Key Points
Combining cellular and plasma biomarkers with clinical features into a diagnostic classifier for pediatric cGVHD achieves a high AUC of 0.89.
This diagnostic classifier may help clinicians differentiate clinical manifestations as being due to cGVHD vs non-cGVHD causes.
Abstract
The National Institutes of Health Consensus criteria for chronic graft-versus-host disease (cGVHD) diagnosis can be challenging to apply in children, making pediatric cGVHD diagnosis difficult. We aimed to identify diagnostic pediatric cGVHD biomarkers that would complement the current clinical criteria and help differentiate cGVHD from non-cGVHD. The Applied Biomarkers of Late Effects of Childhood Cancer (ABLE) study, open at 27 transplant centers, prospectively evaluated 302 pediatric patients after hematopoietic cell transplant (234 evaluable). Forty-four patients developed cGVHD. Mixed and fixed effect regression analyses were performed on diagnostic cGVHD onset blood samples for cellular and plasma biomarkers, with individual markers declared relevant if they met 3 criteria: an effect ratio ≥1.3 or ≤0.75; an area under the curve (AUC) of ≥0.60; and a P value <5.814 × 10−4 (Bonferroni correction) (mixed effect) or <.05 (fixed effect). To address the complexity of cGVHD diagnosis in children, we built a machine learning–based classifier that combined multiple cellular and plasma biomarkers with clinical factors. Decreases in regulatory natural killer cells, naïve CD4 T helper cells, and naïve regulatory T cells, and elevated levels of CXCL9, CXCL10, CXCL11, ST2, ICAM-1, and soluble CD13 (sCD13) characterize the onset of cGVHD. Evaluation of the time dependence revealed that sCD13, ST2, and ICAM-1 levels varied with the timing of cGVHD onset. The cGVHD diagnostic classifier achieved an AUC of 0.89, with a positive predictive value of 82% and a negative predictive value of 80% for diagnosing cGVHD. Our polyomic approach to building a diagnostic classifier could help improve the diagnosis of cGVHD in children but requires validation in future prospective studies. This trial was registered at www.clinicaltrials.gov as #NCT02067832.
Introduction
Allogeneic hematopoietic cell transplant (HCT) is performed as part of the management of high-risk leukemias and several nonmalignant disorders in children. For HCT survivors, chronic graft-versus-host disease (cGVHD) remains a major long-term complication that negatively impacts the quality of life and increases morbidity and mortality.1-8 Historically, pediatric cGVHD has been an understudied disease because of the reduced incidence of cGVHD in children relative to adults and the inherent difficulty in researching rare disorders using a multiinstitutional study design.
cGVHD has complex pathogenesis. Multiple arms of the innate and adaptive immune systems operate in parallel to produce an alloreactive disease characterized by tissue injury, chronic inflammation, dysregulated immunity, aberrant tissue repair, and fibrosis.9,10 The clinical manifestations of cGVHD vary among patients, including the timing of onset, organs affected, severity, and natural history. To address this, the 2005 and 2014 National Institutes of Health Consensus criteria (NIH-CC) were created to impart minimal diagnostic criteria for cGVHD.11,12 Despite these criteria, clinicians still experience challenges in diagnosing cGVHD, particularly early in its onset (when signs and symptoms are in development and less specific) or when faced with atypical manifestations.13 Our work has shown that experienced pediatric transplant physicians misclassified cGVHD in 28% of cases, with manifestations initially thought related to cGVHD being better classified as due to either late-acute GVHD (L-aGVHD) or an alternative non-GVHD diagnosis (eg, infections and drug reactions) following central study adjudication.14 Clinically useful diagnostic biomarkers could therefore greatly aid clinicians in the diagnosis of cGVHD, particularly at its onset and early stages of development.15 To date, no cGVHD biomarkers are validated or available for routine clinical use.16
The Applied Biomarkers of Late Effects of Childhood Cancer (ABLE)/Pediatric Blood and Marrow Transplant Consortium (PBMTC) 1202 study (www.clinicaltrials.gov, #NCT02067832) evaluated diagnostic biomarkers at the onset of pediatric cGVHD using a prospective study design at 27 HCT centers.14,17,18 Using clinical cohorts of pediatric patients who had undergone extensive clinical adjudication of their GVHD status,14 we analyzed several individual cellular and plasma markers in patients with and without cGVHD. Given the heterogeneous nature of cGVHD, we further developed a machine learning–based classifier that combines multiple cellular and plasma biomarkers with clinical factors for diagnosing pediatric cGVHD. Theis study aimed to (1) define diagnostic biomarkers at the onset of cGVHD in children that would complement NIH-CC, (2) develop a diagnostic classifier that could help clinicians differentiate cGVHD at its onset from other non-cGVHD manifestations, and (3) lay the foundation for clinically applicable diagnostic biomarkers of pediatric cGVHD in the future. To our knowledge, this is the largest cohort of pediatric patients with cGVHD reported in a prospective, multiinstitutional study design.
Methods
Patients and study design
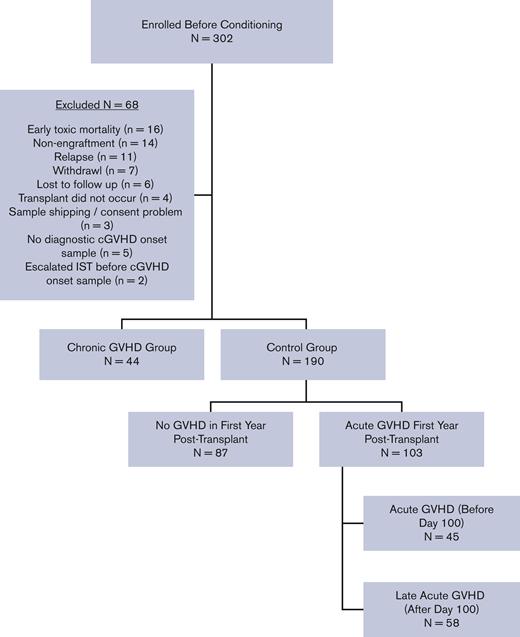
Between August 2013 and February 2017, allogeneic HCT patients aged ≤18 years were enrolled before the start of conditioning and prospectively followed for 1 year post-HCT for the development of acute GVHD (aGVHD; onset before day +100), L-aGVHD (onset after day +100), and cGVHD (onset at any time after HCT). All 27 transplant centers (6 Canadian, 20 American, and 1 Austrian) had research ethics board approval for the study and informed consent/assent was obtained for all participants in accordance with the Declaration of Helsinki. Any transplant indication (except for second transplants) and all graft sources, conditioning regimens, and GVHD prophylaxis regimens were included. Detailed clinical assessments were performed, and case report forms were completed on day +100 (±14 days), 6 months (±1 month), and 12 months (±1 month), with emphasis on GVHD status. Of the 302 patients enrolled in the ABLE/PBMTC 1202, 234 were eligible for this analysis, including 44 with cGVHD and 190 controls. Figure 1 depicts a consort diagram. The baseline characteristics of patients are presented in supplemental Table 1.
GVHD definitions and groups
aGVHD was defined as an erythematous rash, nausea, vomiting, diarrhea, and hyperbilirubinemia occurring before day +100 and was staged and graded according to the modified Glucksberg criteria.19 Updated Mount Sinai Acute GVHD International Consortium grading criteria were not published when the ABLE study opened.20 L-aGVHD was defined as the same manifestations after day +100 in the absence of cGVHD. cGVHD was defined according to the 2005 NIH-CC, since the 2014 NIH-CC were not yet published when the study opened.11 Centers completed a detailed cGVHD case report form in near real time after cGVHD diagnosis, documenting the clinical manifestations of cGVHD and severity according to NIH-CC, with a follow-up form at 1 year post-HCT. Each submitted cGVHD case was reviewed by the site principal investigator, centrally by the study principal investigator, and, when necessary, by a central study adjudication committee of cGVHD experts. Six of the 44 (13.6%) patients with cGVHD did not meet the formal NIH-CC for cGVHD diagnosis; however, after a central review by the adjudication committee, they were assessed as having manifestations that would be considered reasonably due to cGVHD and were included as cGVHD cases. Details have previously been published by our group.14 At the end of the study, patients were divided into (1) a cGVHD group, consisting of patients developing cGVHD in the first year post-HCT; or (2) a control group, consisting of patients with either no evidence of any GVHD or aGVHD and/or L-aGVHD only in the first year. Patients with overlap syndrome (concurrent aGVHD and cGVHD features at the onset of cGVHD) were included in the cGVHD group. The study groups were chosen to emphasize the clinical scenario of attempting to differentiate clinical and/or laboratory manifestations as being either due to cGVHD or a non-cGVHD cause (whether aGVHD, L-aGVHD, or a nonalloreactive etiology).
Blood samples
Peripheral blood samples were collected on day +100 (±14 days), 6 months (±1 month), and 12 months (±1 month) post-HCT in all patients, and at the onset of a new cGVHD diagnosis before the escalation of immunosuppression, specifically to treat cGVHD (diagnostic cGVHD onset blood sample). Details of study blood sampling have previously been published.17
Immunophenotyping and cytokine measurement
Six flow cytometric antibody panels, consisting of a combination of cell-surface markers, were used to delineate 76 subpopulations of T cells, regulatory T cells (Tregs; CD4+CD127lowCD25+), B cells (including T1 transitional B cells: CD19+CD38highCD10high; T2 transitional B cells: CD19+CD38intermediateCD10intermediate; T3 transitional B cells. CD19+CD38dimCD10low; mature naïve B cells: CD19+CD38−CD10− immunoglobulin D+ [IgD+] CD27−; unswitched memory B cells: CD19+IgD+CD27+; and class-switched memory B cells CD19+IgD−CD27+), natural killer (NK) cells (including regulatory NK cells [NKregs]), and myeloid cells. Cell subsets were measured as a percentage of their parent cell type. The flow cytometric methods and additional details of the analyzed cellular subpopulations are available in the supplemental Tables 2 and 3. Seven plasma cytokines and chemokines were analyzed using enzyme-linked immunosorbent assay (ELISA) (measured as concentrations), including soluble BAFF, soluble CD25 (soluble interleukin 2 receptor α), ICAM-1, CXCL10 (IP10), TIM-3, ST2, and MMP-3. CXCL9 and CXCL11 levels were measured using an electrochemiluminescence dual-plex plate (Meso Scale Diagnostics, Gaithersburg, MD). Soluble CD13 (sCD13, aminopeptidase N) was measured in the plasma using an enzymatic assay. Reg3α could not be performed because of hemolysis in some samples, which affected the accuracy of the assay. Details of all assays have been previously published.17 Completeness of analyzable control samples was 91.6% (day +100), 96.3% (6 months), and 87.7% (12 months) for cellular analysis by flow cytometry; and 95.8% (day +100), 90.4% (6 months) and 82.8% (12 months) for plasma cytokines and chemokines.
Statistical analysis
To represent clinical practice needs (differentiating true cGVHD from aGVHD, L-aGVHD, and other causes that mimic GVHD manifestations), we compared biomarkers from blood samples collected at the onset of cGVHD (experimental group) with those from the patients in the control group. Regularly scheduled blood samples from patients with cGVHD drawn before cGVHD onset (ie, at day +100 and 6 months, if cGVHD had not yet developed at this time point) were treated as control samples, provided that the blood samples were collected >14 days before the diagnosis of cGVHD.
First, we evaluated the individual cellular and plasma biomarkers at the onset of cGVHD diagnosis. For the main analysis, we applied a mixed effect linear regression model to compare the marker values of the cGVHD onset samples against those of the control samples at all the measured time points. Patient-specific intercepts and the number of days post-HCT for blood collection were included as random effects to account for within-patient correlations. The confounding factors considered in this analysis included recipient age, malignant vs nonmalignant disease, graft type (peripheral blood stem cells [PBSCs], bone marrow, or umbilical cord), sibling vs unrelated donors, donor and recipient sex, HLA and ABO match/mismatch, myeloablative vs reduced intensity conditioning, use of serotherapy (antithymocyte globulin and alemtuzumab), and total body irradiation. An individual biomarker was considered relevant if 3 criteria were met (all had to be present): (1) an effect ratio ≥1.3 or ≤0.75, (2) receiver operating characteristic (ROC) area under the curve (AUC) ≥0.60, and (3) a P value less than the Bonferroni-corrected threshold (P < 5.814 × 10−4). The effect ratio was estimated as the mean marker value of the cGVHD samples compared with that of the control group. The ROC AUC was computed by estimating the true positive rate (proportion of cGVHD correctly classified) against the false-positive rate (proportion of controls falsely classified as cGVHD) for the different marker thresholds.
A secondary analysis of individual biomarkers using fixed effect linear regression models was performed to explore the effect of the different days of cGVHD onset post-HCT. For this analysis, patients with cGVHD were divided into early-onset (<4 months), mid-onset (4-8 months), and late-onset (>8 months), and compared against time-matched control samples at day 100, 6 months, and 12 months, respectively. Identical criteria for defining relevant markers were applied in the fixed effect analysis, as in the mixed effect analysis, except for P < .05, which was used to provide a more encompassing view of the plasma and cellular biomarker patterns (considering the lower statistical power with a reduced sample size in the fixed effect analysis).
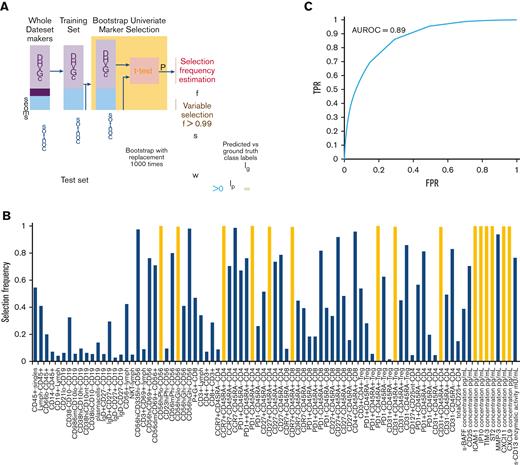
Given the pleomorphic nature of cGVHD (hence, a single marker might not be adequate to capture the variability across all patients for cGVHD diagnosis), we developed a clinically applicable machine learning–based classifier that combines multiple cellular and plasma markers, along with clinical factors, to determine whether a patient has cGVHD. Clinical factors included malignant vs nonmalignant disease, recipient age, graft type (PBSC, bone marrow, or umbilical cord), donor and recipient sex, HLA and ABO match/mismatch, donor source (sibling or unrelated), myeloablative vs reduced intensity conditioning, use of serotherapy, total body irradiation, and days post-HCT. The classifier training and evaluation procedures are summarized in Figure 4A, and the details are provided in the supplement. In brief, we first divided the samples into a test set (10 cGVHD and 10 control samples, randomly selected) and a training set (remaining samples), and performed marker selection on the training set using a bootstrapping approach. A marker was selected if it reached nominal significance for 99% of the 1000 bootstrap samples (ie, selection frequency >0.99). We then trained a support vector machine (SVM), which finds a linear weighting of the selected markers and clinical factors that best separates cGVHD from the control samples of the training set. Lastly, we applied the trained SVM to remove the test samples for classification evaluation. The procedure was performed 1000 times to assess the variability in performance across different sample splits. We also tested the addition of metabolomic markers from our recent study to the classifier.18 All analyses were performed using MATLAB (MathWorks, Natwick, MA).
Results
Cellular and plasma diagnostic biomarkers of cGVHD
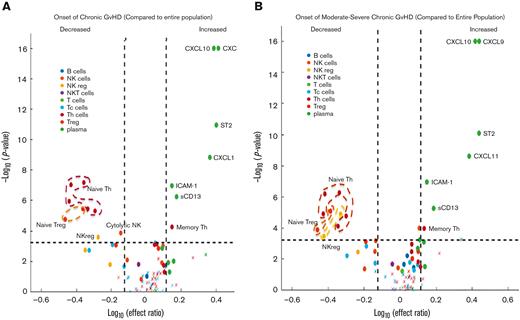
Contrasting the onset samples of pediatric patients with cGVHD against the control group using mixed effect analysis, patients with cGVHD exhibited decreased proportions of CD56+ NK cells (as a percentage of total lymphocytes), noncytolytic NKregs (CD56brightCD3−Perforin−), 5 populations of naïve T helper (Th) cells (CD4+CD45RA+ as a percentage of CD4+; and CD4+CD45RA+CCR7+, CD4+CD45RA+PD1−, CD4+CD45RA+CD27+, CD4+CD45RA+CD31+, all as a percentage of CD4+CD45RA+), and 2 populations of naïve Tregs (CD45RA+PD1− Tregs and CD45RA+CD31+ Tregs as a percentage of total Tregs), along with increased proportions of effector memory Th cells (CD4+CD45RA−CCR7− as percentage of CD4+). Elevated concentrations of CXCL9, CXCL10, CXCL11, ST2, and ICAM-1, and greater enzyme activity of sCD13 in patients with cGVHD relative to controls were also present (Figure 2A; supplemental Table 4).
Mixed effect regression results of individual cellular and plasma biomarkers. Biomarker values at the onset of cGVHD were compared against blood samples from patients without cGVHD across all time points, combined with blood samples from patients with cGVHD before the onset of cGVHD. The dashed horizontal lines correspond to the Bonferroni-corrected P value threshold. Dashed vertical lines indicate the log10 of the lower and upper limits of the effect ratio criterion. A dot (as opposed to a “x”) indicates the ROC AUC is above 0.6. (A) Onset of cGVHD of all severities (mild, moderate, or severe) according to the NIH-CC. Various populations of naïve Th cells, naïve Treg cells, NKreg cells, and cytolytic NK cells were decreased in cGVHD, whereas various cytokines and chemokines, including CXCL9, CXCL10, CXCL11, ST2, ICAM-1, and enzymatic activity in sCD13 (aminopeptidase N) were increased at the onset of cGVHD (detailed in supplemental Table 4). (B) Onset of cGVHD restricted to cases meeting the NIH-CC for moderate to severe cGVHD (mild cases removed). Similar patterns of cellular and plasma biomarkers are present in moderate to severe cGVHD, with the exception that an additional population of NKregs is decreased (CD56brightCD3−Granzyme B−), and decreased cytolytic NK cells are no longer significant (detailed in supplemental Table 5). Tc, cytotoxic T cell.
Mixed effect regression results of individual cellular and plasma biomarkers. Biomarker values at the onset of cGVHD were compared against blood samples from patients without cGVHD across all time points, combined with blood samples from patients with cGVHD before the onset of cGVHD. The dashed horizontal lines correspond to the Bonferroni-corrected P value threshold. Dashed vertical lines indicate the log10 of the lower and upper limits of the effect ratio criterion. A dot (as opposed to a “x”) indicates the ROC AUC is above 0.6. (A) Onset of cGVHD of all severities (mild, moderate, or severe) according to the NIH-CC. Various populations of naïve Th cells, naïve Treg cells, NKreg cells, and cytolytic NK cells were decreased in cGVHD, whereas various cytokines and chemokines, including CXCL9, CXCL10, CXCL11, ST2, ICAM-1, and enzymatic activity in sCD13 (aminopeptidase N) were increased at the onset of cGVHD (detailed in supplemental Table 4). (B) Onset of cGVHD restricted to cases meeting the NIH-CC for moderate to severe cGVHD (mild cases removed). Similar patterns of cellular and plasma biomarkers are present in moderate to severe cGVHD, with the exception that an additional population of NKregs is decreased (CD56brightCD3−Granzyme B−), and decreased cytolytic NK cells are no longer significant (detailed in supplemental Table 5). Tc, cytotoxic T cell.
Evaluation of biomarkers in NIH moderate to severe pediatric cGVHD
Given the clinical importance of moderate to severe NIH-CC cGVHD, we repeated the mixed effect analysis by including only patients who developed moderate to severe cGVHD in the first year post-HCT. Similar patterns of cellular populations were found, except for an additional population of NKregs (CD56brightCD3−Granzyme B−), which decreased in cGVHD, and cytolytic NK cells no longer met these criteria. When the analysis included only NIH-CC moderate to severe cGVHD, CXCL9, CXCL10, CXCL11, ST2, ICAM-1, and sCD13 remained elevated (Figure 2B; supplemental Table 5).
Evaluation of time dependence of cGVHD biomarker expression
To evaluate whether biomarker changes were dependent on the timing of onset of cGVHD after HCT, we divided the patients with cGVHD into 3 groups (early-, mid-, and late-onset) and performed a fixed effect analysis for each time point. Similar patterns of cellular biomarkers were found, although not all biomarkers retained their significance at all the 3 time points. CXCL9, CXCL10, and CXCL11 maintained their significance across all 3 time points of cGVHD onset; however, ICAM-1 and sCD13 were only elevated at the early and late time points, whereas ST2 was elevated at the mid and late time points (Figure 3; supplemental Table 6).
Fixed effect regression of individual cellular and plasma biomarkers. Patients with cGVHD were divided into groups based on days post-HCT of cGVHD onset and compared with time-matched controls. (A) Early-onset of cGVHD before 4 months post-HCT were compared with control biomarkers at day +100. (B) Mid-onset of cGVHD between 4 to 8 months post-HCT were compared with control biomarkers at 6 months post-HCT. (C) Late-onset of cGVHD between 8 and 12 months post-HCT were compared with control biomarkers at 12 months post-HCT. Dashed horizontal lines correspond to a nominal P value threshold of .05. Dashed vertical lines indicate the log10 of the lower and upper limits of the effect ratio criterion. Circled and labeled dots represent cell and plasma biomarkers that met our criteria in both the mixed effect and across all 3 time points in the fixed effect models. Details are presented in supplemental Table 6.
Fixed effect regression of individual cellular and plasma biomarkers. Patients with cGVHD were divided into groups based on days post-HCT of cGVHD onset and compared with time-matched controls. (A) Early-onset of cGVHD before 4 months post-HCT were compared with control biomarkers at day +100. (B) Mid-onset of cGVHD between 4 to 8 months post-HCT were compared with control biomarkers at 6 months post-HCT. (C) Late-onset of cGVHD between 8 and 12 months post-HCT were compared with control biomarkers at 12 months post-HCT. Dashed horizontal lines correspond to a nominal P value threshold of .05. Dashed vertical lines indicate the log10 of the lower and upper limits of the effect ratio criterion. Circled and labeled dots represent cell and plasma biomarkers that met our criteria in both the mixed effect and across all 3 time points in the fixed effect models. Details are presented in supplemental Table 6.
Combinations of cellular, plasma cytokines and chemokines and clinical factors in developing a cGVHD diagnostic classifier
Nine cellular markers, including populations of NKregs, naïve Th cells, naïve Tregs, and naïve CD8+ Tc cells (all as percentages of their parent cell types), and 6 plasma cytokine and chemokine markers, including CXCL9, CXCL10, CXCL11, ICAM-1, TIM-3, and ST2, attained a selection frequency >0.99 (Figure 4B; Table 1). The SVM classifier weight for each variable is presented in supplemental Table 7. Combining these cellular and plasma markers with 11 clinical factors (see statistical analysis) into a cGVHD diagnostic classifier achieved an average AUC (more than 1000 random test sets) of 0.89 (±0.07) (Figure 4C), average positive predictive value (PPV) of 82% (±11%) and average negative predictive value (NPV) of 80% (±11%). As some cellular and plasma cytokine/chemokine markers are interrelated (and may therefore not be required), we tested only representative markers by removing potentially redundant markers from the classifier. Using 4 plasma markers (CXCL10 representing the CXCL family, ICAM-1, TIM-3, and ST2, all measured by ELISA) and 2 cellular markers (CD56brightCD3−Perforin− representing NK-regs and CD45RA+PD1– Treg representing naïve T cells) along with all clinical factors retained an AUC of 0.89 (±0.07), a PPV of 81%, and an NPV of 81%. Hence, 2 plasma markers (CXCL9 and CXCL11) and 7 cellular markers were excluded from the analysis. We also tested the addition of metabolomic markers from our recent study to the classification pipeline,18 but the average AUC did not improve (AUC, 0.88).
cGVHD diagnostic classifier. (A) Samples were first divided into training and test sets. Bootstrap univariate marker selection was then performed by applying t test to the training samples of each marker and estimating the percentage of bootstraps over which a given marker has P < .05, referred to as selection frequency (f). A set of markers (S) with selection frequency >0.99 were used for classifier training. Labels of test samples (Ip) were then predicted using the trained classifier weights (w) and compared against the ground truth labels (lg) to evaluate the classifier’s performance. This procedure was repeated 1000 times with random sample splits to assess variability in performance. (B) Selection frequency of cellular and plasma markers based on all samples plotted. Markers with selection frequency >0.99 indicated in yellow. (C) The classifier achieved an average ROC AUC of 0.89 over the 1000 random sample splits. FPR, false-positive rate.
cGVHD diagnostic classifier. (A) Samples were first divided into training and test sets. Bootstrap univariate marker selection was then performed by applying t test to the training samples of each marker and estimating the percentage of bootstraps over which a given marker has P < .05, referred to as selection frequency (f). A set of markers (S) with selection frequency >0.99 were used for classifier training. Labels of test samples (Ip) were then predicted using the trained classifier weights (w) and compared against the ground truth labels (lg) to evaluate the classifier’s performance. This procedure was repeated 1000 times with random sample splits to assess variability in performance. (B) Selection frequency of cellular and plasma markers based on all samples plotted. Markers with selection frequency >0.99 indicated in yellow. (C) The classifier achieved an average ROC AUC of 0.89 over the 1000 random sample splits. FPR, false-positive rate.
Cellular and plasma cGVHD markers with a selection frequency greater than or equal to 0.99 in the diagnostic cGVHD classifier
| Cell type or plasma protein . | Immune phenotype or plasma protein . | Selection frequency . |
|---|---|---|
| NKreg noncytolytic | CD56bright Perforin− | 0.999 |
| NKreg noncytolytic | CD56bright Granzyme B− | 0.999 |
| Naïve Th cells | CD4+ CD45RA+ PD1− | 1 |
| Naïve Th cells | CD4+ CD45RA+ CCR7+ | 1 |
| Naïve Th cells | CD4+ CD45RA+ CD27+ | 1 |
| Naïve Th cells (RTE) | CD4+ CD45RA+ CD31+ | 1 |
| Naïve Tc cells | CD8+ CD45RA+ CCR7+ | 0.994 |
| Naïve Treg cells | CD45RA+ PD1− Treg | 1 |
| Naïve Treg cells (RTE) | CD45RA+ CD31+ Treg | 1 |
| Plasma protein | CXCL9 | 1 |
| Plasma protein | CXCL10 (IP10) | 0.998 |
| Plasma protein | CXCL11 | 1 |
| Plasma protein | ICAM-1 | 1 |
| Plasma protein | TIM-3 | 0.999 |
| Plasma protein | ST2 | 1 |
| Cell type or plasma protein . | Immune phenotype or plasma protein . | Selection frequency . |
|---|---|---|
| NKreg noncytolytic | CD56bright Perforin− | 0.999 |
| NKreg noncytolytic | CD56bright Granzyme B− | 0.999 |
| Naïve Th cells | CD4+ CD45RA+ PD1− | 1 |
| Naïve Th cells | CD4+ CD45RA+ CCR7+ | 1 |
| Naïve Th cells | CD4+ CD45RA+ CD27+ | 1 |
| Naïve Th cells (RTE) | CD4+ CD45RA+ CD31+ | 1 |
| Naïve Tc cells | CD8+ CD45RA+ CCR7+ | 0.994 |
| Naïve Treg cells | CD45RA+ PD1− Treg | 1 |
| Naïve Treg cells (RTE) | CD45RA+ CD31+ Treg | 1 |
| Plasma protein | CXCL9 | 1 |
| Plasma protein | CXCL10 (IP10) | 0.998 |
| Plasma protein | CXCL11 | 1 |
| Plasma protein | ICAM-1 | 1 |
| Plasma protein | TIM-3 | 0.999 |
| Plasma protein | ST2 | 1 |
RTE, recent thymic emigrant.
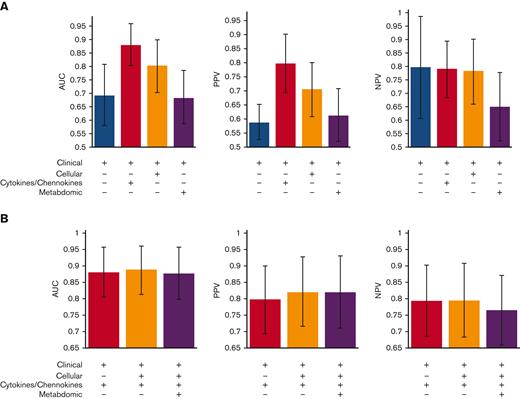
In clinical practice, rapid turnaround is critical. Therefore, we compared the benefits of including each data type in the classification performance (Figure 5A). Using clinical factors alone attained an AUC of 0.69. Adding the 6 plasma cytokine and chemokine markers with selection frequency >0.99 (measured by combinations of ELISA and Meso Scale) to the clinical factors resulted in the largest classification improvement, with an AUC of 0.88. Furthermore, including the 9 cellular markers with a selection frequency of >0.99, provided only marginal benefits (Figure 5B). However, using only 4 (supposedly) nonredundant plasma cytokine and chemokine markers (CXCL10, ICAM-1, TIM-3, and ST2; all measured by ELISA), along with clinical factors, resulted in a notable decrease in the AUC to 0.86 and PPV to 75%. Considering the time and cost of assaying plasma cytokines and chemokines by both ELISA and the Meso Scale, compared with assaying cellular markers, which can be done quickly and inexpensively by flow cytometry, the nonredundant combination of 4 plasma cytokines and chemokines (CXCL10, ICAM-1, TIM-3, and ST2) and 2 cellular markers (CD56brightCD3−Perforin− NKregs and CD45RA+PD1– Treg) might thus be preferred over 6 plasma cytokine and chemokine markers. We noted that older patients who received PBSC grafts were more often misclassified (supplemental Figure 1).
Classifier performance on various combinations of clinical factors and selected cellular, plasma cytokines/chemokines, and metabolomics markers. (A) Separately adding 1 data type (cellular markers, plasma cytokines/chemokines, or metabolomics) to clinical factors. (B) Combining multiple data types. + indicates the data types used for building the classifier. Average AUC, PPV, and NPV over 1000 random train-test sample splits plotted. Error bars correspond to standard deviation.
Classifier performance on various combinations of clinical factors and selected cellular, plasma cytokines/chemokines, and metabolomics markers. (A) Separately adding 1 data type (cellular markers, plasma cytokines/chemokines, or metabolomics) to clinical factors. (B) Combining multiple data types. + indicates the data types used for building the classifier. Average AUC, PPV, and NPV over 1000 random train-test sample splits plotted. Error bars correspond to standard deviation.
cGVHD subset analysis
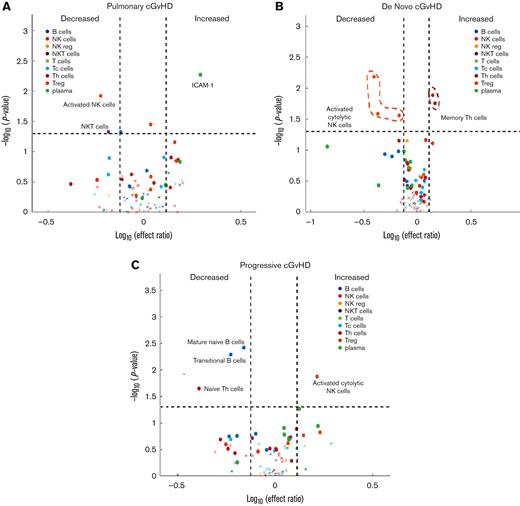
To better understand the clinically relevant subtypes of cGVHD, a post hoc exploratory analysis was performed by applying mixed effect modeling to individual biomarkers using diagnostic samples from patients with cGVHD with a pulmonary cGVHD phenotype (n = 12). Our group previously published the challenges in diagnosing pediatric pulmonary cGVHD in this cohort.14 Therefore, patients were included if they met NIH-CC or if they did not, they were still highly suspected of having pulmonary cGVHD. Decreases in NK T cells and activated cytolytic CD56bright NK cells (CD56brightCD69+) and increases in ICAM-1 were observed in pulmonary cGVHD (Figure 6A; supplemental Table 8). We also explored the biomarkers observed in de novo cGVHD (no previous history of aGVHD) (n = 7) and progressive cGVHD (aGVHD progressing into cGVHD, including overlap syndrome) (n = 18). De novo cGVHD was characterized by decreased activation of cytolytic NK cell populations (CD56brightCD69+, CD56brightPerforinhigh, and Granzyme Bhigh) and increased in memory Th cells (Figure 6B; supplemental Table 8). Progressive cGVHD was characterized by decreased percentages of B cells, reduction in mature naïve B cells (CD19+CD38−CD10−IgD+CD27−), T3 transitional B cells (CD19+CD38dimCD10low), and naïve Th cells (CD4+CD45RA+CCR7+), with increased activation of CD56dim cytolytic NK cells (CD56dimCD69+) (Figure 6C; supplemental Table 8). Other cGVHD subsets, including specific organ systems, were not possible owing marked heterogeneity in clinical presentations and small patient numbers.
Mixed effect regression of individual cellular and plasma biomarkers in subsets of pediatric cGVHD. (A) Pulmonary cGVHD (n = 12). (B) De novo cGVHD (n = 7). (C) Progressive cGVHD (including all cases of overlap syndrome) (n = 18).
Mixed effect regression of individual cellular and plasma biomarkers in subsets of pediatric cGVHD. (A) Pulmonary cGVHD (n = 12). (B) De novo cGVHD (n = 7). (C) Progressive cGVHD (including all cases of overlap syndrome) (n = 18).
Discussion
Using a prospective, multiinstitutional study design, a well-characterized cohort of pediatric HCT survivors with central adjudication of cGVHD status, and strict biomarker criteria, the ABLE/PBTMC 1202 study demonstrated that cellular and plasma diagnostic cGVHD biomarkers were present at the onset of cGVHD in children and adolescents. Relevant biomarkers included decreased number of noncytolytic NKregs, naïve Th cells, and naïve Tregs; increased effector memory Th cells, and increased levels of CXCL9, CXCL10, CXCL11, ICAM-1, ST2, and sCD13. These markers are present at the onset of cGVHD in patients who develop moderate to severe cGVHD according to the NIH-CC in the first year after HCT. Some of these markers appear to be independent of the time post-HCT when cGVHD is diagnosed (eg, CXCL9, CXCL10, and CXCL11), whereas others (eg, ST2, sCD13, and ICAM-1) may be time dependent. We also identified novel markers for pulmonary, de novo, and progressive cGVHD. However, given the small number of patients included in these subgroup analyses, an independent validation cohort is required before making definitive conclusions.
Given the complex immunopathology and clinical heterogeneity of cGVHD, a single marker is unlikely to be sufficient for diagnosing all cGVHD cases. Therefore, we developed a machine learning–based cGVHD diagnostic classifier that incorporates multiple cellular, plasma, and clinical factors. The high AUC (0.89) suggests that this classifier could aid clinicians in differentiating cGVHD at its initial onset from aGVHD, L-aGVHD, and other non-cGVHD manifestations. However, using the classifier alone could result in ∼10% of the patients being misclassified on average. This emphasizes the necessity for clinicians to perform comprehensive cGVHD clinical assessments and use clinical judgment, both at the time of suspected cGVHD diagnosis and thereafter, while considering the non-GVHD causes of various symptoms, signs, and investigations. Previous data from the ABLE study showed that pediatric transplant physicians still experience challenges in diagnosing cGVHD (particularly when the signs and symptoms are early and nonspecific), with 28% of cases initially thought to have cGVHD found later to have alternative explanations or not meet the NIH-CC.14 Therefore, this classifier could be useful for ∼25% to 30% of pediatric patients where signs and symptoms are suggestive of cGVHD, but not definitive. Given the insidious nature of cGVHD in general, adult physicians might also find a similar classifier in adult patients with bone marrow transplants if developed. Importantly, the intent was not to use the classifier when there was no clinical concern for cGVHD (ie, it was not a monitoring tool), nor when the diagnosis was obvious based on the diagnostic and distinctive signs of cGVHD according to NIH-CC. Instead, the classifier provided further evidence for clinical evaluation, helping achieve a more accurate cGVHD diagnosis when additional clarity is required. Interestingly, although the clinical factors used in the classifier were mostly risk factors for cGVHD (eg, use of PBSC vs other graft sources or HLA match vs mismatch) as opposed to diagnostic or distinctive cGVHD signs, they improved the classification performance. These clinical factors can easily be obtained in routine practice. Moreover, despite our previous publication showing elevated α-ketoglutaric acid levels both before and at the onset of cGVHD,18 adding metabolomic markers to the classifier did not improve the AUC. Furthermore, removing plasma markers and including only clinical factors and cellular markers reduced the AUC to 0.80, suggesting that plasma markers are important for cGVHD classification. Our diagnostic classifier requires further validation in a new pediatric cohort before clinical application. The ABLE 2.0/PTCTC GVH-1901 study (#NCT04372524) is currently open, enrolling pediatric patients, and will attempt to do this. The secondary objective of this study was to test the feasibility of performing both the plasma and cellular cGVHD biomarker assays with a 10-day turnaround time from receipt of blood samples to return the results to the clinician, which is an important consideration in developing a real-world application.
Important for clinical application is that when cGVHD developed, most of the patients in our cohort were either receiving GVHD prophylaxis or had recently been or were being treated with systemic immune suppression for aGVHD/L-aGVHD. As patients are often immunosuppressed when cGVHD develops, the clinical application of the diagnostic classifier is independent of this fact. For proper clinical translation, however, blood samples must be drawn when NIH-CC cGVHD is initially suspected or diagnosed and before the further escalation of immunosuppression therapy to treat cGVHD (as was done in our data analysis).
Many diagnostic cGVHD biomarkers in this study have been previously described, lending validity to our findings. CD56bright NK cells are mostly noncytolytic NKregs, expressing low levels of granzyme B and perforin and appearing to regulate innate and adaptive immunity.21-23 Consistent with our finding of decreased NKregs, low percentages of CXCR3+CD56bright NKregs and elevations in CXCL10 (a chemokine important for trafficking CXCR3+ effector cells, including CD56bright NKregs) have been documented in adult cGVHD.24 Lower proportions of CD56bright NKregs in peripheral blood donor grafts have also been associated with higher cGVHD rates.25
The interferon γ–inducible CXCR3-binding chemokines CXCL9, CXCL10, and CXCL11 recruit Th1 cells and Tc cells (Tc1) to sites of inflammation and have well-appreciated roles in cGVHD.9,26-28 Elevated levels of these chemokines have been increasingly reported as reproducible early diagnostic biomarkers of cGVHD.24,26,29-32 Our results from the ABLE/PBMTC 1202 study are consistent with this. Adults who later developed severe cGVHD had elevated CXCL9 levels by day +100 post-HCT,30,31 suggesting its importance in the early inflammatory stages of cGVHD. ST230 and sCD1333 levels have also been reported to be elevated at the time of cGVHD diagnosis.
One observation from the ABLE/PBMTC 1202 study was that pediatric cGVHD might be associated with diminished thymopoeisis. RTEs (CD4+CD45RA+CD31+)34,35 and Tregs recently emigrated from the thymus (CD31+CD45RA+ Treg)36,37 decreased in our cGVHD cohort, and both were selected as diagnostic classifiers. Numerous factors influence thymopoeisis after HCT, including age, sex, genetic factors, GVHD, and intensity of the conditioning regimen.34 Because most of our patients received myeloablative regimens and we controlled for conditioning intensity and the use of total body irradiation, we hypothesized that the impact of preexisting aGVHD on the thymus may be an explanation.38,39 In this same cohort of pediatric ABLE patients, grades 2 to 4 aGVHD and age ≥12 years (where thymic rebound post-HCT might be less robust) were the 2 most important risk factors for developing cGHVD.14 This suggests that efforts to protect thymic function in children and adolescents after HCT may be particularly important for cGVHD prevention.
The ABLE study has several strengths, including its prospective study design, near real-time adjudication of cGVHD clinical features according to the NIH-CC (ensuring proper classification of patient cohorts and avoiding recall bias), inclusion of multiple centers with low to high transplant volumes (real-world representation), and blood samples drawn in the early stages of cGVHD diagnosis before immune suppression is escalated. However, this classifier requires validation with a new independent cohort before broad clinical application. The next-generation ABLE 2.0 study, occurring in collaboration with the Pediatric Transplant and Cellular Therapy Consortium, will serve as a validation cohort for the diagnostic classifier developed here, potentially bringing diagnostic biomarkers of pediatric cGVHD closer to clinical utility.
Acknowledgments
The authors thank the patients and their families, transplant physicians, nurses, research nurses, and coordinators at all the sites to ensure completion of the study. The authors also thank Anat Halevy who helped coordinate the study.
This study was supported by the grants from the Canadian Institutes of Health Research (grant 255075). Pediatric Blood and Marrow Transplant Consortium efforts in this study were supported in part by a grant from the National Heart, Lung, and Blood Institute, National Institutes of Health (grant U01HL069254) and the Johnny Christopher Fund/St. Baldrick’s Foundation.
Authorship
Contribution: G.D.E.C. recruited study participants, wrote the manuscript, designed the study, evaluated the results, and was the principal investigator for the clinical aspects of the trial; K.R.S. recruited study participants, wrote the manuscript, designed the study, evaluated the results, and was the principal investigator for the laboratory aspects of the trial; B.N. performed all the statistical analyses and wrote the manuscript; S.A. performed and interpreted all laboratory studies; E.R.N., C.L.K., V.A.L., D.A.J., and A.C.H. were site principal investigators, recruited study participants, reviewed and wrote the manuscript, and were members of the study adjudication committee; A.M., T.S., H.B., S.W.C., E.H.C., K.A.K., M.B., B.R.O., S.C., D.C., J.H.C., M.J., S.S., A.B.P., G.C.M., D.M., A.C.C., and A.L., were site principal investigators, recruited study patients, reviewed, and wrote the manuscript; M.A.P. provided Pediatric Blood and Marrow Transplant Consortium infrastructure, recruited study patients, and reviewed and wrote the manuscript; and E.O. helped interpret data and edited the manuscript.
Conflict-of-interest disclosure: G.D.E.C. has received consultancy fees from Miltenyi Biotec. E.R.N. has served on advisory boards for Novartis, Medexus, and Atara Biotherapeutics. C.L.K. has served on an advisory board for Horizon Therapeutics. M.A.P. has served on the advisory boards of Novartis, Medexus, Equillium, Mesoblast; has received clinical study support from Adaptive and Miltenyi Biotec; and has received financial support for educational lectures for Miltenyi Biotec and Novartis. E.H.C. has served on an advisory board for Pfizer. The remaining authors declare no competing financial interests.
Correspondence: Geoffrey Cuvelier, Pediatric Blood and Marrow Transplantation, Manitoba Blood and Marrow Transplant Program, CancerCare Manitoba, University of Manitoba, ON2021a-675 McDermot Ave, Winnipeg, MB R3E 0V9, Canada; e-mail: gcuvelier@cancercare.mb.ca.
References
Author notes
Presented in oral abstract form at the 62nd annual meeting of the American Society of Hematology, 6 December 2020 (virtual meeting).
Data are available on request from the corresponding author, Geoffrey D. E. Cuvelier (gcuvelier@cancercare.mb.ca).
The full-text version of this article contains a data supplement.