Key Points
APC is a pleiotropic coagulation protease with anticoagulant, anti-inflammatory, and cytoprotective activities.
An APC nonactive site–specific nanobody library is developed to selectively modulate APC for potential therapeutic purpose.
Abstract
Activated protein C (APC) is a pleiotropic coagulation protease with anticoagulant, anti-inflammatory, and cytoprotective activities. Selective modulation of these APC activities contributes to our understanding of the regulation of these physiological mechanisms and permits the development of therapeutics for the pathologies associated with these pathways. An antibody library targeting the nonactive site of APC was generated using llama antibodies (nanobodies). Twenty-one nanobodies were identified that selectively recognize APC compared with the protein C zymogen. Overall, 3 clusters of nanobodies were identified based on the competition for APC in biolayer interferometry studies. APC functional assays for anticoagulant activity, histone H3 cleavage, and protease-activated receptor 1 (PAR1) cleavage were used to understand their diversity. These functional assays revealed 13 novel nanobody-induced APC activity profiles via the selective modulation of APC pleiotropic activities, with the potential to regulate specific mechanisms for therapeutic purposes. Within these, 3 nanobodies (LP2, LP8, and LP17) inhibited all 3 APC functions. Four nanobodies (LP1, LP5, LP16, and LP20) inhibited only 2 of the 3 functions. Monofunction inhibition specific to APC anticoagulation activity was observed only by 2 nanobodies (LP9 and LP11). LP11 was also found to shift the ratio of APC cleavage of PAR1 at R46 relative to R41, which results in APC-mediated biased PAR1 signaling and APC cytoprotective effects. Thus, LP11 has an activity profile that could potentially promote hemostasis and cytoprotection in bleedings associated with hemophilia or coagulopathy by selectively modulating APC anticoagulation and PAR1 cleavage profile.
Introduction
Activated protein C (APC) is a serine protease in the blood with physiological functions in anticoagulation and cytoprotection.1,2 APC circulates at a very low concentration of 40 pM in human blood, whereas its zymogen, protein C (PC), circulates at a much higher concentration of 65 nM.3 Thrombin/thrombomodulin activates PC via proteolytic cleavage at Arg169 and the release of a 12–amino acid peptide. APC downregulates coagulation by inactivating factor Va (FVa) and FVIIIa to terminate thrombin generation as soon as hemostasis is achieved. This role in anticoagulation makes APC a potential therapeutic target for bleeding. Patients with hemophilia with the APC-resistant FV Leiden variant were observed to have reduced bleeding.4-6 Mice with acute traumatic coagulopathy experienced significantly less bleeding when treated with an APC-resistant superFVa variant.7 These findings show that APC inhibition promotes a procoagulant state. In addition, inhibitory antibodies specific for the anticoagulant activity of APC are efficacious in hemophilic monkeys8 and mice.9,10 Furthermore, an engineered KRK-α1-antitrypsin specific for inhibiting APC is also showing efficacy in reducing the annualized bleed rates in hemophilia trials.11,12
APC also induces regenerative effects, including neurogenesis13,14 and wound healing.15 In addition, APC exerts protection from extracellular histones16 released from immune or necrotic cells, which in pathological amounts can lead to systemic inflammation, organ failure, and even mortality.17 Protease-activated receptor 1 (PAR1) is a major mediator of APC cytoprotective activities. PAR1 has 2 cleavage sites for thrombin and APC. The cleavage at R41 by thrombin induces a proinflammatory response. In contrast, the cleavage at R46 by APC elicits biased PAR1 signaling and the activation of cytoprotection pathways.18,19 Oligonucleotide-based aptamers, such as APC-167,20 HS02,21,22 and G-NB323 have been developed to target APC nonactive sites for inhibiting anticoagulant activity without interfering with cytoprotective functions. Recombinant APC (drotrecogin-α [activated]) was developed as a treatment for severe sepsis.24 However, its initial efficacy was not replicated in a subsequent trial.25 With further understanding of the mechanism of action, it has been proposed that, with optimized dosing regimens for maximizing cytoprotective signaling, APC could provide benefit in a septic situation.2 More recently, 3K3A-APC (K191A/K192A/K193A), which has <10% APC anticoagulant activity while retaining cytoprotective activity, has advanced to clinical trials for ischemic stroke.26,27 Importantly, these APC cytoprotective roles are found to be essential because the complete blockade of the APC active site by antibodies leads to many adverse effects, including death.8,28,29 Therefore, the development of modalities that target APC should be judicious of their effects on these essential pathways.
For the modulation of the pleiotropic activity of APC, selective targeting of APC epitopes is essential. One type of camelid antibody is heavy chain immunoglobulin G (IgG). It is composed of 2 heavy chains, each with 1 single variable binding domain and 2 constant domains, CH2 and CH3.30,31 Antibodies with only the variable binding domain region (also known as nanobodies) represent the smallest size of antibodies among the different antibody formats. The application of nanobodies in the modulation of the coagulation pathway has been explored over the years.32 Nanobodies have high stability against pH, temperature, and proteases and have a fast clearance property suitable for an acute treatment setting. The small size allows for high tissue penetration, with some nanobodies reported to cross the blood-brain barrier.33,34 The small prolate shape of nanobodies with a long complementarity determining region 3 (CDR3) and a convex paratope provides the advantage of targeting epitopes that are potentially inaccessible or whose 3-dimensional structure is not recognized by conventional antibody formats.35-37 In an attempt to target the different properties of APC selectively, a nanobody phage library was constructed using peripheral blood mononuclear cells (PBMCs) of llamas immunized with active site–blocked APC (APC-PPACK). Here, a set of 21 llama nanobodies specific to nonactive site APC was identified as having the potential of selectively regulating the pleiotropic activity of APC for therapeutic use.
Methods
Generation of llama nanobodies against the nonactive site of human APC
Two male llamas were immunized with APC-PPACK on days 1 and 77. PBMCs were isolated 5 days after the last immunization. Total RNA was extracted using the RNeasy kit (Qiagen). Complementary DNA was synthesized from RNA using oligonucleotides primers that contain a segment of repeating deoxythymidines and SuperScript III First-Strand Synthesis for reverse transcription polymerase chain reaction (Thermo Fisher Scientific). A nanobody phage library was constructed by directly amplifying the nanobody using framework region 1 and hinge region primers and cloning into a vector with V5 and His-tag. APC-PPACK binders were enriched by panning against APC-PPACK and negatively deselected with FXa-PPACK and PC (supplemental Table 1). The immunization of llamas described in the manuscript was approved by the institutional animal care and use committee of Capralogics (Hardwick, MA).
Phage and bacterial periplasmic extracts enzyme-linked immunosorbent assay screening
Plates coated overnight at 4°C with 2 μg/mL APC-PPACK, PC, or FXa-PPACK and blocked with 2% bovine serum albumin (BSA) at room temperature for 1 hour were used to screen clones from panning rounds R2, R3, and R3′ (supplemental Table 1). The phages were blocked in 4% BSA at room temperature for 1 hour and detected with 1:5000 horseradish peroxidase–anti-M13 monoclonal antibody. Plates were developed with tetramethylbenzidine and quenched with 2N sulfuric acid. Clones with an optical density with a wavelength of 450 nm threefold higher in antigen-coated wells than blocked wells were selected for sequencing. Phage and bacterial periplasmic extracts generated from individual clones in panning round R4 were screened similarly by enzyme-linked immunosorbent assays with mouse anti-V5 monoclonal antibody (1:5000) and secondary goat horseradish peroxidase–anti-mouse IgG (1:5000).
Expression and purification of nanobodies
Escherichia coli in 2YTGC supplemented with 0.1% glucose were inoculated with plasmids of nanobodies containing 6× His-tag and induced with 1 mM isopropyl β-d-1-thiogalactopyranoside at an optical density with a wavelength of 600 nm of 0.5 (25°C). Nanobodies in the periplasm were purified with 1 mL of immobilized metal chelate affinity chromatography resin at 4°C. The resin was washed with 3× 1 mL 10 mM imidazole/phosphate-buffered saline (PBS) followed by elution with 3 mL 100 mM imidazole/PBS. The buffer of nanobodies was exchanged into PBS containing 1 mM calcium chloride (CaCl2). The purity of nanobodies was characterized by sodium dodecyl sulfate polyacrylamide gel electrophoresis (supplemental Figure 1). LP11 was also expressed in mammalian Chinese hamster ovary cells and purified via immobilized metal chelate affinity chromatography, as described earlier.
Binding kinetics analysis by BLI
The Octet HTX (Sartorius, Fremont, CA) sensor surface with anti-mouse IgG was loaded with anti-V5 mouse antibody (10 μg/mL). The loaded sensor was immersed in a nanobody solution (5 μg/mL in the first analysis and 10 μg/mL in the second) to immobilize to the V5-tagged nanobody. The sensor with the immobilized nanobody was immersed in APC dilutions (78.12-5000 nM in PBS, 1 mM CaCl2, 0.1% weight by volume ratio Tween 20, with or without 0.5 mg/mL BSA) for 60 seconds. After APC binding, the surface was immersed in assay buffer for 120 seconds to allow for dissociation. Biolayer interferometry (BLI) measurements were collected during the binding and dissociation phases (25°C). The surface was regenerated between samples with 10 mM glycine buffer and pH 1.7. Kinetic constants (kon, koff, and KD) were calculated using a monovalent (1:1) binding model. Each nanobody was tested against 5 different concentrations of APC.
Effect of nanobodies on APC anticoagulant activity
The PC-activating snake venom reagent, Protac, was used to generate APC in healthy human plasma, and the activated partial thromboplastin time (APTT) clotting time was used to test the effects of the nanobody candidates on the anticoagulant activity of APC. Healthy plasma samples (50 μL) with anti-APC nanobodies were mixed with Protac (25 μL; 1 U/mL) and Stago STA-PTT reagent (75 μL) at 37°C. After a 4-minute incubation, 75 μL of 25 mM CaCl2 solution was added to initiate clotting. The Protac-APTT clotting time of healthy plasma serially diluted with PC-depleted plasma was used as a standard curve. For FVIII inhibitor plasma, 0.5 μg/mL (16 BU/mL) of the FVIII inhibitory antibody GMA-8015 (Green Mountain, Burlington, VT) was added to healthy plasma and incubated for 2 hours before the Protac-APTT assay.
Effect of nanobodies on APC-mediated histone H3 cleavage
APC (50 nM) and anti-APC nanobodies (500 nM) were preincubated in N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid buffered saline (20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 147 mM sodium chloride, and 3 mM potassium chloride; pH 7.4) with 100 μg/mL BSA and 2 mM CaCl2. After 30 minutes, the mixtures were added to 100 μg/mL H3. Over a period of 2 hours, the mixtures were subsampled at different time points and quenched with reducing sample buffer. The samples were heated for 15 minutes at 95°C, centrifuged for 5 minutes at 4000 revolutions per minute, vortexed briefly, and loaded onto a 12% Bis-Tris gel (Bio-Rad) with 2-(N-morpholino)ethanesulfonic acid running buffer (30 μL per well; 750 ng H3). The 20 kDa marker in the Odyssey standard (10 μL) was used as a reference for normalization between gels. Gels were stained overnight with Biotium One-Step Blue Protein Stain (50 mL per gel). Protein bands were scanned on a Li-cor Odyssey imaging system (700 channel; intensity 6; 1 mm offset; and 169 μm resolution) and analyzed with Image Studio version 5.2.
Effect of nanobodies on APC-mediated SEAP-PAR1 cleavage
The effects of nanobodies on APC-mediated PAR1 cleavage were determined using HEK293 cells expressing wild-type–endothelial protein C receptor (EPCR) and a wild-type–, R41Q-, or R46Q-PAR1 cleavage reporter construct with an N-terminal secreted embryonic alkaline phosphatase (SEAP)–PAR1 as described.18,38
Endothelial barrier integrity assay
The effects of nanobodies on APC-mediated endothelial barrier integrity were measured by transendothelial electrical resistance assay (iCelligence system, ACEA, San Diego, CA), as previously described.18,39
Additional experimental details can be found in supplemental Data.
Results
Generation of nanobodies specific for the nonactive site of APC using a phage display immune library
APC-PPACK was used as an immunogen for generating nanobodies against the nonactive site of APC in llamas. After immunization with 2 doses of APC-PPACK, PBMCs were collected, and nanobody-binding regions were cloned to create a nanobody library. The library was panned against APC-PPACK and PC to select clones specific for APC but not PC. After panning round R1, panning was continued in 2 arms differing in the stringency of negative selection for PC (supplemental Table 1). Both the first arm (R1, R2, R3, and R4) and the second arm (R1, R2′, and R3′) led to the enrichment of APC-specific clones. Of the total 1128 phage clones selected from R2, R3, R4, and R3′ (supplemental Figure 2), 725 clones were identified as selective APC-PPACK binders that lacked nonspecific binding to PC or FXa-PPACK (supplemental Figure 3). After sequencing, 158 were found to be unique. Based on their CDR1, CDR2, and CDR3 sequences, these 158 clones were grouped into 18 families, from which 21 nanobodies (named LP1 to LP21) with the highest binding within each family were selected. Additional nonspecific binding studies against FVIIa, FIXa, and FXIa were performed and further demonstrated the selectivity of the LP1 to LP21 nanobodies toward APC (supplemental Figure 4).
Binding kinetics and epitope binning of nanobodies against APC by BLI
To determine if any of the nanobodies bind to the same epitope as the previously reported nonactive site–specific anti-APC TPP-4885 Fab antibody,8 a BLI-based epitope binning study was performed. The nanobodies were tested pairwise with TPP-4885 Fab. Thirteen clones blocked TPP-4885 Fab binding and binned into cluster 1 (Figure 1A). Nanobodies that did not block TPP-4885 Fab binding to APC were selected for additional binning experiments with BLI, except for LP11 because it has >90% CDRs sequence identity in comparison to LP9 (supplemental Table 2). Thus, LP9 was considered as a surrogate for LP11 and belonged to the same epitope bin. Based on the response measurements with BLI, LP7, LP9, LP12, and LP18, they did not compete with LP2, LP3, and LP10 (Figure 1B). LP7 had a weak response measurement as a ligand but appeared to bind well as an analyte. This might have been due to the faster koff of LP7. Clustering analysis showed the clones could be binned into clusters 2 and 3 (Figure 1C). Most nanobodies (62%) are in cluster 1. The lower number of nanobodies for clusters 2 (24%) and 3 (14%) implies that these epitope bins were less accessible and/or more complex for immune recognition and antibody development. These epitope bins also appear to be targetable only by nanobodies with longer CDR3. Unlike cluster 1 nanobodies, which had a median CDR3 length of 9.5 amino acids, the CDR3 of clusters 2 and 3 nanobodies had a median length of 13.5 and 17 amino acids, respectively (Figure 1D; supplemental Table 2). These nanobodies had kons ranging from 1.26 × 105 to 6.22 × 105 M-1s-1 and koffs of 2.59 × 10-4 to 1.37 × 10-3 s-1, with nanobody LP9 having a very low koff, estimated to be ∼6.33 × 10-7 s-1. Two nanobodies had subnanomolar KDs, whereas 15 nanobodies had single-digit nanomolar KDs, and 4 nanobodies had KDs ranging from 16.5 to 27.5 nM (Figure 1E; supplemental Table 3).
Epitope binning and affinity analysis of LPs 1 to 21 nanobody clones with regard to human APC via BLI. (A) In a BLI epitope binning experiment, the binding of human APC to nanobody was detected based on the spectral shift of the reflected light at the sensor surface loaded with LPs ranging from 1 to 21. The biosensor tips with nanobody-APC complexes were then dipped into TPP-4885 Fab solution. Based on spectral shifts, 8 clones were found to not block TPP-4885 Fab binding to APC, whereas 13 clones were found to compete with TPP-4885 Fab for the same epitope bin and blocked it from binding to APC. (B) Nanobodies that did not block TPP-4885 Fab binding to human APC were selected for an additional binning experiment by BLI. LP7, LP9, LP12, and LP18 were found to not compete with LP2, LP3, and LP10. Values in the table represent binding responses over background (red, competition between analyte and ligand for APC; orange, competition but with measurable analyte binding; and green, no competition). (C) Clustering analysis showed 2 clusters of nanobodies at an approximately unbiased (AU) P value cutoff of 95% (red, AU P values; green, bootstrap probability [bp]; and purple, edge number). (D) Summary of clusters and median length of CDR3. (E) KD measurements of LPs ranging from 1 to 21 via BLI. Each nanobody was tested against 5 different concentrations of APC. R2 of these binding kinetic measurements ranged from 0.92 to 0.99.
Epitope binning and affinity analysis of LPs 1 to 21 nanobody clones with regard to human APC via BLI. (A) In a BLI epitope binning experiment, the binding of human APC to nanobody was detected based on the spectral shift of the reflected light at the sensor surface loaded with LPs ranging from 1 to 21. The biosensor tips with nanobody-APC complexes were then dipped into TPP-4885 Fab solution. Based on spectral shifts, 8 clones were found to not block TPP-4885 Fab binding to APC, whereas 13 clones were found to compete with TPP-4885 Fab for the same epitope bin and blocked it from binding to APC. (B) Nanobodies that did not block TPP-4885 Fab binding to human APC were selected for an additional binning experiment by BLI. LP7, LP9, LP12, and LP18 were found to not compete with LP2, LP3, and LP10. Values in the table represent binding responses over background (red, competition between analyte and ligand for APC; orange, competition but with measurable analyte binding; and green, no competition). (C) Clustering analysis showed 2 clusters of nanobodies at an approximately unbiased (AU) P value cutoff of 95% (red, AU P values; green, bootstrap probability [bp]; and purple, edge number). (D) Summary of clusters and median length of CDR3. (E) KD measurements of LPs ranging from 1 to 21 via BLI. Each nanobody was tested against 5 different concentrations of APC. R2 of these binding kinetic measurements ranged from 0.92 to 0.99.
Effect of nanobodies on APC-mediated cleavage of S-2366 chromogenic substrate
Eight nanobodies had no effect on APC-mediated S-2366 cleavage (supplemental Figure 5). Five nanobodies induced partial inhibition, and 8 nanobodies showed an enhancement effect. Among the inhibitory nanobodies, 2 were noncompetitive inhibitors, and 3 were competitive inhibitors for S-2366 cleavage (supplemental Figure 6; supplemental Table 4). Of the 8 enhancing nanobodies, 4 enhanced S-2366 cleavage by increasing the maximum velocity (Vmax), and the other 4 by decreasing KM. The lack of complete inhibition of S-2366 cleavage demonstrated that these 21 APC binders are nonactive site–targeting nanobodies.
Effect of nanobodies on Protac-APTT in healthy human plasma
The effect of nanobodies on APC anticoagulant activity was screened using a Protac-modified APTT assay. Protac, a PC activator derived from snake venom, was added to the plasma to generate APC. Protac prolonged the APTT clotting time from an average of 40.7 to 262.0 seconds depending on the concentration of PC in the plasma (Figure 2A). The nanobodies were screened twice at 300 nM with consistent potency rank order between screens (Figure 2B). All nanobodies reversed the prolonged clot time, at least partially. Among these nanobodies, LP2, LP8, LP9, LP11, LP17, and LP20 inhibited APC by ≥90%, with the prolonged clotting time reversed to below 80 seconds. LP20 was the most potent, with clotting times almost fully corrected (Figure 2C). These data demonstrate that there are at least 3 different epitopes (clusters 1, 2, and 3) on APC recognized by nanobodies for inhibiting anticoagulant activity.
Effect of nanobodies on APC anticoagulant activity. (A) Standard curve of Protac-APTT clotting time in PC-depleted plasma diluted in healthy human plasma (o) to achieve a range of PC levels from 0.78% to 100%. The APTT clotting time of healthy human plasma (•) and PC-depleted plasma (∇) in the absence of Protac are also shown. (B) Twenty-one nanobodies were screened at 300 nM in 2 independent experiments (screens 1 and 2). A consistent rank order was observed in the shortening of the Protac-APTT clotting by the 21 nanobodies in both screens. (C) Nanobodies showed a wide variety of effects on the shortening of the Protac-APTT clotting time (duplicates in 2 independent experiments). Among these nanobodies, LP11, LP8, and LP20 exhibited the highest potency to inhibit anticoagulant activity.
Effect of nanobodies on APC anticoagulant activity. (A) Standard curve of Protac-APTT clotting time in PC-depleted plasma diluted in healthy human plasma (o) to achieve a range of PC levels from 0.78% to 100%. The APTT clotting time of healthy human plasma (•) and PC-depleted plasma (∇) in the absence of Protac are also shown. (B) Twenty-one nanobodies were screened at 300 nM in 2 independent experiments (screens 1 and 2). A consistent rank order was observed in the shortening of the Protac-APTT clotting by the 21 nanobodies in both screens. (C) Nanobodies showed a wide variety of effects on the shortening of the Protac-APTT clotting time (duplicates in 2 independent experiments). Among these nanobodies, LP11, LP8, and LP20 exhibited the highest potency to inhibit anticoagulant activity.
Effect of nanobodies on APC-mediated histone H3 cleavage
APC inhibits histone-induced cytotoxicity via proteolytic cleavage.16 In this assay, APC was incubated with H3 in the presence of nanobodies and the cleavage of H3 was monitored by sodium dodecyl sulfate polyacrylamide gel electrophoresis (Figure 3A). Nine APC-specific nanobodies had minimal impact on APC-mediated H3 cleavage (Figure 3B). Among them, LP14 enhanced H3 cleavage (Figure 3C). Twelve nanobodies inhibited APC-mediated H3 cleavage, resulting in <75% residual activity (Figure 3D), with LP21 being the most potent (Figure 3E).
Effect of nanobodies on APC-mediated histone H3 cleavage. APC (50 nM) and anti-APC nanobody (500 nM) were preincubated for 30 minutes. After the incubation, the mixtures were added to histone H3 (100 μg/mL). Over a period of 2 hours, samples were taken at different time points and added to reducing sample buffer for SDS-PAGEs. The 20 kDa molecular marker was used as a reference for normalization between gels. (A) Time course of H3 cleavage by APC in the absence nanobody (representative of 3 experiments). (B) Nine APC-specific nanobodies have minimal effects on APC-mediated H3 cleavage. The percent of H3 cleavage was calculated by normalizing with the H3 band intensity at time = 0 minute: 100% (intensityt=x min/intensityt=0 minute). The 20 kDa molecular weight (MW) marker was used as reference for normalization between gels. (C) SDS-PAGE of the enhancement of H3 cleavage by LP14. (D) Twelve nanobodies inhibited APC-mediated H3 cleavage. (E) SDS-PAGE of the potent inhibition of H3 cleavage by LP21. SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Effect of nanobodies on APC-mediated histone H3 cleavage. APC (50 nM) and anti-APC nanobody (500 nM) were preincubated for 30 minutes. After the incubation, the mixtures were added to histone H3 (100 μg/mL). Over a period of 2 hours, samples were taken at different time points and added to reducing sample buffer for SDS-PAGEs. The 20 kDa molecular marker was used as a reference for normalization between gels. (A) Time course of H3 cleavage by APC in the absence nanobody (representative of 3 experiments). (B) Nine APC-specific nanobodies have minimal effects on APC-mediated H3 cleavage. The percent of H3 cleavage was calculated by normalizing with the H3 band intensity at time = 0 minute: 100% (intensityt=x min/intensityt=0 minute). The 20 kDa molecular weight (MW) marker was used as reference for normalization between gels. (C) SDS-PAGE of the enhancement of H3 cleavage by LP14. (D) Twelve nanobodies inhibited APC-mediated H3 cleavage. (E) SDS-PAGE of the potent inhibition of H3 cleavage by LP21. SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Effect of nanobodies on APC-mediated SEAP-PAR1 cleavage
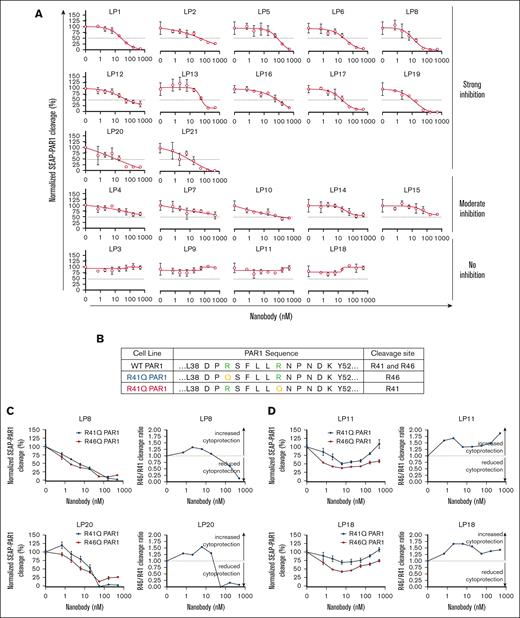
Nanobodies that inhibited PAR1 cleavage by APC could be found in all 3 clusters, with LP1, LP8, LP17, LP19, and LP21 showing the most inhibition. In contrast, the nanobodies LP3, LP9, LP11, and LP18 showed minimal inhibition even at their highest concentration of 500 nM (Figure 4A). At a lower concentration range of 0.7 to 6 nM, these 4 nanobodies had a trend in lowering PAR1 cleavage down to 73% to 82% of controls. These nanobodies belonged to cluster 2 (LP9, LP11, and LP18) and cluster 3 (LP3), respectively. All nanobodies in cluster 1 inhibited PAR1 cleavage. APC can cleave PAR1 at both R41 and R46, with the latter cleavage resulting in cytoprotective signaling. Therefore, 2 PAR1 cleavage-inhibitory nanobodies and 2 PAR1 cleavage-sparing nanobodies were evaluated for their effects on the APC cleavage pattern using R41Q-SEAP-PAR1 and R46Q-SEAP-PAR1 cell lines (Figure 4B). In the presence of inhibitory nanobodies LP8 and LP20, the cleavages at both sites by APC (at 50 nM) were inhibited in a dose-dependent manner. At high concentrations of LP8 and LP20, the cleavage ratio shifted to <1, a less cytoprotective PAR1 cleavage profile (Figure 4C). In comparison, LP11 and LP18 appeared to induce a curvilinear cleavage response for both R46 and R41. At 0.7 to 6 nM, the cleavage of R41 was suppressed to a larger extent than R46. At 6 nM and higher, the suppression of cleavage was reversed. Interestingly, the R46 to R41 cleavage ratio was >1 for the entire concentration range of the assay, suggesting an enhanced cytoprotection effect by LP11 and LP18 (Figure 4D).
Effect of nanobodies on the APC-mediated cleavage of SEAP-PAR1. (A) APC and nanobodies were added to HEK293 cells expressing SEAP-PAR1 and endothelial protein C receptor. Nanobodies were found to modulate PAR1 cleavage to different degrees. LP8, LP17, LP19, and LP21 showed the most potent inhibition. In contrast, LP3, LP9, LP11, and LP18 showed minimal inhibition of APC-mediated PAR1 cleavage even at the highest concentration of 500 nM (n = 3 independent experiments). PAR1 cleavage were expressed as the percentage of the total SEAP activity present on the cells vs background. (B) R41Q-SEAP-PAR1 and R46Q-SEAP-PAR1 cell line designs used to evaluate the cleavage pattern of APC (50 nM) at R46 and R41, respectively, in the presence of cleavage-inhibiting and cleavage-sparing nanobodies. (C) PAR1 cleavage-inhibiting nanobodies, LP8 and LP20, inhibited the cleavage at both R46 and R41 sites by APC in a dose-dependent manner. At high concentrations of LP8 and LP20, the cleavage ratio shifted to below 1, a less cytoprotective PAR1 cleavage profile (n = 3 independent experiments). (D) PAR1 cleavage-sparing nanobodies LP11 and LP18 induced a curvilinear cleavage response at both R46 and R41. The R46 to R41 cleavage ratios were above 1 for the entire concentration range of the assay, suggesting LP11 and LP18 may enhance cytoprotection by APC (n = 3 independent experiments).
Effect of nanobodies on the APC-mediated cleavage of SEAP-PAR1. (A) APC and nanobodies were added to HEK293 cells expressing SEAP-PAR1 and endothelial protein C receptor. Nanobodies were found to modulate PAR1 cleavage to different degrees. LP8, LP17, LP19, and LP21 showed the most potent inhibition. In contrast, LP3, LP9, LP11, and LP18 showed minimal inhibition of APC-mediated PAR1 cleavage even at the highest concentration of 500 nM (n = 3 independent experiments). PAR1 cleavage were expressed as the percentage of the total SEAP activity present on the cells vs background. (B) R41Q-SEAP-PAR1 and R46Q-SEAP-PAR1 cell line designs used to evaluate the cleavage pattern of APC (50 nM) at R46 and R41, respectively, in the presence of cleavage-inhibiting and cleavage-sparing nanobodies. (C) PAR1 cleavage-inhibiting nanobodies, LP8 and LP20, inhibited the cleavage at both R46 and R41 sites by APC in a dose-dependent manner. At high concentrations of LP8 and LP20, the cleavage ratio shifted to below 1, a less cytoprotective PAR1 cleavage profile (n = 3 independent experiments). (D) PAR1 cleavage-sparing nanobodies LP11 and LP18 induced a curvilinear cleavage response at both R46 and R41. The R46 to R41 cleavage ratios were above 1 for the entire concentration range of the assay, suggesting LP11 and LP18 may enhance cytoprotection by APC (n = 3 independent experiments).
Activity profile of nanobodies specific for nonactive site of APC
These 21 nanobodies could be categorized into 13 different activity profiles based on their clusters and effects on the 3 APC functional assays (Table 1). Three nanobodies (LP2, LP8, and LP17) strongly inhibited all 3 APC functional activities. A few nanobodies were found to inhibit only 2 of the 3 functions: LP16 and LP20 inhibit anticoagulation and PAR1 cleavage, and LP1 and LP5 inhibit H3 and PAR1 cleavage. By sparing only 1 of 3 APC functions, the selective inhibition of APC functions by these nanobodies could be useful for dissecting the pleiotropic effects of APC, especially in an in vivo setting. No nanobody was identified for strongly inhibiting only PAR1 cleavage or histone H3 cleavage. Strong monofunction inhibition of APC anticoagulant activity was observed only by LP9 and LP11. Such monofunction inhibition of APC anticoagulant activity may help attenuate bleeding. For instance, LP11 inhibited APC in a dose-dependent manner and shortened the prolonged clotting time in Protac-APTT clotting assays of FVIII inhibitor plasma (supplemental Figure 7).
Characterization of LP11 on endothelial cell barrier function
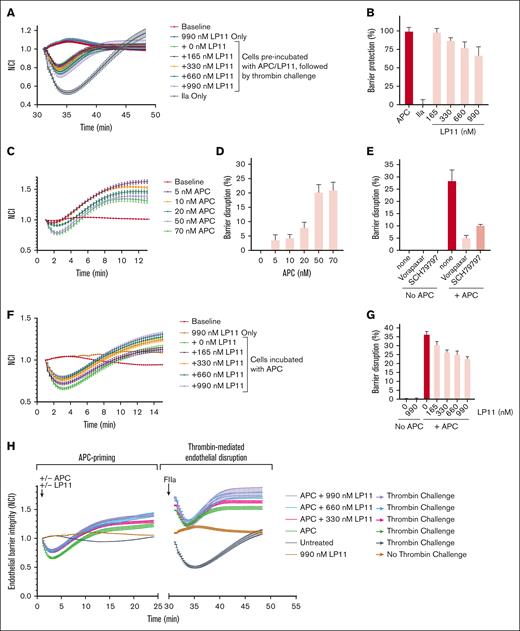
The inhibition of anticoagulation and the enhancement of PAR1 R46 to R41 cleavage ratio by LP11 could lead to prohemostatic activity and enhanced APC-mediated cytoprotection effect at the same time. LP11 was expressed in mammalian cells to permit endothelial barrier function assays on cells. Characterization of Chinese hamster ovary cell–derived LP11 demonstrated that its properties are indistinguishable from E coli–derived LP11 (ie, inhibition of APC anticoagulant activity and minimal effect on R46 cleavage by APC in SEAP-PAR1 cells while significantly inhibiting the cleavage of R41 in PAR1 by APC; supplemental Figure 8). The classical endothelial barrier function assay on EA.hy926 cells for APC barrier protective effects against thrombin-induced barrier disruption18,39 showed minimal effects of LP11 on APC barrier protective effects (Figure 5A-B), consistent with the minimal effect of LP11 on APC-mediated PAR1 cleavage at R46. To evaluate the potential effects of LP11 on the priming of endothelial cells by APC through PAR1 cleavage, we evaluated the endothelial cell layer integrity before barrier disruption is induced by thrombin. APC priming was observed to induce a dose-dependent transient barrier disruption (Figure 5C-D) that could be inhibited by the PAR1 antagonists, vorapaxar and SCH-79797 (Figure 5E), consistent with the notion that APC priming encompasses the effects of APC-mediated cleavage of PAR1 at R41. LP11 inhibited the transient barrier disruption induced by APC priming in a dose-dependent manner. This was consistent with the inhibition of APC-mediated PAR1 cleavage at R41 by LP11 (Figure 5F-G). Most importantly, analysis of the overall barrier function profile showed that the normalized cell index for APC in the presence of LP11 reached higher values compared with APC in the absence of LP11 (Figure 5H), supporting a net enhancement of APC barrier protective effects by LP11. LP11 alone did not change the electrical resistance of the endothelial layer. This data set demonstrates that preferential inhibition of APC-mediated R41 cleavage by LP11 over R46 cleavage during APC priming exerted a cytoprotective effect on endothelial cells.
Effect of LP11 on APC-mediated endothelial barrier function. (A) APC (40 nM) barrier protective effect in the presence of LP11 against barrier permeability induced by thrombin (0.25 nM). Shown is the cell index normalized (NCI) to the time of thrombin addition (n = 4). (B) Quantification of APC barrier protective effects the presence of LP11 shown in panel A was based on the lowest NCI value of each time course, with the APC effect in the absence of LP11 set to 100% and the thrombin alone group as 0%. (C) Transient endothelial barrier disruption shortly after the APC priming of the EA.hy926 cells (n = 4). (D) Quantification of the transient barrier disruption upon APC priming in panel C. (E) Inhibition of the transient barrier disruption by APC priming (40 nM) using PAR1 antagonists, vorapaxar (1 μM) or SCH79797 (1 μM; n = 3). (F) Inhibition of the transient endothelial cell barrier disruption upon APC priming (40 nM) in the presence of LP11 (n = 4). (G) Quantification of the transient barrier disruption upon APC priming in panel F. (H) Overall profile of the changes in the NCI to the time of APC addition to the cells (APC priming).
Effect of LP11 on APC-mediated endothelial barrier function. (A) APC (40 nM) barrier protective effect in the presence of LP11 against barrier permeability induced by thrombin (0.25 nM). Shown is the cell index normalized (NCI) to the time of thrombin addition (n = 4). (B) Quantification of APC barrier protective effects the presence of LP11 shown in panel A was based on the lowest NCI value of each time course, with the APC effect in the absence of LP11 set to 100% and the thrombin alone group as 0%. (C) Transient endothelial barrier disruption shortly after the APC priming of the EA.hy926 cells (n = 4). (D) Quantification of the transient barrier disruption upon APC priming in panel C. (E) Inhibition of the transient barrier disruption by APC priming (40 nM) using PAR1 antagonists, vorapaxar (1 μM) or SCH79797 (1 μM; n = 3). (F) Inhibition of the transient endothelial cell barrier disruption upon APC priming (40 nM) in the presence of LP11 (n = 4). (G) Quantification of the transient barrier disruption upon APC priming in panel F. (H) Overall profile of the changes in the NCI to the time of APC addition to the cells (APC priming).
Discussion
The APC pathway, which includes thrombin/thrombomodulin, PC, protein S, endothelial protein C receptor, and PARs, mediates multiple physiological functions, including anticoagulant, anti-inflammatory, and cytoprotective activities. The targeting of these proteins may provide clinical benefits for the myriad of pathologies involving this pathway. Currently, KRK-α1-antitrypsin and 3K3A-APC are under clinical development for the treatment of hemophilia and ischemic stroke, respectively, by targeting different aspects of this pathway.40 However, much of the therapeutic potential of APC and its substrates awaits further exploration. Studies based on single-molecule fluorescence resonance energy transfer and small-angle radiograph scattering have shown that there are limited changes in the overall architecture of PC upon activation that mostly involve conformational changes within the active site region.41,42 Thus, the identification of antibodies specific only to nonactive sites of APC but not the zymogen presents a challenge. We sought to overcome this challenge by constructing a llama antibody library. This approach allowed the discovery of small-sized nanobodies for the interrogation of APC in their activated conformation. Twenty-one nanobodies belonging to 18 families were further characterized for their binding diversity. In epitope-binding studies, only 3 clusters of nanobodies specific for 3 epitope bins were found. The low number of epitope bins points to limited conformational differences present between nonactive site regions of APC and its zymogen, PC, and corresponds with the data in previous reports.41,42
With APC functional studies, all 3 epitope bins appear to play multiple roles in anticoagulation, H3 cleavage, and PAR1 cleavage simultaneously because none of the bins upon blockade showed an exclusive loss of only one role. Nanobodies within cluster 1 competed with a previously reported antibody, TPP-4885, which targets the APC autolysis loop and potentially also amino acid L429-N431.8 Thus, nanobodies within cluster 1 are expected to bind to a similar region. The autolysis loop is a positively charged loop that plays a role in FVa cleavage,43 FVIIIa cleavage,44 and the sensitivity of APC to serpin inhibition.45,46 Residues R306, K308, K311, R312, and R314 in the autolysis loop are important for FVa cleavage, whereas R306, K311, and R314 are important for FVIIIa cleavage. Because all cluster 1 nanobodies show an effect in reversing the prolonged Protac-APTT clotting time, they are all likely binding to the important arginine and lysine residues on the autolysis loop. The observation of some cluster 1 nanobodies being able to inhibit H3 and/or PAR1 cleavage could suggest previously unreported interactions between the autolysis loop with H3 and PAR1. In contrast, these nanobodies could be binding to regions important for H3 and PAR1 interaction that are proximal but not within the autolysis loop.
Clusters 2 and 3 comprise nanobodies that did not compete with TPP-4885 Fab. Among them, LP9 and LP11 inhibited the anticoagulant activity of APC without interfering with H3 and PAR1 cleavage. This activity profile of LP9- or LP11-bound APC is similar to that of APC mutants designed to have minimal anticoagulation activity. 3K3A-APC (K191A/K192A/K193A)47 has amino acid substitutions in loop 37, and the 5A-APC (K191A/K192A/K193A/R229A/R230A)48 mutant has substitutions in both loop 37 and the calcium-binding loop. The cytoprotective functions of APC are based on its biased agonism of PAR1 by cleaving at R46. In R46 to R41 cleavage ratio studies on SEAP-PAR1 cell lines, LP11 increased the R46 to R41 cleavage ratio by up to 2.5-fold, leading to an overall improvement of endothelial cell barrier function. The preclinical hemophilia and coagulopathy studies have shown that the prohemostatic effect of APC inhibition should be elicited without perturbing the cytoprotective function of APC to provide improved outcomes in hemophilic arthropathy9 and survival outcome in traumatic coagulopathy.28 The novel property of LP11, enhancing the R46 to R41 cleavage ratio to provide additional cytoprotection signaling, may potentially further improve the outcome of these conditions. Further in vivo studies are warranted to elucidate the extent to which LP11 can fulfill these anticipated improved outcomes.
In summary, this llama nanobody library yielded 21 nanobodies that can selectively modulate APC anticoagulation and cytoprotection pathways. These nanobodies represent a tool kit of 13 categories of antibodies based on their cluster and activity profile and offer the opportunity to develop novel therapeutics for the treatment of an array of APC-associated indications, including acute bleeding, hemophilia, ischemia, and sepsis.
Acknowledgments
This work was supported by research funding from Coagulant Therapeutics. L.O.M. was supported by research funding from Coagulant Therapeutics and grants from the National Heart, Lung and Blood Institute, National Institutes of Health (HL142975 and HL104165).
Authorship
Contribution: D.S.S., C.R.M., T.W.H., and L.O.M. contributed to writing of the original draft; D.S.S., M.S., C.R.M., D.S., M.B., T.W.H., and L.O.M. contributed to reviewing and editing of the final manuscript; and all authors designed and performed experiments and analyzed data.
Conflict-of-interest disclosure: D.S.S., C.R.M., and T.W.H. are employees of Coagulant Therapeutics. D.S. and M.B. are consultants for Coagulant Therapeutics. M.B. was employed at Bayer HealthCare for a part of this study. D.S.S., C.R.M., M.B., and T.W.H. have stock ownerships of Coagulant Therapeutics. L.O.M. received research funding from Coagulant Therapeutics. D.S.S., D.S., M.B., and T.W.H. are listed as coinventors on patents filed by Coagulant Therapeutics related to this study. The remaining authors declare no competing financial interests.
Correspondence: Derek S. Sim, Coagulant Therapeutics Corporation, 2630 Bancroft Way, Berkeley, CA 94720; e-mail: dsim@coagulanttherapeutics.com.
References
Author notes
Data are available on request from the corresponding author, Derek S. Sim (dsim@coagulanttherapeutics.com).
The full-text version of this article contains a data supplement.


![Epitope binning and affinity analysis of LPs 1 to 21 nanobody clones with regard to human APC via BLI. (A) In a BLI epitope binning experiment, the binding of human APC to nanobody was detected based on the spectral shift of the reflected light at the sensor surface loaded with LPs ranging from 1 to 21. The biosensor tips with nanobody-APC complexes were then dipped into TPP-4885 Fab solution. Based on spectral shifts, 8 clones were found to not block TPP-4885 Fab binding to APC, whereas 13 clones were found to compete with TPP-4885 Fab for the same epitope bin and blocked it from binding to APC. (B) Nanobodies that did not block TPP-4885 Fab binding to human APC were selected for an additional binning experiment by BLI. LP7, LP9, LP12, and LP18 were found to not compete with LP2, LP3, and LP10. Values in the table represent binding responses over background (red, competition between analyte and ligand for APC; orange, competition but with measurable analyte binding; and green, no competition). (C) Clustering analysis showed 2 clusters of nanobodies at an approximately unbiased (AU) P value cutoff of 95% (red, AU P values; green, bootstrap probability [bp]; and purple, edge number). (D) Summary of clusters and median length of CDR3. (E) KD measurements of LPs ranging from 1 to 21 via BLI. Each nanobody was tested against 5 different concentrations of APC. R2 of these binding kinetic measurements ranged from 0.92 to 0.99.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/13/10.1182_bloodadvances.2022008740/7/m_blooda_adv-2022-008740-gr1.jpeg?Expires=1767733371&Signature=h5OvDw4gaMzk5NRTqfJHNMzKX~F-U98TIbhNBsuDzsRA3BtLa2W6kdnp-hUMGsRoEQWsIiMvR0XpYCw1Ndo6MUkzxUzRrJsNNdZN4-FVJt6HmqjE43lV-X9hyDGYnP3P0m6crOKkXHQLY4THepoVu86zIfDvttajkRzZ9x3OV2Fsr4DMSx-SoySENhTtksif8nhtF4yqTJQ40JVto3nH8T7rxcQg~zv7VNsQzuNVL5RRWTWZj-K1dWEyE6rB-F4kkPn7dPjUte1m0od6VWWo3AWrrSqxBsiPz~GXjKVD0QcGDnkZjR0dc7B-VDBPwGfkxLoC-hBhi9dlKYr9TNpmTA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)