Key Points
Variants at a locus comprising GSDMA quantitatively influence neutrophil counts in older individuals.
Aging and cardiometabolic diseases undermine hereditary factors in neutrophil counts and may contribute to age-related myeloid skewing.
Abstract
Blood cell production is a complex process, partly genetically determined and influenced by acquired factors. However, there is a paucity of data on how these factors interplay in the context of aging, which is associated with a myeloid proliferation bias, clonal hematopoiesis (CH), and an increased incidence of myeloid cancers. We investigated hereditary and acquired factors underlying blood cell trait variability in a cohort of 2996 related and unrelated women from Quebec aged from 55 to 101 years. We performed a genome-wide association study, evaluated the impact of chronic diseases, and performed targeted deep sequencing of CH driver genes and X-chromosome inactivation (XCI)–based clonality analyses. Multivariable analyses were conducted using generalized linear mixed models. We document that aging is associated with increasing neutrophil and monocyte counts and decreasing lymphocyte counts. Neutrophil counts were influenced by the variants in the region of GSDMA and PSMD3-CSF3, but this association decreased with age; in parallel, older individuals with cardiometabolic comorbidities exhibited significantly higher neutrophil counts (4.1 × 109/L vs 3.83 × 109/L; P < .001) than younger individuals. These age-related diseases were also associated with an increase in other myeloid-derived cells. Neither CH nor XCI clonality correlated with neutrophil counts. In conclusion, we show that neutrophil counts are genetically influenced, but as individuals age, this contribution decreases in favor of acquired factors. Aging is associated with a myeloid proliferation bias which is greater in the presence of cardiometabolic comorbidities but not of CH. These findings support that cell-extrinsic factors may contribute to the myeloid shift possibly through low-grade inflammation.
Introduction
Hematopoiesis is a complex and tightly orchestrated process. It enables the lifelong replenishment of blood cells to ensure homeostasis, in addition to allowing the modulation of production in response to various stressors.1,2 Nevertheless, there is important interindividual variation in blood cell counts.3
It has been estimated that 40% to 60% of the variance observed in blood cell traits is heritable.4,5 In fact, large genome-wide association studies (GWASs) have recently allowed to identify thousands of germline genetic variants associated with quantitative traits of specific blood lineages or blood cell indices.6-10 Deciphering the relative contribution of these numerous loci and their interrelation is a daunting task,7 yet certain loci may have a predominant effect and can be prioritized for further investigation. Blood counts are also influenced by nonhereditary factors. It is well recognized that blood and immune cell production is altered in response to insults such as infection, inflammation, therapy2,11 and exogenous factors, such as smoking.12,13 Furthermore, hematopoiesis is also subjected to several alterations with aging, such as a functional decline of hematopoietic stem cells (HSC) with a reduction in self-renewal potential, a lymphoid-to-myeloid bias,14-19 the acquisition of somatic mutation(s) leading to clonal hematopoiesis (CH),20-23 and an increased incidence of myeloid blood cancers.24
A better understanding of how germline and acquired factors modulate blood cell production may yield insight into the vulnerability of aging hematopoiesis to develop premalignant and malignant hematopoiesis. Accordingly, we performed a GWAS study and assessed factors such as lifestyle factors, chronic disease status, and CH to capture the hereditary and acquired determinants significantly associated with peripheral blood cell counts in a cohort of older women of French ancestry from Quebec. We document that aging is associated with a myeloid proliferation bias, which may be partly mediated by cardiometabolic diseases. The contribution of aging and these acquired age-related diseases undermined the effect of genetic hereditary factors on neutrophil count variability.
Methods
Study population
The study population comprised 2996 related and unrelated women with 4 French-speaking grandparents born in Quebec, recruited from the general population through advertising. Inclusion criteria were age ≥55 years at the time of recruitment and absence of blood disorder or active malignancy. The family cohort included 2172 subjects from 321 families, whereas the unrelated cohort was composed of 824 women. All subjects answered a medical questionnaire to gather demographic data and information related to medical history, lifestyle factors, and medications. All subjects provided written informed consent. The study was approved by the ethics committee of the Maisonneuve Rosemont Hospital Research Centre.
Blood cell indices
Peripheral blood samples were obtained from each subject via venipuncture. Complete blood counts with white blood cell differential were performed with GenS automated cell counter (Beckman Coulter, Brea, CA). The following 7 blood cell traits were studied as continuous phenotypic traits of interest for the GWAS: hemoglobin (g/L) and mean corpuscular volume (fL), lymphocyte count (109/L), monocyte count (109/L), neutrophil count (109/L), total white blood cell count (109/L), and platelet count (109/L). Absolute leucocyte counts were used for analyses to prevent biased results related to the interdependence of relative frequencies.
Clonality determination
Clonal hematopoiesis of indeterminate potential (CHIP) status was obtained via targeted next-generation sequencing of DNA from polymorphonuclear cells using an Ion Proton sequencer (Thermo Fisher Scientific, MA). The Ampliseq AML panel (Thermo Fisher Scientific, MA) was used to analyze the following 19 recurrently mutated genes in myeloid cancers: DNMT3A, TET2, ASXL1, TP53, JAK2, BRAF, CBL, CEBPA, FLT3, GATA2, IDH1, IDH2, KIT, KRAS, NPM1, NRAS, PTPN11, RUNX1, and WT1. CHIP was diagnosed in the presence of a predicted deleterious variant with allele frequency ≥2%, as described in the study by Buscarlet et al.25 X-chromosome inactivation patterns were determined at the HUMARA locus, as previously described.26
Genotyping and genome-wide association analysis
Genomic DNA samples were derived from peripheral blood samples. Genome-wide genotyping was achieved using 200 ng of genomic DNA in a GLP-environment at the Beaulieu-Saucier Pharmacogenomics Centre (Montreal, QC, Canada) for the included 2996 individuals. Genotyping was performed per the manufacturer’s specifications on the Illumina Infinium Global Array v3-MD (Illumina, San Diego, CA), which targets 700 625 genomic markers. Imputation and phasing were accomplished using IMPUTE2 (version 2.3.2) and the SHAPEIT2 algorithm (version 2r790), with the reference data released by 1000 Genomes reference on 16 June 2014, including all population samples (distributed via the IMPUTE2 website).27-29 This allowed us to profile 11 345 359 variants with an imputation probability ≥ .80 and completion rate ≥ 98%. A total of 4 499 752 genetic variants with a minor allele frequency >5% and 6 103 606 variants with a minor allele frequency >1% were included in the GWAS. The position of genetic variants was derived from the reference genome GRCh37 (hg19).
Genome-wide association testing was performed with SAIGE_0.43.3, which enables adjustment for sample relatedness by constructing a genetic relationship matrix.30 It fits a null linear mixed model to identify independent markers and subsequently performs single-variant association tests. P = 5 × 10-8 significance threshold was used to identify genome-wide associations. Genotype clustering quality was verified and confirmed at the region of top findings. Markers from pseudoautosomal regions were analyzed as autosomal variants.
We also tested the colocalization between the neutrophil signal on chromosome 17 (covering GSDMA and PSMD3-CSF3) and the inflammatory-related phenotypes (allergy, arthritis, asthma, and hay fever) in our cohort, using the COLOC R package version 5.1.0 with approximate bayes factor analysis to derive the posterior probabilities for the shared genetic architecture of the traits (H4).31
Statistical analyses
Categorical variables are presented as frequencies and percentages. Continuous variables are reported as means ± standard deviation. Outliers for continuous blood cell traits (identified as lying >1.5 times the interquartile range above and below the third and first quartile, respectively) were excluded after manual verification. This led to the exclusion of 10 cases for each phenotype. Normality was assessed using the Kolmogorov-Smirnov and Shapiro-Wilk tests. When applicable, logarithmic and inverse normal transformation were performed to normalize nonnormal continuous variables (the neutrophil count was normalized using logarithmic transformation, and inverse normal transformation was required for all other phenotypes). χ2 tests or Fisher exact probability tests were used to assess between group differences in categorical variables. Student t tests and analysis of variance were used for continuous variables with a normal distribution, whereas Mann-Whitney U and Kruskal-Wallis H tests were performed in the setting of non-Gaussian variables. All statistical analyses were 2-sided, with a significance level of .05 or as otherwise specified in the setting of multiple testing. Generalized linear mixed models (GLMMs) were used to assess for factors associated with peripheral blood counts, accounting for family structure as a random effect. We used an additive genetic model to score the genotype of each variant using imputation dosage coded for the alternative allele. For variants related to the same gene and on the same linkage disequilibrium block (for which haplotypes were shared among subjects), only 1 genetic variant, selected on the basis of the smallest P value, was considered in this analysis. These analyses were performed with R version 4.032 on 2465 individuals whose genetic and clinical information was available.
Results
The demographic and clinical characteristics of our study population are presented in Table 1. The mean age of participants was 69.2 years (standard deviation, 9.1 years; range, 55-101 years). Of these, 14.0% had CHIP. Twenty-six percent of the individuals had hypertension, 8.8% had coronary artery disease, 18.0% had dyslipidemia, and 4.5% had diabetes. Mean blood cell counts are presented in Table 1.
Demographic and clinical characteristics of the study cohort (n = 2465)
| General characteristics . | . |
|---|---|
| Age, mean (SD) | 69.2 (9.1) |
| Smoking, n (%) | 241 (9.8%) |
| Comorbidities, n (%) | |
| Eczema, n (%) | 64 (2.6%) |
| Coronary artery disease, n (%) | 218 (8.8%) |
| Hypertension, n (%) | 634 (26.0%) |
| Diabetes, n (%) | 110 (4.5%) |
| Thyroid disease, n (%) | 517 (21%) |
| Dyslipidemia, n (%) | 445 (18.0%) |
| Venous thrombosis, n (%) | 123 (5.0%) |
| Asthma, n (%) | 183 (7.4%) |
| Lupus, n (%) | 9 (0.4%) |
| Previous cancer, n (%) | 259 (11.0%) |
| CH status | |
| CHIP, n (%) | 333 (14.0%) |
| TET2 mutation, n (%) | 103 (4.2%) |
| DNMT3A mutation, n (%) | 195 (7.9%) |
| Peripheral blood counts | |
| Total white blood count, 109/L, mean (SD) | 6.43 (1.63) |
| Absolute lymphocyte count, 109/L, mean (SD) | 1.86 (0.57) |
| Absolute neutrophil count, 109/L, mean (SD) | 3.93 (1.29) |
| Absolute monocyte count, 109/L, mean (SD) | 0.46 (0.16) |
| Hemoglobin, g/L, mean (SD) | 131.2 (9.67) |
| Mean corpuscular volume, fL, mean (SD) | 91.1 (3.9) |
| Platelet count, 109/L, mean (SD) | 249.2 (57.6) |
| General characteristics . | . |
|---|---|
| Age, mean (SD) | 69.2 (9.1) |
| Smoking, n (%) | 241 (9.8%) |
| Comorbidities, n (%) | |
| Eczema, n (%) | 64 (2.6%) |
| Coronary artery disease, n (%) | 218 (8.8%) |
| Hypertension, n (%) | 634 (26.0%) |
| Diabetes, n (%) | 110 (4.5%) |
| Thyroid disease, n (%) | 517 (21%) |
| Dyslipidemia, n (%) | 445 (18.0%) |
| Venous thrombosis, n (%) | 123 (5.0%) |
| Asthma, n (%) | 183 (7.4%) |
| Lupus, n (%) | 9 (0.4%) |
| Previous cancer, n (%) | 259 (11.0%) |
| CH status | |
| CHIP, n (%) | 333 (14.0%) |
| TET2 mutation, n (%) | 103 (4.2%) |
| DNMT3A mutation, n (%) | 195 (7.9%) |
| Peripheral blood counts | |
| Total white blood count, 109/L, mean (SD) | 6.43 (1.63) |
| Absolute lymphocyte count, 109/L, mean (SD) | 1.86 (0.57) |
| Absolute neutrophil count, 109/L, mean (SD) | 3.93 (1.29) |
| Absolute monocyte count, 109/L, mean (SD) | 0.46 (0.16) |
| Hemoglobin, g/L, mean (SD) | 131.2 (9.67) |
| Mean corpuscular volume, fL, mean (SD) | 91.1 (3.9) |
| Platelet count, 109/L, mean (SD) | 249.2 (57.6) |
Entries correspond to frequency (%) for categorical data and mean (SD) for continuous data.
SD, standard deviation.
Genome-wide association analysis
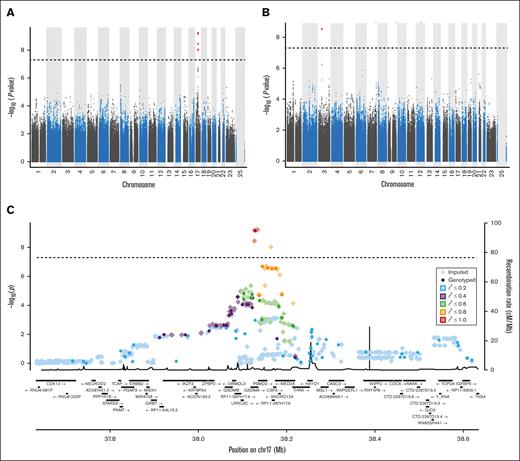
Genetic variants that reached genome-wide significance with GWAS analysis for blood cell traits in our general cohort are detailed in Table 2 and illustrated in Figure 1. Ten variants were significantly associated with the total white blood cell count. These are located in the intronic, exonic, 3′UTR, or downstream region of the GSDMA gene on chromosome 17. These were also significantly associated with the neutrophil count. As variants at this locus have previously been associated with inflammatory traits, colocalization analysis of the traits of neutrophil-allergy, neutrophil-arthritis, neutrophil-asthma, and neutrophil-hay fever at the chromosome 17 locus were performed and resulted in H4 posterior probabilities of .029, .019, .021, and .028, respectively, suggesting no shared causal variants with inflammatory traits at this locus. In addition, variants in the intergenic region of genes PSMD3-CSF3 on chromosome 17 also met the genome-wide significance threshold for the neutrophil count. Finally, rs1354034, which is intronic to gene ARHGEF3 (chromosome 3) was significantly associated with platelet count. In our cohort, no variants met genome-wide requirements for monocyte and lymphocyte counts or hemoglobin levels and mean corpuscular volume.
Genome-wide significant variants associated with blood cell traits
| Blood cell trait . | Variant . | Chromosome . | Position∗ . | Gene . | Reference/alternative allele . | Alternative allele frequency . | Beta (in SD units of the trait) . | P . | Annotation . |
|---|---|---|---|---|---|---|---|---|---|
| White blood count (n = 2842) | rs150060808 | 17 | 38 126 911 | GSDMA | AACACACAC/A | 0.4070 | 0.1657 (0.0285) | 5.80 × 10–9 | Intronic |
| rs4239225 | 17 | 38 127 112 | GSDMA | G/A | 0.4555 | 0.1626 (0.0280) | 6.0 × 10–9 | Intronic | |
| rs3859192 | 17 | 38 128 648 | GSDMA | C/T | 0.4555 | 0.1616 (0.0279) | 6.6 × 10–9 | Intronic | |
| rs3859191 | 17 | 38 128 714 | GSDMA | G/A | 0.4554 | 0.1617 (0.0279) | 6.5 × 10–9 | Intronic | |
| rs4065876 | 17 | 38 129 506 | GSDMA | G/A | 0.4557 | 0.1618 (0.0279) | 6.5 × 10–9 | Intronic | |
| rs60137005 | 17 | 38 129 996 | GSDMA | A/T | 0.4558 | 0.1619 (0.0279) | 6.4 × 10–9 | Intronic | |
| rs56326707 | 17 | 38 130 139 | GSDMA | C/T | 0.4559 | 0.1619 (0.0279) | 6.4 × 10–9 | Intronic | |
| rs56030650 | 17 | 38 131 187 | GSDMA | C/A | 0.4562 | 0.1621 (0.0279) | 6.2 × 10-9 | Exonic | |
| rs60134943 | 17 | 38 133 792 | GSDMA | G/T | 0.4569 | 0.1623 (0.0279) | 6.1 × 10–9 | 3′UTR | |
| rs3907022 | 17 | 38 134 889 | GSDMA | T/C | 0.4571 | 0.1622 (0.0279) | 6.3 × 10–9 | Downstream | |
| Neutrophils (n = 2847) | rs150060808 | 17 | 38 126 911 | GSDMA | AACACACAC/A | 0.4073 | 0.1679 (0.0284) | 3.5 × 10–9 | Intronic |
| rs4239225 | 17 | 38 127 112 | GSDMA | G/A | 0.4557 | 0.1729 (0.0280) | 6.2 × 10–10 | Intronic | |
| rs3859192 | 17 | 38 128 648 | GSDMA | C/T | 0.4557 | 0.1718 (0.0278) | 6.8 × 10–10 | Intronic | |
| rs3859191 | 17 | 38 128 714 | GSDMA | G/A | 0.4557 | 0.1720 (0.0278) | 6.6 × 10–10 | Intronic | |
| rs4065876 | 17 | 38 129 506 | GSDMA | G/A | 0.4559 | 0.1721 (0.0279) | 6.6 × 10–10 | Intronic | |
| rs60137005 | 17 | 38 129 996 | GSDMA | A/T | 0.4561 | 0.1722 (0.0279) | 6.5 × 10–10 | Intronic | |
| rs56326707 | 17 | 38 130 139 | GSDMA | C/T | 0.4561 | 0.1723 (0.0279) | 6.4 × 10–10 | Intronic | |
| rs56030650 | 17 | 38 131 187 | GSDMA | C/A | 0.4564 | 0.1725 (0.0279) | 6.2 × 10–10 | Exonic | |
| rs60134943 | 17 | 38 133 792 | GSDMA | G/T | 0.4571 | 0.1729 (0.0279) | 5.9 × 10–10 | 3′UTR | |
| rs3907022 | 17 | 38 134 889 | GSDMA | T/C | 0.4573 | 0.1728 (0.0279) | 6.0 × 10–10 | Downstream | |
| rs8068260 | 17 | 38 163 650 | PSMD3-CSF3 | T/C | 0.5199 | 0.1608 (0.0280) | 9.8 × 10–9 | Intergenic | |
| rs8067817 | 17 | 38 163 936 | PSMD3-CSF3 | C/T | 0.5208 | 0.1613 (0.0281) | 9.1 × 10–9 | Intergenic | |
| rs8072548 | 17 | 38 164 025 | PSMD3-CSF3 | T/G | 0.5208 | 0.1613 (0.0281) | 9.1 × 10–9 | Intergenic | |
| rs139156536 | 17 | 38 164 432 | PSMD3-CSF3 | TTTTA/T | 0.5198 | 0.1610 (0.0280) | 9.4 × 10–9 | Intergenic | |
| rs7222039 | 17 | 38 165 541 | PSMD3-CSF3 | T/C | 0.5207 | 0.1612 (0.0281) | 9.2 × 10–9 | Intergenic | |
| Platelets (n = 2841) | rs1354034 | 3 | 56 849 749 | ARHGEF3 | T/C | 0.5906 | 0.1727 (0.0291) | 3.0 × 10–9 | Intronic |
| Blood cell trait . | Variant . | Chromosome . | Position∗ . | Gene . | Reference/alternative allele . | Alternative allele frequency . | Beta (in SD units of the trait) . | P . | Annotation . |
|---|---|---|---|---|---|---|---|---|---|
| White blood count (n = 2842) | rs150060808 | 17 | 38 126 911 | GSDMA | AACACACAC/A | 0.4070 | 0.1657 (0.0285) | 5.80 × 10–9 | Intronic |
| rs4239225 | 17 | 38 127 112 | GSDMA | G/A | 0.4555 | 0.1626 (0.0280) | 6.0 × 10–9 | Intronic | |
| rs3859192 | 17 | 38 128 648 | GSDMA | C/T | 0.4555 | 0.1616 (0.0279) | 6.6 × 10–9 | Intronic | |
| rs3859191 | 17 | 38 128 714 | GSDMA | G/A | 0.4554 | 0.1617 (0.0279) | 6.5 × 10–9 | Intronic | |
| rs4065876 | 17 | 38 129 506 | GSDMA | G/A | 0.4557 | 0.1618 (0.0279) | 6.5 × 10–9 | Intronic | |
| rs60137005 | 17 | 38 129 996 | GSDMA | A/T | 0.4558 | 0.1619 (0.0279) | 6.4 × 10–9 | Intronic | |
| rs56326707 | 17 | 38 130 139 | GSDMA | C/T | 0.4559 | 0.1619 (0.0279) | 6.4 × 10–9 | Intronic | |
| rs56030650 | 17 | 38 131 187 | GSDMA | C/A | 0.4562 | 0.1621 (0.0279) | 6.2 × 10-9 | Exonic | |
| rs60134943 | 17 | 38 133 792 | GSDMA | G/T | 0.4569 | 0.1623 (0.0279) | 6.1 × 10–9 | 3′UTR | |
| rs3907022 | 17 | 38 134 889 | GSDMA | T/C | 0.4571 | 0.1622 (0.0279) | 6.3 × 10–9 | Downstream | |
| Neutrophils (n = 2847) | rs150060808 | 17 | 38 126 911 | GSDMA | AACACACAC/A | 0.4073 | 0.1679 (0.0284) | 3.5 × 10–9 | Intronic |
| rs4239225 | 17 | 38 127 112 | GSDMA | G/A | 0.4557 | 0.1729 (0.0280) | 6.2 × 10–10 | Intronic | |
| rs3859192 | 17 | 38 128 648 | GSDMA | C/T | 0.4557 | 0.1718 (0.0278) | 6.8 × 10–10 | Intronic | |
| rs3859191 | 17 | 38 128 714 | GSDMA | G/A | 0.4557 | 0.1720 (0.0278) | 6.6 × 10–10 | Intronic | |
| rs4065876 | 17 | 38 129 506 | GSDMA | G/A | 0.4559 | 0.1721 (0.0279) | 6.6 × 10–10 | Intronic | |
| rs60137005 | 17 | 38 129 996 | GSDMA | A/T | 0.4561 | 0.1722 (0.0279) | 6.5 × 10–10 | Intronic | |
| rs56326707 | 17 | 38 130 139 | GSDMA | C/T | 0.4561 | 0.1723 (0.0279) | 6.4 × 10–10 | Intronic | |
| rs56030650 | 17 | 38 131 187 | GSDMA | C/A | 0.4564 | 0.1725 (0.0279) | 6.2 × 10–10 | Exonic | |
| rs60134943 | 17 | 38 133 792 | GSDMA | G/T | 0.4571 | 0.1729 (0.0279) | 5.9 × 10–10 | 3′UTR | |
| rs3907022 | 17 | 38 134 889 | GSDMA | T/C | 0.4573 | 0.1728 (0.0279) | 6.0 × 10–10 | Downstream | |
| rs8068260 | 17 | 38 163 650 | PSMD3-CSF3 | T/C | 0.5199 | 0.1608 (0.0280) | 9.8 × 10–9 | Intergenic | |
| rs8067817 | 17 | 38 163 936 | PSMD3-CSF3 | C/T | 0.5208 | 0.1613 (0.0281) | 9.1 × 10–9 | Intergenic | |
| rs8072548 | 17 | 38 164 025 | PSMD3-CSF3 | T/G | 0.5208 | 0.1613 (0.0281) | 9.1 × 10–9 | Intergenic | |
| rs139156536 | 17 | 38 164 432 | PSMD3-CSF3 | TTTTA/T | 0.5198 | 0.1610 (0.0280) | 9.4 × 10–9 | Intergenic | |
| rs7222039 | 17 | 38 165 541 | PSMD3-CSF3 | T/C | 0.5207 | 0.1612 (0.0281) | 9.2 × 10–9 | Intergenic | |
| Platelets (n = 2841) | rs1354034 | 3 | 56 849 749 | ARHGEF3 | T/C | 0.5906 | 0.1727 (0.0291) | 3.0 × 10–9 | Intronic |
UTR, untranslated region.
Genome assembly GRCh37 – hg19
Manhattan and regional plots summarizing GWAS results. Manhattan plots summarizing GWAS results for neutrophil counts (A) and platelet counts (B). The x-axis represents the chromosomal positions of genetic variants, and the y-axis represents the –log10 of P values. (C) Regional plot of genetic variants associated with neutrophil count with GWAS analysis in our cohort. The first vertical axis shows the negative log10 of P values for genotyped genetic variants whereas the second axis displays the recombination rate from the HapMap reference samples.
Manhattan and regional plots summarizing GWAS results. Manhattan plots summarizing GWAS results for neutrophil counts (A) and platelet counts (B). The x-axis represents the chromosomal positions of genetic variants, and the y-axis represents the –log10 of P values. (C) Regional plot of genetic variants associated with neutrophil count with GWAS analysis in our cohort. The first vertical axis shows the negative log10 of P values for genotyped genetic variants whereas the second axis displays the recombination rate from the HapMap reference samples.
We also investigated whether genome-wide significant variants modulated the risk of relevant clinical outcomes. The significant variants identified by GWAS were not associated (P < .05) with coronary artery disease, hypertension, diabetes, dyslipidemia, lupus, cancer, and venous thromboembolism (supplemental Tables 1-3).
Environmental and acquired factors associated with peripheral blood counts
Acquired factors contributing to peripheral blood cell counts in our cohort are shown in supplemental Table 4. Smoking was positively significantly associated with neutrophil, monocyte, lymphocyte counts, and hemoglobin levels. Neutrophil and monocyte counts increased with age, and lymphocyte and platelet counts decreased with age. A statistically significant increase in neutrophil and monocyte counts was observed among patients with coronary artery disease, hypertension, dyslipidemia, and diabetes. As these cardiometabolic comorbidities often cooccur, these were also analyzed as a single outcome, and the association remained significant. Platelet count was also significantly higher among individuals with a history of hypertension, dyslipidemia, and diabetes and for combined cardiometabolic risk factors. Similar results were found when analyses were restricted to participants aged 65 and more, as per the conventional definition of aging and when the relative frequencies of different leucocyte subtypes were considered (data not shown). Peripheral blood cell counts were not significantly altered by CHIP or by clonal hematopoiesis, as assessed by X-chromosome inactivation clonality analysis.
Integration of genetic variants identified with GWAS and acquired factors
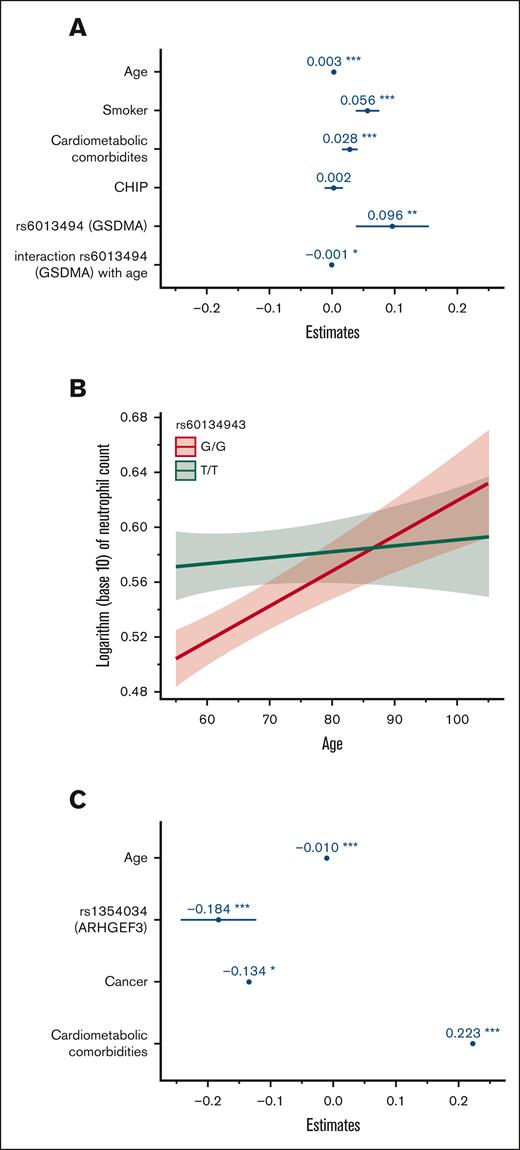
To gain a further understanding of the relative contribution of hereditary and acquired factors associated with blood cell indices in our population, GLMMs were computed including genome-wide significant variants and acquired factors in a multivariable analysis (Table 3). For neutrophil count, we fitted a GLMM adjusting for age, smoking status, cardiometabolic comorbidities (CAD, dyslipidemia, hypertension, or diabetes), CHIP status, variant genotypes, and family structure. We also sought to identify potential interactions between hereditary and environmental factors. Coefficient estimates for this model are shown in Figure 2A. All factors remained significantly associated with neutrophil count except for single nucleotide polymorphisms in the PSMD3-CSF3 region. We also identified a significant interaction between age and variants in GSDMA (rs60134943) as shown in Figure 2B. Although homozygosity for the minor allele (TT) was generally associated with a higher neutrophil count, this effect was curtailed in older individuals. No other significant interaction variable was identified. A GLMM was also fitted for platelet count adjusting for age, prior cancer, combined cardiometabolic diseases, rs1354034 (ARHGEF3) genotype, and family structure. All variables remained statistically significantly associated with this peripheral blood trait. Coefficient estimates are plotted in Figure 2C. No significant interaction variable between hereditary and acquired factors was identified for platelet count. The variance explained by GLMM models, as indicated by the full model R2, remained modest at 5.4% and 3.1% for neutrophil and platelet count, respectively.
Integration of hereditary and acquired factors associated with the neutrophil and platelet counts in multivariable GLMM
| Predictors . | Logarithm (base 10) of neutrophil count . | Inverse normal transformation of platelet count . | ||||||
|---|---|---|---|---|---|---|---|---|
| Estimate . | SE . | P . | R2 . | Estimate . | SE . | P . | R2 . | |
| (Intercept) | 0.27 | 0.04 | < .001 | 0.73 | 0.17 | < .001 | ||
| Age (y) | 0.003 | 0.001 | < .001 | 0.028 | −0.01 | 0.002 | < .001 | 0.004 |
| Smoker | 0.06 | 0.009 | < .001 | 0.012 | ||||
| Cardiometabolic comorbidities | 0.03 | 0.006 | < .001 | 0.014 | 0.22 | 0.05 | < .001 | 0.007 |
| rs60134943 (GSDMA) (T vs G) | 0.1 | 0.03 | .003 | 0.014 | 0.014 | |||
| CHIP | 0.002 | 0.008 | .780 | 0.012 | 0.012 | |||
| Interaction variable of age and rs60134943 (GSDMA) | −0.001 | 0.000 | .013 | |||||
| rs1354034 (ARHGEF3) (C vs T) | −0.18 | 0.03 | < .001 | 0.015 | ||||
| Cancer | −0.13 | 0.06 | .03 | 0.002 | ||||
| Full model R2 | 0.054 | 0.031 | ||||||
| Predictors . | Logarithm (base 10) of neutrophil count . | Inverse normal transformation of platelet count . | ||||||
|---|---|---|---|---|---|---|---|---|
| Estimate . | SE . | P . | R2 . | Estimate . | SE . | P . | R2 . | |
| (Intercept) | 0.27 | 0.04 | < .001 | 0.73 | 0.17 | < .001 | ||
| Age (y) | 0.003 | 0.001 | < .001 | 0.028 | −0.01 | 0.002 | < .001 | 0.004 |
| Smoker | 0.06 | 0.009 | < .001 | 0.012 | ||||
| Cardiometabolic comorbidities | 0.03 | 0.006 | < .001 | 0.014 | 0.22 | 0.05 | < .001 | 0.007 |
| rs60134943 (GSDMA) (T vs G) | 0.1 | 0.03 | .003 | 0.014 | 0.014 | |||
| CHIP | 0.002 | 0.008 | .780 | 0.012 | 0.012 | |||
| Interaction variable of age and rs60134943 (GSDMA) | −0.001 | 0.000 | .013 | |||||
| rs1354034 (ARHGEF3) (C vs T) | −0.18 | 0.03 | < .001 | 0.015 | ||||
| Cancer | −0.13 | 0.06 | .03 | 0.002 | ||||
| Full model R2 | 0.054 | 0.031 | ||||||
Because continuous traits were not normally distributed, logarithmic and inverse normal transformation were performed for neutrophil counts and platelet counts, respectively.
Family structure was accounted for as a random effect. Combined cardiometabolic comorbidities include coronary artery disease, dyslipidemia, hypertension, or diabetes.
SE, standard error.
Coefficient estimates of factors associated with neutrophil and platelet count using a generalized linear mixed model. (A) Coefficient estimates of factors associated with neutrophil count with multivariable analysis. The horizontal axis represents coefficient estimates values and the factors are included in the vertical axis. Coefficient estimates represent the expected change in neutrophil count per 1 unit increase in factor (continuous variable such as age) or in the presence of factor (categorical variable such as smoking). (B) Plot depicting the interaction effect of age and variant rs60134943 (GSDMA) on the neutrophil count. Age is represented on the horizontal axis and the logarithm of neutrophil count is represented on the vertical axis. (C) Coefficient estimates of factors associated with platelet count. The horizontal axis represents coefficient estimate values and the factors are included in the vertical axis. Coefficient estimates represent the expected change in platelet count per 1 unit increase in factor (continuous variable, such as age) or in the presence of factor (categorical variable, such as cancer). ∗ P < .05; ∗∗ P < .01; ∗∗∗ P < .001.
Coefficient estimates of factors associated with neutrophil and platelet count using a generalized linear mixed model. (A) Coefficient estimates of factors associated with neutrophil count with multivariable analysis. The horizontal axis represents coefficient estimates values and the factors are included in the vertical axis. Coefficient estimates represent the expected change in neutrophil count per 1 unit increase in factor (continuous variable such as age) or in the presence of factor (categorical variable such as smoking). (B) Plot depicting the interaction effect of age and variant rs60134943 (GSDMA) on the neutrophil count. Age is represented on the horizontal axis and the logarithm of neutrophil count is represented on the vertical axis. (C) Coefficient estimates of factors associated with platelet count. The horizontal axis represents coefficient estimate values and the factors are included in the vertical axis. Coefficient estimates represent the expected change in platelet count per 1 unit increase in factor (continuous variable, such as age) or in the presence of factor (categorical variable, such as cancer). ∗ P < .05; ∗∗ P < .01; ∗∗∗ P < .001.
Discussion
In this study, we assessed some of the inherited and acquired factors that contribute to the variability of peripheral blood traits in the older population. We performed a GWAS and assessed comorbidities and lifestyle factors in a cohort that included related and unrelated women from a population enriched for French ancestry from the province of Quebec, Canada. We identified genetic variants significantly associated with neutrophil and total white blood cell counts as well as those associated with platelet counts. A statistical interaction effect between age and genetic variants in the region of the GSDMA gene with neutrophil counts was found. In addition, aging was associated with higher neutrophil and monocyte counts and lower lymphocyte counts, reflecting a myeloid proliferation bias in hematopoiesis. This myeloid skewing was also present with cardiometabolic comorbidities. CH was not associated with peripheral blood cell traits and the myeloid proliferation bias.
Through genome-wide associated analysis, we identified 15 genetic variants significantly associated with the neutrophil count in a region of strong linkage disequilibrium within the 17q21.1 locus which includes, among others, the GSDMA and PSMD3-CSF3 genes.
GSDMA encodes gasdermin A, a member of the gasdermin family of proteins, which is involved in inflammasome signaling. Inflammasome is a protein complex that orchestrates host immune responses to pathogens and danger signals.33 Inflammasome activation leads to the cleavage and activation of gasdermins by inflammatory caspases. In turn, gasdermins drive pyroptosis execution, a form of inflammatory cell death, through their pore-forming activity.34-36 The inflammasome also activates interleukin-1β (IL-1β) and IL-18 and feeds forward the inflammatory response by permitting their release in the extracellular environment through pores formed by gardermins.37-40 Although the role of gasdermin D has been more deeply acknowledged, it has now been recognized that most members of the gasdermin family have pore-forming activity.41 Interestingly, a host of studies have suggested the potential involvement of GSDMA in inflammatory conditions, such as asthma, systemic sclerosis, and inflammatory bowel disease.42-48 Asthma-associated polymorphisms at 17q21 have also been associated with higher GSDMA (and ORMDL3) gene expression and elevated IL-17 secretion in the cord blood.49 Shi et al also suggested a role of Gsdma3 in the modulation of autophagy and noted enhanced unrestrained autophagy leading to inflammation in mice with gain-of-function Gsdma3 mutations.50PSMD3, also located within the 17q21.1 locus, is a member of the 19S regulatory complex in the 26S proteasome. Similar to variants near the GSDMA gene, polymorphisms in this gene have also been linked to asthma susceptibility.43,51,52 Because variants at the locus on chromosome 17 including GSDMA and PSMD3 have previously been associated with asthma predisposition, and asthma has also been linked to increased neutrophil counts,53 it was important to exclude that the association identified via GWAS between neutrophil counts and variants at this locus had not been biased by subjects with a diagnosis of asthma in our cohort. Accordingly, colocalization analyses were performed and did not suggest shared causal variants between neutrophil counts and inflammatory traits at this locus, thus arguing against a confounding effect brought about by asthma.
Our findings are further supported by several studies that have also emphasized the contribution of genes within the 17q21.1 locus to white blood cell and neutrophil counts, in addition to their role in modulating immune system function and susceptibility to several diseases. In fact, GWASs have yielded an association between variants within this region and several white blood cell traits in the general population.7,8,10,54-56 In the disease setting, a genetic variant in the intronic region of PSMD3 has previously been associated with interferon-induced neutropenia in patients with chronic hepatitis in a study by Iio et al.57 In addition, Glisovic et al found that in a cohort of children with acute lymphoblastic leukemia, individuals harboring the minor T allele of rs3859192 (GSDMA) carried a higher risk of prolonged chemotherapy-induced neutropenia and a higher risk of requiring hospitalization for infection associated with longer hospital admissions. Even though CSF3, which encodes granulocyte colony-stimulating factor, lies in the vicinity of this region, the association was independent of CSF3 polymorphisms.58
Although several studies have suggested the role of the 17q21.1 locus in immune responses and modulation of neutrophil counts in health and disease, causal genes and variants within this locus and whether these are independent of CSF3 remain elusive. Nonetheless, this locus appears to have a predominant effect in shaping neutrophil count variability among individuals as it has consistently been associated with this trait in large and smaller-scale studies.7,8,10,54-56,59-66 Importantly, a role of GSDMA in the biology of neutrophils has also recently been illustrated by Deng et al in a study that demonstrated that Gsdma1 deficiency is associated with a reduction in neutrophil recruitment at infection sites in Gsdma1 knockout mice subjected to Group A Streptococcus infection.67 Further investigation is required to expand on these findings and delineate causal variants within this locus and the mechanisms by which they modulate neutrophil counts in the general population. This may provide additional insight into the biology of neutrophils and potential mechanistic correlations with pyroptosis. Additional work could also explore whether variants within this region contribute to cases of idiopathic neutrophilia or modulate peripheral blood counts in patients with hematologic malignancies, such as myeloproliferative neoplasms.
In our study population, the variant rs1354034 was significantly associated with platelet count. It lies in an intronic region of the ARHGEF3 gene, coding for a Rho guanine nucleotide exchange factor involved in the regulation of megakaryopoiesis. In line with our findings, previous genome-wide associated analyses have reported this variant to be associated with platelet count and volume.6-8,10,55,68,69 Vuckovic et al identified that ARHGEF3 could transregulate a subnetwork of genes involved in platelet function (including GP9, GB1BB, and ITGA2B).7 Although genetic variants in the ARHGEF10 gene have previously been linked to stroke predisposition in the Japanese70 and Han Chinese populations,71 no association between the different alleles of rs1354034 and arterial or venous thrombosis risk was identified in our study.
In our cohort, no variant met genome-wide association thresholds with regard to hemoglobin levels and mean cell volume. Nonetheless, our top findings for these phenotypic traits largely replicated those found by previous groups which analyzed cohorts of European ancestry at sites near the TMPRSS6 and HFE genes (data not shown).7-10,72-75 The smaller sample size of our cohort likely limited the power of our GWAS with red blood cell traits.
We also assessed the impact of acquired factors on peripheral blood counts. We document that an increase in neutrophil and monocyte counts with a concurrent decrease in lymphocyte counts is observed as individuals age. This aligns with the phenomenon of a lymphoid-to-myeloid bias in hematopoiesis, which has been well-described in preclinical models of aging.76-78 Interestingly, we also document that age-related chronic cardiometabolic diseases further promote myeloid proliferation as affected individuals exhibited significantly higher neutrophil, monocyte, and platelet counts. Furthermore, in our cohort, smoking was associated with increased neutrophil counts as previously reported;12,13 however it did not contribute to myeloid skewing because a concurrent increase in lymphocyte counts was also identified.
To this date, the phenomenon of age-related myeloid skewing remains incompletely understood. Several underlying causes have been posited including a state of chronic, low-grade, sterile inflammation associated with aging, commonly referred to as inflammaging.79 Inflammaging is characterized by increased levels of cytokines and echoing the phenomenon of emergency granulopoiesis observed in acute inflammation, these inflammatory mediators may promote HSC proliferation and myelopoiesis.78 In addition, inflammatory exposure has been associated with the functional attrition of murine HSCs and accelerated hematopoietic aging phenotypes with myeloid-skewed hematopoiesis80 Inflammaging is a major risk factor for the development of cardiometabolic diseases as endothelial damage, alteration of vascular remodeling, atherosclerosis, and insulin resistance are adverse events induced by increased proinflammatory cytokines in the setting of inflammaging.81 In turn, cardiac ischemic damage, hypertension, and the metabolic syndrome may further the production of damage-associated molecular patterns by necrotic cells thus increasing the proinflammatory cytokine burden (such as IL-1β, IL-6, and tumor necrosis factor α).82,83 We hypothesize that the increased level of neutrophil, monocyte, and platelet counts observed in patients with diverse cardiometabolic comorbidities in our study may reflect a state of low-grade inflammation associated with these diseases and be entwined in the phenomenon of inflammaging. In accordance with this hypothesis, in a large-scale GWAS, Vuckovic et al have suggested that abnormal blood counts may be the result of chronic diseases.7 Alternatively, it remains possible that the observed increase in myelopoiesis could feedforward an inflammatory response and promote or perpetuate aging-related diseases. Our findings are in line with a model proposed by Salminen et al that states that age-related peripheral changes lead to the release of proinflammatory factors, which then stimulate myelopoiesis and hamper lymphopoiesis. Subsequently, this fuels a vicious cycle by which enhanced myeloid output can foster peripheral tissue damage.78
Aging hematopoiesis is also characterized by the appearance of CH. We thus sought to evaluate whether CH could participate in the variability of peripheral blood cell counts. We found that it did not modulate blood cell indices in our cohort and importantly, did not contribute to the hematopoietic proliferation myeloid bias. This suggests that age-related myeloid skewing is a phenomenon mediated by cell-extrinsic factors such as proinflammatory age-related diseases and not by mutation-driven hematopoietic clones. In fact, we have previously shown that the mutations in the TET2 gene are associated with a modest but significant decrease in neutrophil counts.25 However, whether the preferential promotion of myelopoiesis in aging could subsequently favor the stochastic occurrence of CHIP remains conceivable, yet our cross-sectional study design precluded addressing this question.84
Our study has some limitations worth highlighting. Firstly, a small sample size might have limited our power to detect genome-wide significant associations. Although a confirmatory replication study was not performed, our results are supported by the fact that the genetic variants identified in our GWAS have been associated with blood cell traits in previous studies. Functional studies to assess the mechanisms by which GSDMA and variants within GSDMA modulate variability of neutrophil counts in the general population were not performed, and these should be explored in subsequent studies. In addition, as inflammatory biomarkers were not assessed in our cohort, we were unable to evaluate the true extent of inflammation in determining peripheral blood counts. Nonetheless, chronic systemic diseases may stand as a relevant indicator of enduring inflammation. Indeed, Cesari et al have reported increased IL-6 and tumor necrosis factor α levels in the setting of subclinical and clinical cardiovascular diseases.85 In addition, an association between insulin resistance and increased levels of IL-6, tumor necrosis factor α, and other inflammatory biomarkers have been documented by Abbatecola et al.86 Furthermore, obesity and the microbiome, which are known to contribute to low-grade systemic inflammation, lymphoid-to-myeloid bias in aging mice, and HSC inflammaging,87-89 were not assessed. The effect of epigenetic alterations and transcriptional alterations, which are hallmarks of aging90,91 and have been implicated in the biology of aging HSCs,92,93 was also not evaluated. The age-associated epigenetic drift may also promote inflammatory pathways94 and contribute to biased lineage potential of aging HSCs.76,95,96 In all, the integration of additional acquired factors, combined with a larger cohort size to achieve more power for genome-wide associated analysis could allow explaining a higher fraction of variance in blood cell traits. Finally, the cross-sectional design of our study limited our capacity to draw causality inferences.
In conclusion, our study draws a portrait of the determinants underlying peripheral blood cell counts in the aging population of women. We identified variants in the region of the GSDMA and PSMD3-CSF3 genes which are associated with neutrophil counts, and the association decreased with increasing age. In parallel, neutrophil counts increased in the setting of acquired chronic cardiometabolic comorbidities. Taken together, these observations suggest that the effect of genetic variants may be undermined by aging and acquired factors, which explain increasing proportions of trait variability. More broadly, we document that aging is associated with increased levels of neutrophils and monocytes and reduced lymphocyte counts, indicating a shift toward myelopoiesis. This myeloid-biased proliferation is further increased among individuals with chronic cardiometabolic diseases, possibly through the perpetuation of a state of chronic low-grade inflammation. Our study shows that CH does not contribute to the age-associated myeloid shift, in further support of this phenomenon being mediated by cell-extrinsic factors. Prospective studies are nonetheless required to evaluate if the cardiometabolic-driven myeloid proliferation bias is a risk factor for CH acquisition or clonal expansion.
Acknowledgments
The authors thank the study participants, research nurses, and research assistants.
This research was funded in part by the Canadian Institutes of Health Research and the Leukemia Lymphoma Society of Canada.
Authorship
Contribution: L.B. designed and provided financial support for the study. M.-F.G. wrote the first version of the manuscript; S.P., M.-F.G., M.S., and M.-P.D. performed bioinformatics and statistical analyses; M.-F.G., S.P., M.S., S.A., M.B., L.M., N.S., M.-P.D., and L.B. interpreted the data; and all authors reviewed and approved the final draft of the manuscript.
Conflict-of-interest disclosures: The authors declare no competing financial interests.
Correspondence: Lambert Busque, Hôpital Maisonneuve-Rosemont, Montréal, QC, Canada H1T 2M4; e-mail: lambert.busque.med@ssss.gouv.qc.ca.
References
Author notes
Data are available upon request from the corresponding author, Lambert Busque (lambert.busque.med@ssss.gouv.qc.ca).
The full-text version of this article contains a data supplement.