM1, NRAS, PHF6, PTPN11, RUNX1, SETBP1, SF3B1, SRSF2, TET2, TP53, U2AF1, WT1, and ZRSR2 | 13 | 29 | 29 |
| ABL1, CALR, CEBPA, GATA1, MYD88, NOTCH1, and STAG2 | 12 | 7 | 36 |
| BCORL1, BTK, DDX41, FBXW7, NF1, and STAT3 | 11 | 6 | 42 |
| ATRX, CD79B, CDKN2A, CREBBP, CXCR4, GNAS, IKZF1, JAK3, MAP2K1, MYC, NOTCH2, PDGFRA, PPM1D, PTEN, RAD21, and SMC1A | 10 | 16 | 58 |
| ATM, KMT2A, SH2B3, SMC3, and XPO1 | 9 | 5 | 63 |
| CARD11, CD79A, EP300, IL7R, JAK1, STAT5B, and U2AF2 | 8 | 7 | 70 |
| BCL2, CBLB, CUX1, ETNK1, GNA13, HRAS, KDM6A, MEF2B, PLCG2, POT1, PRPF8, SETD2, and TNFRSF14 | 7 | 13 | 83 |
| ALK, BIRC3, BRCC3, CCND1, CDKN2B, CHEK2, CRLF2, CSF1R, CTCF, EED, FANCL, ID3, KLHL6, KMT2D, PAX5, PIK3CA, PIM1, PRPF40B, RHOA, RIT1, SF1, SF3A1, SOCS1, STAT6, TERT, and TNFAIP3 | 6 | 26 | 109 |
| B2M, BCL6, CBLC, CCND3, DDX3X, FOXO1, GATA3, GNB1, IKZF2, IKZF3, KMT2C, LUC7L2, NFE2, NFKBIE, NT5C2, PDGFRB, PIGA, PLCG1, PRDM1, RARA, RB1, RPS15, RRAGC, SUZ12, TCF3, TERC, TRAF3, TYK2, and ZMYM3 | 5 | 29 | 138 |
| ABL2, ANKRD26, ARID1A, ARID1B, ATR, BCL11B, BTG1, CCND2, CD28, CDKN1B, CDKN2C, CHD2, CIITA, CTNNB1, DIS3, DNM2, EBF1, EGR2, ELANE, FAM46C, FAS, HNRNPK, IRF4, IRF8, KLF2, MAPK1, MED12, NFKBIA, NSD1, PIK3CD, PIK3CG, PTPRD, REL, SAMD9, SAMHD1, SMARCA4, SPEN, SYK, TBL1XR1, TPMT, and WHSC1 | 4 | 41 | 179 |
| Genes . | Number of panels with listed genes (N = 13) . | Number of genes . | Cumulative panel size . |
|---|---|---|---|
| ASXL1, BCOR, BRAF, CBL, CSF3R, DNMT3A, ETV6, EZH2, FLT3, GATA2, IDH1, IDH2, JAK2, KIT, KRAS, MPL, NPM1, NRAS, PHF6, PTPN11, RUNX1, SETBP1, SF3B1, SRSF2, TET2, TP53, U2AF1, WT1, and ZRSR2 | 13 | 29 | 29 |
| ABL1, CALR, CEBPA, GATA1, MYD88, NOTCH1, and STAG2 | 12 | 7 | 36 |
| BCORL1, BTK, DDX41, FBXW7, NF1, and STAT3 | 11 | 6 | 42 |
| ATRX, CD79B, CDKN2A, CREBBP, CXCR4, GNAS, IKZF1, JAK3, MAP2K1, MYC, NOTCH2, PDGFRA, PPM1D, PTEN, RAD21, and SMC1A | 10 | 16 | 58 |
| ATM, KMT2A, SH2B3, SMC3, and XPO1 | 9 | 5 | 63 |
| CARD11, CD79A, EP300, IL7R, JAK1, STAT5B, and U2AF2 | 8 | 7 | 70 |
| BCL2, CBLB, CUX1, ETNK1, GNA13, HRAS, KDM6A, MEF2B, PLCG2, POT1, PRPF8, SETD2, and TNFRSF14 | 7 | 13 | 83 |
| ALK, BIRC3, BRCC3, CCND1, CDKN2B, CHEK2, CRLF2, CSF1R, CTCF, EED, FANCL, ID3, KLHL6, KMT2D, PAX5, PIK3CA, PIM1, PRPF40B, RHOA, RIT1, SF1, SF3A1, SOCS1, STAT6, TERT, and TNFAIP3 | 6 | 26 | 109 |
| B2M, BCL6, CBLC, CCND3, DDX3X, FOXO1, GATA3, GNB1, IKZF2, IKZF3, KMT2C, LUC7L2, NFE2, NFKBIE, NT5C2, PDGFRB, PIGA, PLCG1, PRDM1, RARA, RB1, RPS15, RRAGC, SUZ12, TCF3, TERC, TRAF3, TYK2, and ZMYM3 | 5 | 29 | 138 |
| ABL2, ANKRD26, ARID1A, ARID1B, ATR, BCL11B, BTG1, CCND2, CD28, CDKN1B, CDKN2C, CHD2, CIITA, CTNNB1, DIS3, DNM2, EBF1, EGR2, ELANE, FAM46C, FAS, HNRNPK, IRF4, IRF8, KLF2, MAPK1, MED12, NFKBIA, NSD1, PIK3CD, PIK3CG, PTPRD, REL, SAMD9, SAMHD1, SMARCA4, SPEN, SYK, TBL1XR1, TPMT, and WHSC1 | 4 | 41 | 179 |
Most laboratories reported panels well designed for myeloid processes. Participants were queried about the number of additional genes they would add to better assess myeloid neoplasms. The vast majority were satisfied with their panels, with 15 of 18 laboratories (83.3%) having no more than 5 genes on their wish list (Figure 1C). Few laboratories (2 of 18 [11.1%]) reported needing no more than 20 additional genes. One laboratory (1 of 18 [5.6%]) reported poor myeloid targeting, with panels designed for other tumor types. An analysis limited to GOAL panels showed a similar distribution. Fewer laboratories reported excellent gene panels appropriate for lymphomas (7 of 17 [41.2%]), with more laboratories reporting good (7 of 17 [41.2%]) and poor (3 of 17 [17.6%]) selection of lymphoid-related genes (Figure 1D). GOAL panels showed a balance of excellent (6 of 11 [54.5%]) and good (5 of 11 [45.5%]) satisfaction for lymphoid-related genes. Only a few participants reported panels with hotspot gene coverage for most genes: 2 of 17 laboratories (11.8%) for 51% to 75% of genes and 1 of 17 (5.9%) for 76% to 100%. Most (14 of 17 [82.4%]) reported hotspot coverage for only 0% to 25% of genes. GOAL probes provide whole coding sequence coverage.
Sequencing protocols and metrics
Most laboratories (15 of 17 [88.2%]) reported the use of Illumina sequencers (Illumina Inc., San Diego, CA), including the NovaSeq (7 of 17 [41.2%]), NextSeq (3 of 17 [17.6%]), and MiSeq (5 of 17 [29.4%]) (supplemental Table 1). One laboratory reported both NovaSeq and HiSeq sequencers. Ion Torrent S5/S5 XL sequencer (Thermo Fisher Scientific Inc., Waltham, MA) use was reported by 2 laboratories (2 of 17 [11.8%]). All GOAL panels will be Illumina sequencer-based, with 7 of 11 (63.6%) run on NovaSeq. Most respondents (11 of 15 [73.3%]) reported using in-house bioinformatics pipelines built from publicly available software tools.
Depth of coverage varied widely from 251 to 350× to >2500×, with most institutions reporting between 501 to 1500× coverage (11 of 14 [78.6%]; supplemental Table 2). A similar coverage ranging from 501 to 1500× was reported for most GOAL panels (7 of 8 [87.5%]). A smaller proportion (4 of 14 [28.6%]) reported the use or planned use of unique molecular identifiers (UMIs), which decreases apparent assay read depth because of UMI-based deduplication of polymerase chain reaction duplicates. Laboratories using UMIs, therefore, reported lower mean consensus coverage for appropriate technical reasons.
Assay characteristics
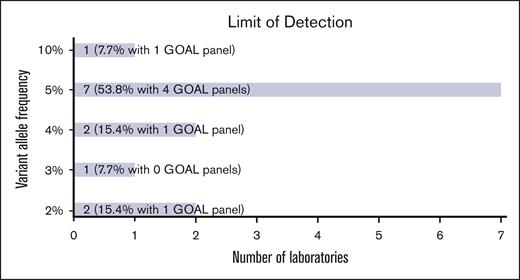
The limit of detection (LOD) among all participant assays ranged from 2% to 10%, with most (12 of 13 [92.3%]) at 5% or lower (Figure 2) for single nucleotide variants. GOAL panels showed a similar range. Several laboratories reported lower detection limits for hotspot and recurrent mutations. One laboratory reported a LOD of <1% for certain individual loci through UMI use.
NGS panel characteristics. Bar chart showing the LOD of variant allele frequency reported for NGS panels.
NGS panel characteristics. Bar chart showing the LOD of variant allele frequency reported for NGS panels.
A slight majority of laboratories were or will be reporting copy number variants (7 of 13 [53.8%]), whereas fewer laboratories were or will be reporting translocations (4 of 15 [26.7%]) and copy neutral loss of heterozygosity (3 of 13 [23.1%]) from their NGS panel.
Most laboratories reported using reference genome hg19 (12 of 16 [75.0%]) vs hg38 (4 of 16 [25.0%]). A more even distribution was seen for GOAL panels regarding hg19 (6 of 10 [60.0%]) vs hg38 (4 of 10 [40.0%]) use.
Operations
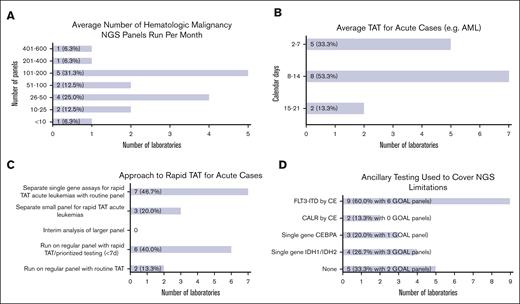
Overall laboratory testing volume varied from <10 cases to 401 to 600 cases per month (Figure 3A). Slightly less than half of the respondents (7 of 16 [43.8%]) reported >100 cases per month. Most reported shorter average TATs for acute cases (eg, acute myeloid leukemia) ranging from 2 to 7 (5 of 15 [33.3%]) and from 8 to 14 (8 of 15 [53.3%]) calendar days, whereas a few reported 15 to 21 (2 of 15 [13.3%]) calendar days (Figure 3B). TAT for routine cases was higher, with just 2 laboratories (of 15 [13.3%]) reporting 2 to 7 calendar days, and the rest, 8 to 14 calendar days (7 of 15 [46.7%]) and 15 to 21 calendar days (6 of 15 [40.0%]).
Laboratory operations. (A) Bar chart showing the average number of hematologic malignancy NGS panels run per month by responding institutions ranging from <10 cases to 401 to 600 cases per month. (B) Bar chart showing average TAT for acute cases ranging from 2 to 7 calendar days to 15 to 21 calendar days. (C) Bar chart showing different approaches for achieving rapid turnaround times for acute cases. Some institutions reported >1 approach. (D) Bar chart showing ancillary testing methods used to cover NGS limitations. Some institutions reported >1 method.
Laboratory operations. (A) Bar chart showing the average number of hematologic malignancy NGS panels run per month by responding institutions ranging from <10 cases to 401 to 600 cases per month. (B) Bar chart showing average TAT for acute cases ranging from 2 to 7 calendar days to 15 to 21 calendar days. (C) Bar chart showing different approaches for achieving rapid turnaround times for acute cases. Some institutions reported >1 approach. (D) Bar chart showing ancillary testing methods used to cover NGS limitations. Some institutions reported >1 method.
Approaches for dealing with rapid TAT requirements for acute cases included separate single gene assays for rapid TAT (see “Hematologic malignancy NGS development plans”) with routine NGS panels (7 of 15 [46.7%]), regular panels with rapid TAT/prioritized testing (6 of 15 [40.0%]), and separate small panels for rapid TAT (3 of 15 [20.0%]) (Figure 3C). No laboratories used interim analysis performed on data acquired before the end of a sequencing run. Two laboratories (of 15 [13.3%]) reported routine TAT for acute cases. Most laboratories were not staffed on weekends for specimen processing (10 of 16 [62.5%]) or reporting of case results (11 of 16 [68.8%]).
Many laboratories reported using ancillary testing to cover NGS limitations, including FLT3-ITD via capillary electrophoresis (CE; 9 of 15 [60.0%]), CALR via CE (2 of 15 [13.3%]), single gene CEBPA (3 of 15 [20.0%]), and single gene IDH1/IDH2 (4 of 15 [26.7%]) testing (Figure 3D). One-third reported no ancillary testing (5 of 15 [33.3%]). These single gene assays were reported for use with GOAL panels, except for CALR via CE. Two laboratories (of 9 [22.20%]) reported no ancillary testing for their GOAL panels.
Case reimbursement
Most laboratories (13 of 16 [81.3%]) reported testing mostly for internal patients seen at their institutions (76%-100% of cases). The remaining performed a larger percentage of external client testing: 1 with 26% to 50% and 2 with 51% to 75% of cases from internal patients. Reimbursement was reported to cover 21% to 40% of cases by a quarter of respondents (4 of 16 [25.0%]), 61% to 80% by 2 respondents (2 of 16 [12.5%]), and 81% to 100% by 1 respondent (1 of 16 [6.3%]). The remaining respondents (9 of 16 [56.3%]) reported not knowing the answer to this question.
Hematologic malignancy NGS development plans
Most laboratories (10 of 15 [66.7%]) expected hospital/institution-based NGS testing to continue to be viable in the future despite the availability of send out testing to large commercial companies, whereas the remaining either were uncertain (4 of 15 [26.7%]) or disagreed (1 of 15 [6.7%]). The most common reason supporting viable in-house testing was faster TAT, especially for acute cases. Other reasons included patient-specific tailoring of discussions with clinicians, availability of data to support clinical studies and research endeavors, special specimen handling and processing, and input on NGS panel content. Concerns about viability included reimbursement, lower costs at large commercial companies, and US Food and Drug Administration involvement. No laboratory was performing B- and T-cell clonality via NGS testing, although most (11 of 16 [68.8%]) were interested in running this testing, with 10 of 16 (62.5%) planning to perform this testing within 3 years. Interest in fusion assay development was indicated through free-text responses by 3 institutions (of 17 [17.6%]). Leading topics of interest for future discussion included reimbursement issues (12 of 15 [80.0%]), comparative performance across laboratories using standardized materials (11 of 15 [73.3%]), guidelines on minimum specifications for reporting (10 of 15 [66.7%]), and guidelines on minimum testing repertoire and testing algorithms (10 of 15 [66.7%]; supplemental Table 3).
Discussion
We report the current and future plans for hematologic NGS assays of 18 institutions participating in the GOAL consortium. To our knowledge, this is the first survey of NGS practices especially for academic genomic laboratories, including benchmarks for gene content and TAT. We believe such surveys are important not only to help laboratories stay current and address operational solutions to meet clinical requirements but also as a first step toward practice standards. With the ever-expanding list of potential gene targets and disease associations, maintaining gene content currency for clinical testing is challenging. The operational workflows required for timely reporting in acute cases and inconsistent reimbursement add further complexity to laboratory management. These issues were discussed (after survey) by a subset of survey participants as part of a group working on NGS for hematologic malignancies.
This survey identifies 1 laboratory subset using a panel specifically targeting hematologic malignancies and another using a comprehensive cancer panel covering both hematologic malignancies and solid tumors. The advantages of a pan-cancer panel include a single laboratory workflow and the ability to batch multiple specimen types in the same run for a combined higher case volume and decreased batch frequency. This strategy improves TAT, lowers reagent costs, and reduces laboratory staffing requirements, which can be critical for institutions with smaller hematologic malignancy case volumes. Challenges such as sequencing bias when combining specimen types (eg, fixed vs fresh) can be mitigated by differential library loading and other techniques. Given variation in biology, inherently larger pan-cancer panels generate unused sequencing data for genes without established hematologic malignancy relevance. Separate panels targeting only genes associated with hematologic malignancies are more efficient, assuming available support and infrastructure. Other advantages of a separate HMP include simpler validation and updates because of smaller size, faster TAT especially for data review and interpretation, and decreased sequencer usage and cost per run. Smaller panels with potentially easier reimbursement may also be better suited for follow-up testing commonly performed by many institutions.
Panel size variability was seen, ranging from 21 to 50 genes to 201 to 250 genes for comprehensive HMPs and from 101 to 150 genes to >300 genes for comprehensive pan-cancer panels. Panel size can be affected by reimbursement codes, with a 50 gene cutoff distinguishing small from large panels. Most laboratories reported NGS panels with excellent gene targeting for myeloid processes (needing no more than 5 genes to be added on), whereas less than half reported excellent targeting for lymphoid processes, reflecting differences in disease group biology and complexity.17-19 Although some laboratories offer more than 1 HMP (eg, separate myeloid and lymphoid panels), concurrent testing for both myeloid and lymphoid processes may sometimes be needed, which becomes less efficient with separate panels.20 A growing need to assess for germline predisposition to hematologic malignancies can further increase panel size.21 One approach being considered by some GOAL institutions is to target germline content at a lower depth of coverage. One institution reported using a smaller non-GOAL panel with ∼100 genes as a rapid diagnostic HMP and a larger GOAL panel (>500 genes) for reflex use based on clinician request and germline testing. This institution noted generally low yield of the larger GOAL panel when reflexed after negative results from a smaller panel. Overall, many laboratories indicated that there were at least some additional genes they would like to add to their NGS panels, which speaks to the rapidly evolving field and the difficulty of staying updated with current findings. For example, the UBA1 gene was not on the submitted gene lists but is currently being added to many panels.22 The availability of the common set of GOAL hybridization probes is expected to facilitate the addition and validation of new genes to updated NGS panels. This probe set can also be used to provide gene content for fusion assay development.23 The consensus gene lists showing genes present in current and planned panels should help provide a basic guide for laboratory and clinical expectations of content for different size panels and support cross-laboratory compatibility. While a recommended gene list based on literature review is outside the scope of this work, the genes in the consensus lists are based on input from multiple institutions and are expected to have high clinical utility.
The LOD for current and forthcoming panels will generally be in the range from 2% to 5%, with some institutions reporting lower levels for recurrent hotspot mutations. Of note, most participants mentioned that they typically report variants at allele frequencies detected below their assay LOD with a disclaimer. One institution reported being able to variably report variants at far lower allele frequencies through the use of UMIs.24 As more studies elucidate the nature and importance of clonal hematopoiesis of indeterminate potential, assays able to detect down to at least 2% variant alle frequency are important to identify this process.25,26 The impact of clonal hematopoiesis in therapy-related myeloid neoplasms is an active research area.27-29 Treatment guidelines incorporate criteria for detecting variants for measurable residual disease (MRD) purposes at even lower levels of disease burden in acute lymphoblastic leukemia,30 and the clinical significance of MRD testing in myelodysplastic syndrome and acute myeloid leukemia has been established.31-33 UMI use to reduce the general LOD of a routine comprehensive HMP to <2% is being evaluated for 1 GOAL panel in development. Some GOAL institutions are considering a small UMI-based MRD panel (<50 genes) with extremely high depth of coverage for myeloid disorders, whereas many are interested in B- and T-cell receptor clonality testing via NGS to evaluate MRD in lymphoid disorders.34-36 Participant laboratories affirmed increasing case volumes that appear to be due to the growing importance of NGS in the screening and monitoring of patients for hematologic malignancies. Therefore, the need for improved analytical sensitivity is being driven by these factors.
Most institutions that were surveyed believe that hospital-/institution-based testing will continue to be viable. Fast TAT for acute leukemia cases will likely remain an important driver, with treatment starting emergently after diagnosis. For example, the Beat Acute Myeloid Leukemia trial aims to have cytogenetic and mutational data within 7 days from sample receipt to treatment selection,8 which highlights a pressure on laboratories to shorten TAT. One-third of laboratories reported a 2-to-7–day TAT for acute cases, and one-half reported an 8-to-14–day TAT. Processing or reporting for 1 or 2 days on weekends was available only at approximately one-third of laboratories. Different methods reported for achieving faster TAT included case prioritization, separate assays for specific mutations, and separate small NGS panels. The need for mutational status to initiate targeted therapies based on hotspot mutations, such as FLT3-ITD11,12 and IDH1/IDH29,10 variants, can be assessed via separate non-NGS assays, with results commonly available within 3 to 5 working days. Although there is constant clinical pressure to reduce the TAT for results on these targets, further reduction in TAT is limited by numerous practical laboratory exigencies. Of note, the European LeukemiaNet 2022 guidelines recommend that FLT3, IDH1, IDH2, and NPM1 results preferably be available within 3 to 5 days.7 Because genes covered by single gene assays are typically also analyzed in NGS panels, billing for the same specimen may be considered duplicate if the remaining genes on the NGS panel are insufficient to meet Current Procedural Terminology code classifications based on their count.37 As official diagnostic categorization and risk stratification of acute myeloid leukemia start to depend more on whole gene information such as TP53 mutation status,7,38-41 information from NGS assays will be needed before therapy initiation. Solutions to provide this information include alternatives such as p53 immunohistochemistry42,43 and small preliminary panels with limited gene coverage and/or sensitivity processed and resulted before a main panel; this latter approach again raises the issue of duplicate billing. A trade-off between gene coverage and TAT can be considered to generally improve TAT. For example, 2 responding institutions reporting a 2-to-7–day TAT for both STAT and routine cases run relatively small (∼100 gene) panels. Although not currently used by GOAL institutions, interim analysis for hybridization-based systems in which data processed before sequencing completion provide preliminary results is an efficient solution using a single panel.44
In addition to TAT, common reasons for maintaining separate but apparently redundant single gene assays include variant types that may not be as easily detectable by NGS (eg, large CALR exon 9 deletions and FLT3-ITDs) or genomic regions with poor NGS coverage (eg, parts of the CEBPA gene). Although fewer non-NGS assays may be needed as sequencing technologies and variant-calling algorithms evolve,45,46 some laboratories may maintain orthogonal testing methods because of concerns about missing or underestimating certain variant types (eg, very large FLT3-ITDs).
Other advantages for hospital/institution-based testing include in-depth access to electronic medical records and interdisciplinary discussions, which allow for the correlation of variants with clinical and pathology findings and the ability to sensitively identify and follow variants across multiple testing points. Tracking variants between institutions is potentially problematic because of outside report availability, differences in genes and gene regions covered, and sequence variant description.47 Competition remains a concern because large commercial enterprises may potentially perform testing at lower costs and be better equipped to obtain federal agency approval if required.48 The operational overhead for running HMPs can be substantial, with most institutions experiencing increasing volumes notably for screening for myeloid processes.49,50 With many panel orders placed as part of an order set before the underlying disease process is known or confirmed, unnecessary testing or refusal of reimbursement by insurance is a concern when testing indications are not readily apparent. Some institutions reported a requirement for insurance preauthorization during discussions. Except for a few institutions including 1 which reported running all orders as received given very high case volume (too many to triage), most institutions manually review requests for molecular testing, commonly with the involvement of a laboratory director. The overhead typically includes review of medical records and clinical communication in cases of changes and cancellations. Systems to delegate some of this review process among staff and the integration of information systems are helpful to avoid user fatigue with growing case volumes.
Reimbursement was a major concern for many laboratories, with variable reimbursement reported. Current testing is generally not well reimbursed, according to respondents, with reimbursement noted to be better for myeloid compared with lymphoid diagnoses during discussions. Of note, slightly more than half of the respondents were not able to answer the survey question about the proportion of cases receiving reimbursement, which may reflect the involvement of most participants with laboratory operations as opposed to hospital finance. In many hospitals, the laboratories send their invoices through the hospital billing department, which also receives reimbursement. The hospital then provides the pathology department with a budget that is separated from the details of reimbursement. Concerns about reimbursement influence future plans and designs for assay development, including panel size and whether to perform follow-up testing using full panels to assess for clonal evolution or targeted assays for previously positive variants.
Of note, the current assessment was limited to academic tertiary-care institutions and may not reflect practice at other venues, such as community medical centers and commercial reference laboratories. The clinical indication for testing was also not addressed. Because pathology laboratories do not typically order testing themselves (because of the Stark Law) but instead receive the requests from the ordering providers, any survey of pathology laboratories that interrogates the intent of testing would necessarily require some inference. An ordering provider survey would provide more robust insight into clinical indications for testing.
The results of this survey elucidate the current practices in NGS of hematologic malignancies at numerous leading academic tertiary-care institutions and provide a reference for peer institutions. The summary presented here establishes some of the requirements, concerns, and challenges of molecular testing. As the choice of which genes to include in NGS testing can be difficult given the growing list of genes associated with hematologic malignancies in the literature, the consensus gene lists provided here should be helpful in establishing basic gene content and further cross-institution compatibility of NGS panels. Operational workflow is a critical factor in successful NGS testing and an area of active discussion between member laboratories.
Acknowledgments
Melvin Limson (GOAL administrator) and Katharine Brien (GOAL project manager) provided editorial and administrative assistance during the preparation of the survey and the manuscript.
Authorship
Contribution: T.D.L. and A.S.K. designed the survey, submitted and analyzed the data, and wrote the manuscript; D.L.A., R.R.X., and J.P.S. critically reviewed the survey, submitted and contributed to the interpretation of data, and critically reviewed the manuscript; and all other authors submitted and/or contributed to the interpretation of data and critically reviewed the manuscript. The current affiliation for L.H. is Diagnostics and Genomics Group, Agilent Technologies, Inc., Santa Clara, CA.
Conflict-of-interest disclosure: D.L.A., C.D.G., D.J., and J.P.S hold leadership roles in GOAL. The remaining authors declare no competing financial interests.
Correspondence: Thomas D. Lee, Department of Pathology and Laboratory Medicine, David Geffen School of Medicine at The University of California, Los Angeles, 10833 Le Conte Ave, Los Angeles, CA 90095; e-mail: tdlee@mednet.ucla.edu.
References
Author notes
Data are available on request from the corresponding author, Thomas D. Lee (tdlee@mednet.ucla.edu).
The full-text version of this article contains a data supplement.