Key Points
Sequence variation in MICA, MICB, and NKG2D genes influences the risk of relapse after haploidentical transplantation.
Transplant outcome is optimal when the features of the patient ligand and donor receptor are both favorable.
Abstract
The recurrence of malignancy after hematopoietic cell transplantation (HCT) is the primary cause of transplantation failure. The NKG2D axis is a powerful pathway for antitumor responses, but its role in the control of malignancy after HCT is not well-defined. We tested the hypothesis that gene variation of the NKG2D receptor and its ligands MICA and MICB affect relapse and survival in 1629 patients who received a haploidentical HCT for the treatment of a malignant blood disorder. Patients and donors were characterized for MICA residue 129, the exon 5 short tandem repeat (STR), and MICB residues 52, 57, 98, and 189. Donors were additionally defined for the presence of NKG2D residue 72. Mortality was higher in patients with MICB-52Asn relative to those with 52Asp (hazard ratio [HR], 1.83; 95% confidence interval [CI], 1.24-2.71; P = .002) and lower in those with MICA-STR mismatch than in those with STR match (HR, 0.66; 95% CI, 0.54-0.79; P = .00002). Relapse was lower with NKG2D-72Thr donors than with 72Ala donors (relapse HR, 0.57; 95% CI, 0.35-0.91; P = .02). The protective effects of patient MICB-52Asp with donor MICA-STR mismatch and NKG2D-72Thr were enhanced when all 3 features were present. The NKG2D ligand/receptor pathway is a transplantation determinant. The immunobiology of relapse is defined by the concerted effects of MICA, MICB, and NKG2D germ line variation. Consideration of NKG2D ligand/receptor pairings may improve survival for future patients.
Introduction
Major advances in the immunobiology of natural killer (NK) cell–mediated eradication of leukemia have significantly increased the curative potential of hematopoietic cell transplantation (HCT) from allogeneic donors.1-4 The successful eradication of malignant cells results from the complex integration of signals from activating and inhibitory NK cell receptors and their ligands.5-7 Among the best-characterized pathways for cancer immunosurveillance is that of the activating NKG2D receptor and its nonclassical HLA class I ligands MICA and MICB.8-12 Engagement of NKG2D with its stress-induced ligands leads to the cytotoxicity of transformed, damaged, or infected cells and is a primary mechanism for averting malignant progression.11,13,14 The expression of NKG2D by NK cells, cytotoxic CD8+ αβ, and γδ T cells places NKG2D at the cornerstone of NK- and T-cell immune responses.14
MICA and MICB ligands are highly polymorphic, and newly discovered alleles are regularly reported.15-17 The dimorphic MICA residue Met129Val has been the subject of extensive research after early studies that showed different binding affinities of NKG2D by MICA-129Met and MICA-129Val.10 Although structurally similar to MICB, MICA has 2 unique sequence features. A short tandem repeat (STR) of 3 nucleotides in exon 5 gives rise to ∼4 to 10 alanine residues (A4, A5, A6, etc) and molecules with long transmembrane regions.15,18,19 A second unique feature is a single base insertion, rs67841474G, in selected MICA alleles (A5.1) that generates a premature stop codon and a truncated short protein devoid of the transmembrane and cytoplasmic tail.15,18-20 The biological ramifications of short and long MICA domains are vast and include differential cell-surface expression, shedding of soluble molecules, and internalization in exosomes, each of which has profound effects on the degree, quality, and sustainability of NKG2D-mediated cytotoxicity.18,20-23 Compared with its ligands, the NKG2D receptor sequence is relatively conserved; the single missense residue Ala72Thr is located outside of the region of MICA contact.24,25 Whether the residue associates with NKG2D expression and/or influences DAP10 binding is not known and remains to be elucidated.13,26,27
MICA-129 has been implicated in graft-versus-host disease (GVHD), relapse, or survival in some but not all studies of T-replete unrelated donor transplantation, using calcineurin inhibitors for prophylaxis of GVHD.28-36 No information on the effect of MICA-STR on the transplantation outcome is available despite its strong effects on expression.18,20-23 New evidence implicates the mismatch of specific MICB residues and NKG2D cytotoxicity haplotypes in the transplantation outcome,37-39 but an understanding of the combined effects of the receptor and its ligands is lacking.
The genes of the NKG2D ligand/receptor axis are diverse, but information regarding the clinical significance of such sequence diversity in haploidentical HCT is rudimentary despite ample evidence that NK cells mediate powerful antileukemia effects in haploidentical HCT. The strength and character of the immune response are strongly shaped by the transplantation regimen, including the removal of T cells to prevent GVHD.2,40 In unmanipulated haploidentical allografting using posttransplantation cyclophosphamide (PTCy), alloreactive T cells are effectively abolished and associated with very low rates of GVHD;41-43 however, another consequence may be the elimination of mature NK cells expressing inhibitory killer-cell immunoglobulin-like receptors (KIRs) and loss of protection from leukemia relapse.44,45 PTCy would presumably also eliminate NKG2D-expressing NK cells, but NKG2D-mediated antileukemia responses have not been fully defined in this setting.
Recurrence of the malignancy remains the chief cause of transplantation failure. We sought to understand the implications of MICA, MICB, and NKG2D variations as single genes and pairwise ligand-receptors in relapse and survival after haploidentical HCT using PTCy. Our hypotheses are based on the premise that patient MICA and MICB ligands are recognized by donor NKG2D. As key sequence features are physically linked, notably from MICA-129 to MICA-STR (supplemental Methods),15,16 we decoupled MICA and MICB polymorphisms to assess the role of each feature in clinical outcome. We tested the hypothesis that ligand-receptor pairs provide important clinical information beyond what can be gleaned from ligands alone. Finally, we assessed whether the transplantation outcome can be optimized by maximizing the number of favorable patient and donor sequence features.
Methods
Study population
We conducted a retrospective cohort analysis of patients who received a haploidentical HCT from 2007 to 2019 (supplemental Table 1). Most patients received PTCy for GVHD prophylaxis. The genomic DNA of patients and donors were characterized for coding variation in MICA and MICB. Donors were additionally defined for the presence of NKG2D variants. Transplantations were performed in US centers that reported clinical data to the Center for International Blood and Marrow Transplant Research. Research biospecimens were purchased from the National Marrow Donor Program Research Repository. We studied all patients (1629) who received a haploidentical related donor transplantation for the treatment of a life-threatening blood disorder using PTCy as GVHD prophylaxis and for whom a research biospecimen was available for the patient and donor. There were no exclusion criteria. The primary outcome measurements were relapse (recurrence of the malignancy) and survival after transplantation. The genetic polymorphisms under study were not considered at the time of HCT. The hypotheses were formulated independent of data collection. Protocols were approved by the institutional review boards of the National Institutes of Health Office for Human Research Protections, the Fred Hutchinson Cancer Center, and the National Marrow Donor Program. Informed consent was obtained from participants.
HLA
HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 were typed to two-field resolution at the time of HCT.46 The 2 recognized MICA variants, MICA-129 and the exon 5 STR, were characterized.15,16 MICB missense changes were selected based on 3 criteria: (1) a frequency of 3% or higher in 1000 Genomes Black, Asian, White, and Hispanic populations;47 (2) missense proxies had a global r2 of 0.75 or higher, and (3) missense change has potential biological relevance.37,38,48-50 Four MICB residues met all criteria (residues 52, 57, 98, and 189). MICA-STR, MICA-129 (rs1051792), MICB-52 (rs3131639), MICB-57 (rs1065075), MICB-98 (rs3134900), MICB-189 (rs41293883), and NKG2D-72 (rs2255336) were characterized as described (supplemental Methods; supplemental Table 2).
Hypotheses focused on the contribution of each sequence feature and haplotype toward the transplantation outcome. Residues were expressed as genotypes (eg, MICA-129 MetMet, MetVal, and ValVal). Shared, patient nonshared, and donor nonshared haplotypes were defined for MICA-129/STR and MICB-52/57/98/189. Mismatches were defined in the graft-versus-host (GVH) vector of incompatibility (any GVH mismatch compared with no GVH mismatch). The GVH vector describes a polymorphism in the patient that is absent in the donor.
Statistical analysis
We examined the association of patient genotype, donor genotype, patient/donor mismatching with relapse (studied in patients with malignant diagnoses), disease-free survival, and overall mortality. Cox regression models were fit to compare the cause-specific hazards of failure between appropriate groups. Patients who did not fail by last contact were censored at last contact. Day 0 for all time-to-event outcomes was taken as the day of transplantation. Failure for disease-free survival was taken to be the earlier date of death or relapse. Models adjusted for patient age, donor age, patient sex, donor sex, year of transplantation, intensity of conditioning regimen, disease status, cytomegalovirus sero-status, source of stem cells, use of total body irradiation, use of PTCy, comorbidity index (0, 1, 2, or >3), patient race, family relationship of donor, HLA-B-leader, and HLA-A, -B, -C, -DR, -DQ, or -DP match status, as appropriate.43,46 Covariates with missing data were included in models by creating an additional category to reflect the missing value of the appropriate covariate. Individual patients were excluded from regression analysis if outcome data were missing for the particular end point. Two-sided P values from Cox regression models were obtained from the Wald test. The outcomes examined are highly correlated, minimizing the impact of multiple comparisons that result from the various outcomes. For this reason, no adjustments were made to the P values associated with the fitted regression model.
Results
We tested a series of hypotheses to define the clinical significance of MICA, MICB, and NKG2D sequence features as individual patient and donor polymorphisms and donor-antihost recognition of ligands in 1629 patients and transplantation donors (supplemental Table 1). We further sought to understand the clinical effects of each paired ligand/receptor combination (patient MICA/donor NKG2D; and patient MICB/donor NKG2D). A total of 656 patients died, 560 relapsed, and 813 survived without disease recurrence. Consistent associations were observed for overall survival, relapse, and disease-free survival. Negative results for these 3 clinical end points and GVHD are provided in the supplemental Data.
MICA ligands
We tested the hypothesis that patient and/or donor MICA-129 genotype provides information regarding the transplantation outcome but found no association with any clinical end point for either the patient, the donor, or patient/donor MICA-129 mismatch (supplemental Table 3).
MICA-STR alleles profoundly affect expression.21,22,51,52 Allele frequencies differed significantly across White, Black, Hispanic, and Asian patients and donors (P < .00001 each) and models adjusted for race. Neither patient nor donor MICA-STR correlated with the outcome (supplemental Tables 4 and 5). However, associations between patient/donor STR mismatch and improved disease-free survival and overall survival were observed (Table 1; Figure 1). Furthermore, there was suggestive evidence that the effect of MICA-STR depended on MICA-129 for mortality (P = .07); when match status at both MICA-129 and MICA-STR are considered concurrently, the protective effect of STR mismatching is even stronger with MICA-129 matching (Table 1).
Donor-recipient mismatching for MICA Exon 5 STR is associated with lower mortality
| Group . | Number . | Overall mortality HR (95% CI; P value) . | Relapse HR (95% CI; P value) . | Disease-free survival HR (95% CI; P value) . |
|---|---|---|---|---|
| No STR GVH vector mismatch | 707 | 1.0 | 1.0 | 1.0 |
| STR GVH vector mismatch∗ | 865 | 0.71 (0.60-0.84; .00006) | 0.87 (0.72-1.04; .13) | 0.81 (0.69-0.94; .007) |
| Both residue 129 and STR-matched | 689 | 1.0 | 1.0 | 1.0 |
| Both residue 129 and STR-mismatched | 327 | 0.77 (0.62-0.96; .02) | 0.87 (0.68-1.11; .25) | 0.82 (0.67-1.00; .05) |
| Mismatched only at STR; matched at residue 129 | 534 | 0.66 (0.54-0.79; .00002) | 0.85 (0.69-1.05; .13) | 0.79 (0.67-0.94; .008) |
| Mismatched only at residue 129; matched at STR | 18 | 0.54 (0.22-1.32; .18) | 0.82 (0.33-2.05; .68) | 0.78 (0.38-1.60; .50) |
| Group . | Number . | Overall mortality HR (95% CI; P value) . | Relapse HR (95% CI; P value) . | Disease-free survival HR (95% CI; P value) . |
|---|---|---|---|---|
| No STR GVH vector mismatch | 707 | 1.0 | 1.0 | 1.0 |
| STR GVH vector mismatch∗ | 865 | 0.71 (0.60-0.84; .00006) | 0.87 (0.72-1.04; .13) | 0.81 (0.69-0.94; .007) |
| Both residue 129 and STR-matched | 689 | 1.0 | 1.0 | 1.0 |
| Both residue 129 and STR-mismatched | 327 | 0.77 (0.62-0.96; .02) | 0.87 (0.68-1.11; .25) | 0.82 (0.67-1.00; .05) |
| Mismatched only at STR; matched at residue 129 | 534 | 0.66 (0.54-0.79; .00002) | 0.85 (0.69-1.05; .13) | 0.79 (0.67-0.94; .008) |
| Mismatched only at residue 129; matched at STR | 18 | 0.54 (0.22-1.32; .18) | 0.82 (0.33-2.05; .68) | 0.78 (0.38-1.60; .50) |
Any GVH vector mismatch: unidirectional and bidirectional.
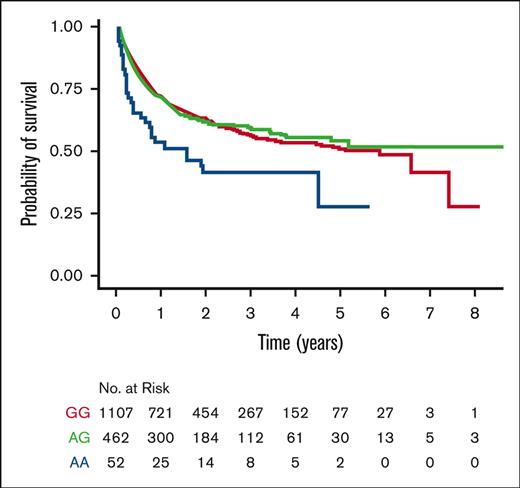
Kaplan-Meier probability of survival. (A) MICA-STR GVH (mis)matching; (B) patient MICB-52 GG (AspAsp), AG (AsnAsp), and AA (AsnAsn); and (C) donor NKG2D-72 GG (AlaAla), AG (ThrAla), and AA (ThrThr).
Kaplan-Meier probability of survival. (A) MICA-STR GVH (mis)matching; (B) patient MICB-52 GG (AspAsp), AG (AsnAsp), and AA (AsnAsn); and (C) donor NKG2D-72 GG (AlaAla), AG (ThrAla), and AA (ThrThr).
Alloreactivity may depend on the specific alleles of the patient and the donor. There were 20 unique mismatch combinations among the 865 MICA-STR–mismatched pairs. Individual group sizes were small and yielded borderline effects. Only mismatches between A5.1 and A6 showed consistent risks for relapse, disease-free survival, and mortality. Interestingly, risks depended on whether the patient or the donor was A5.1- or A6-mismatched. Relative to patient A6/donor A5.1 mismatches, patient A5.1/donor A6 mismatches were associated with an increased risk of relapse (hazard ratio [HR], 1.81; 95% confidence interval [CI], 1.06-3.08; P = .03) and decreased disease-free survival (HR, 1.83; 95% CI 1.21-2.79; P = .004); the HR of mortality was 1.37 (95% CI, 0.87-2.18; P = .18).
MICB ligands
MICB-52, -57, -98, and -189 were examined individually and as 4-residue haplotypes in patients, separately from donors. Patient MICB-52 correlated with outcome, in which MICB-52Asn (rs3131639AA) was associated with a significantly higher risk of mortality and lower disease-free survival compared with MICB-52Asp (rs3131639GG) (Table 2; Figure 1). Patient MICB-52 did not show an association with GVHD (supplemental Table 6). The negative effect of 52Asn is further demonstrated by comparing haplotypes comprised of 57Lys-98Ile-189Thr that differ only for Asp52Asn (Table 2). Variation of patient MICB-57 showed trends for lower relapse and mortality and higher disease-free survival. Variation of patient MICB-98, patient MICB-189, donor genotype, and patient/donor mismatch did not correlate with outcome (supplemental Table 7).
Patient MICB genotype at residue 52 is associated with mortality and disease-free survival
| Patient MICB residue . | Residue (SNP) or 4-residue (SNP) haplotype . | Number . | Overall mortality HR (95% CI; P value) . | Relapse HR (95% CI; P value) . | Disease-free survival HR (95% CI; P value) . |
|---|---|---|---|---|---|
| Residue 52∗ | AspAsp (GG) | 1107 | 1.0 | 1.0 | 1.0 |
| AsnAsp (AG) | 463 | 1.01 (0.84-1.21; .93) | 1.06 (0.87-1.29; .57) | 1.08 (0.92-1.26; .37) | |
| AsnAsn (AA) | 52 | 1.83 (1.24-2.71; .002) | 1.41 (0.84-2.35; .19) | 1.74 (1.20-2.53; .004) | |
| 4-residue haplotype | Asp-Lys-Ile-Thr (GG-AA-CC-CC) | 403 | 1.0 | 1.0 | 1.0 |
| AG-AA-CC-CC | 268 | 0.89 (0.68-1.16; .39) | 0.92 (0.69-1.23; .59) | 0.96 (0.76-1.22; .75) | |
| Asn-Lys-Ile-Thr (AA-AA-CC-CC) | 51 | 1.88 (1.22-2.92; .005) | 1.20 (0.68-2.14; .53) | 1.64 (1.07-2.49; .02) |
| Patient MICB residue . | Residue (SNP) or 4-residue (SNP) haplotype . | Number . | Overall mortality HR (95% CI; P value) . | Relapse HR (95% CI; P value) . | Disease-free survival HR (95% CI; P value) . |
|---|---|---|---|---|---|
| Residue 52∗ | AspAsp (GG) | 1107 | 1.0 | 1.0 | 1.0 |
| AsnAsp (AG) | 463 | 1.01 (0.84-1.21; .93) | 1.06 (0.87-1.29; .57) | 1.08 (0.92-1.26; .37) | |
| AsnAsn (AA) | 52 | 1.83 (1.24-2.71; .002) | 1.41 (0.84-2.35; .19) | 1.74 (1.20-2.53; .004) | |
| 4-residue haplotype | Asp-Lys-Ile-Thr (GG-AA-CC-CC) | 403 | 1.0 | 1.0 | 1.0 |
| AG-AA-CC-CC | 268 | 0.89 (0.68-1.16; .39) | 0.92 (0.69-1.23; .59) | 0.96 (0.76-1.22; .75) | |
| Asn-Lys-Ile-Thr (AA-AA-CC-CC) | 51 | 1.88 (1.22-2.92; .005) | 1.20 (0.68-2.14; .53) | 1.64 (1.07-2.49; .02) |
SNP, single nucleotide polymorphism.
Residue 52: rs3131639 A (Asn)/G (Asp).
Donor NKG2D receptor
Donor NKG2D-72Thr was associated with lower relapse compared with 72Ala (Table 3; Figure 1). Donor NKG2D-72 did not show an association with GVHD (supplemental Table 8). The frequency of the donor NKG2D-72 genotype varies significantly based on the donor race (P < .00001). The frequencies of NKG2D-72 AlaAla, AlaThr, and ThrThr genotypes were: 58%, 35%, and 7%, respectively, in Asian; 37%, 45%, and 18%, respectively, in Black; 63%, 33%, and 4%, respectively, in White; 76%, 22%, and 2%, respectively, in Hispanic donors.
Donor genotype at NKG2D residue 72 is associated with relapse
| Donor NKG2D residue 72 (SNP)∗ . | Number . | Overall mortality HR (95% CI; P value) . | Relapse HR (95% CI; P value) . | Disease-free survival HR (95% CI; P value) . |
|---|---|---|---|---|
| AlaAla (GG) | 954 | 1.0 | 1.0 | 1.0 |
| ThrAla (AG) | 560 | 1.13 (0.95-1.34; .16) | 1.0 (0.83-1.20; .97) | 1.02 (0.87-1.19; .83) |
| ThrThr (AA) | 104 | 0.78 (0.53-1.16; .22) | 0.57 (0.35-0.91; .02) | 0.74 (0.52-1.05; .09) |
| Donor NKG2D residue 72 (SNP)∗ . | Number . | Overall mortality HR (95% CI; P value) . | Relapse HR (95% CI; P value) . | Disease-free survival HR (95% CI; P value) . |
|---|---|---|---|---|
| AlaAla (GG) | 954 | 1.0 | 1.0 | 1.0 |
| ThrAla (AG) | 560 | 1.13 (0.95-1.34; .16) | 1.0 (0.83-1.20; .97) | 1.02 (0.87-1.19; .83) |
| ThrThr (AA) | 104 | 0.78 (0.53-1.16; .22) | 0.57 (0.35-0.91; .02) | 0.74 (0.52-1.05; .09) |
Residue 72: rs2255336 A (Thr)/G (Ala).
Combined patient and donor characteristics
The favorable sequence features associated with lower relapse and improved survival are patient MICB-52Asp, donor NKG2D-72Thr, and patient/donor MICA-STR mismatch. A patient’s genotype cannot be modified, but donors can be selected to optimize outcome. We hypothesized that outcomes for patients with the favorable MICB-52Asp may be further improved when donors have the favorable NKG2D-72Thr and are MICA-STR–mismatched. The lowest relapse and highest survival are observed with STR-mismatched and NKG2D-72Thr donors, followed by STR-matched and NKG2D-72Thr donors (Table 4). These results suggest that the protective effects of NKG2D-72Thr are stronger than those of MICA-STR mismatch. The remaining 4 donor options were generally associated with less favorable outcomes, especially when the donor was both STR-matched and NKG2D-72AlaThr. The beneficial effect of donor MICA-STR mismatch and donor NKG2D-72Thr was less evident for patients with MICB-52Asn or 52AsnAsp (supplemental Table 9).
Outcome of MICB-52Asp patients according to donor MICA-STR–match status and NKG2D-72 genotype
| Donor STR match status/donor NKG2D-72 residue genotype . | Number . | Overall mortality HR (95% CI; P value) . | Relapse HR (95% CI; P value) . | Disease-free survival HR (95% CI; P value) . |
|---|---|---|---|---|
| Matched/Ala | 286 | 1.0 | 1.0 | 1.0 |
| Matched/AlaThr | 162 | 1.30 (0.97-1.74; .08) | 1.09 (0.77-1.53; .63) | 1.12 (0.85-1.48; .41) |
| Matched/Thr | 30 | 0.65 (0.31-1.34; .24) | 0.56 (0.24-1.31; .18) | 0.55 (0.27-1.14; .11) |
| Mismatched/Ala | 331 | 0.79 (0.60-1.03; .08) | 0.89 (0.66-1.18; .41) | 0.83 (0.65-1.05; .12) |
| Mismatched/AlaThr | 226 | 0.88 (0.66-1.18; .40) | 0.99 (0.73-1.34; .95) | 0.93 (0.72-1.20; .57) |
| Mismatched/Thr | 34 | 0.53 (0.25-1.10; .09) | 0.40 (0.17-0.93; .03) | 0.58 (0.32-1.07; .08) |
| Donor STR match status/donor NKG2D-72 residue genotype . | Number . | Overall mortality HR (95% CI; P value) . | Relapse HR (95% CI; P value) . | Disease-free survival HR (95% CI; P value) . |
|---|---|---|---|---|
| Matched/Ala | 286 | 1.0 | 1.0 | 1.0 |
| Matched/AlaThr | 162 | 1.30 (0.97-1.74; .08) | 1.09 (0.77-1.53; .63) | 1.12 (0.85-1.48; .41) |
| Matched/Thr | 30 | 0.65 (0.31-1.34; .24) | 0.56 (0.24-1.31; .18) | 0.55 (0.27-1.14; .11) |
| Mismatched/Ala | 331 | 0.79 (0.60-1.03; .08) | 0.89 (0.66-1.18; .41) | 0.83 (0.65-1.05; .12) |
| Mismatched/AlaThr | 226 | 0.88 (0.66-1.18; .40) | 0.99 (0.73-1.34; .95) | 0.93 (0.72-1.20; .57) |
| Mismatched/Thr | 34 | 0.53 (0.25-1.10; .09) | 0.40 (0.17-0.93; .03) | 0.58 (0.32-1.07; .08) |
Patients with MICB-52Asp who received a transplant from a MICA-STR–mismatched, NKG2D-72Thr donor have lower risk of relapse after transplantation compared with patients who received a transplant from a MICA-STR–matched, NKG2D-72Ala donor.
When only donors with one favorable feature are available, it is of interest to understand whether a STR-mismatched donor is preferable over a NKG2D-72Thr donor. In the study population, far more transplantations were performed from STR-mismatched donors (n = 557) than from NKG2D-72Thr donors (n = 30) (Table 5). Nonetheless, when MICB-52Asp patients lack NKG2D-72Thr donors, transplantation from a STR-mismatched donor lowers mortality and, suggestively, improves disease-free survival. When MICB-52Asp patients lacked STR-mismatched donors, transplantation from NKG2D-72Thr donors yielded HRs of 0.70 for mortality, 0.69 for relapse, and 0.62 for disease-free survival (not significant). Although the number of NKG2D-72Thr donors was a limitation, the results suggest that patients will benefit from transplantation from donors with one favorable feature, either STR mismatch or the NKG2D-72Thr genotype. When donor NKG2D-72 genotype is evaluated for MICB-52AspAsn and 52Asn patients, the numbers of transplantations are limited, particularly for 52Asn patients; however, there is a consistent trend toward lower relapse with NKG2D-72Thr donors (supplemental Table 9).
Outcomes for MICB-52Asp patients when donors have 1 favorable feature
| Donor characteristic . | No. . | Mortality HR (95% CI; P value) . | Relapse HR (95% CI; P value) . | DFS HR (95% CI; P value) . |
|---|---|---|---|---|
| No NKG2D-72Thr donor | ||||
| MICA-STR match | 448 | 1.0 | 1.0 | 1.0 |
| MICA-STR mismatch | 557 | 0.77 (0.62-0.94; .01) | 0.91 (0.72-1.13; .38) | 0.84 (0.70-1.02; .07) |
| No MICA-STR–mismatched donor | ||||
| NKG2D-72Ala | 286 | 1.0 | 1.0 | 1.0 |
| NKG2D-72AlaThr | 162 | 1.32 (0.97-1.79; .08) | 1.11 (0.79-1.57; .55) | 1.13 (0.85-1.50; .39) |
| NKG2D-72Thr | 30 | 0.70 (0.33-1.47; .35) | 0.69 (0.29-1.64; .40) | 0.62 (0.30-1.30; .21) |
| Donor characteristic . | No. . | Mortality HR (95% CI; P value) . | Relapse HR (95% CI; P value) . | DFS HR (95% CI; P value) . |
|---|---|---|---|---|
| No NKG2D-72Thr donor | ||||
| MICA-STR match | 448 | 1.0 | 1.0 | 1.0 |
| MICA-STR mismatch | 557 | 0.77 (0.62-0.94; .01) | 0.91 (0.72-1.13; .38) | 0.84 (0.70-1.02; .07) |
| No MICA-STR–mismatched donor | ||||
| NKG2D-72Ala | 286 | 1.0 | 1.0 | 1.0 |
| NKG2D-72AlaThr | 162 | 1.32 (0.97-1.79; .08) | 1.11 (0.79-1.57; .55) | 1.13 (0.85-1.50; .39) |
| NKG2D-72Thr | 30 | 0.70 (0.33-1.47; .35) | 0.69 (0.29-1.64; .40) | 0.62 (0.30-1.30; .21) |
Models show HRs associated with MICA-STR mismatching relative to matching when no NKG2D-72Thr donor is available (ie, donors are NKG2D-72Ala or AlaThr).
Models show HRs associated with donor NKG2D-72 genotype when no MICA-STR–mismatched donor is available (ie, donors are MICA-STR–matched)
DFS, disease-free survival.
Discussion
Genetic differences across human populations have been extensively cataloged, particularly for the loci that govern histocompatibility (HLA).15,16 The basic premise of HLA in transplantation is donor matching to lower the risks of graft rejection and GVHD.46,53-55 Recently, a role for specific proteins or protein motifs has been observed in a manner similar to classic HLA disease associations in autoimmunity, providing new information about the immunogenicity of HLA and strategies for lowering risk.46,56,57 The principles of classical HLA motivated us to examine the NKG2D axis. Because ligands and receptors may each contribute to the immune response, we studied paired ligands/receptors to understand their combined effects on outcome. Similar to inhibitory KIR,1,3,4 NKG2D ligands and receptors are encoded on different chromosomes and segregate independently, providing an opportunity to leverage each to the fullest potential. As the patient’s ligands cannot be modified, a more complete understanding of germ line variation may inform approaches for selecting the optimal donor.
The implications of MICA structure on function are well-defined and involve 2 highly distinct features: MICA-129, which directly influences the nature and strength of receptor interactions, and MICA-STR, which affects all manner of protein expression.10,21-23,58 We decoupled the 2 sequence features to understand how each might contribute to outcome. Our hypotheses were focused on specific sequence features and did not rely on allele names. This approach uncovered protective effects of STR mismatching on relapse but no association of MICA-129 in either the patient or the donor. We reconstructed the MICA haplotype to test whether STR-mismatch effects depend on MICA-129 and observed the lowest risk of relapse when the donor is STR-mismatched and MICA-129–matched. The underlying mechanisms for MICA-associated relapse risk remain to be elucidated but could involve both the strength of ligand-receptor interactions (in turn influenced by ligand variation and amount of available ligand) and the direct recognition of recipient target cells by the donor. To our knowledge, this genetic model is novel in that allele mismatching has been the classic approach for evaluating the effects of disparity. Yet, the MICA findings show that functionality can be contributed differentially by discrete features of a given allele haplotype. The data strongly point to MICA-STR as the more dominant feature in relapse in our clinical population and raise the possibility that the haplotypic relationship between MICA-STR and MICA-129 may help clarify prior observations.28-36 The well-described implications of MICA-STR (and the rs67841474G insertion) on protein length, cell-surface expression, exosomal sequestration, shedding, and soluble MICA raise intriguing questions regarding their potential contributions to antihost elimination of leukemia. Future analysis of differential surface and soluble MICA protein expression for different MIC-STR alleles might clarify the mechanisms underlying STR-associated relapse risk. Examination of specific MICA-STR mismatch combinations was limited by low numbers; however, increased relapse with patient A5.1/donor A6 suggests that patient A5.1 molecules that are more readily shed might lead to tumor immune escape facilitated by the immunosuppressive effects of high levels of soluble MICA, routes used for ligand release, NKG2D counterregulation, NK cytotoxicity impairment, and CD8+ T-cell costimulation dampening.22,23,52,58-61 The recent demonstration that antibodies targeting the α3 domain of MICA can inhibit MICA shedding and tumor growth provides a promising approach to enhance MICA expression and NKG2D cytotoxicity against the patient’s leukemic cells.62 Recently, a role for donor-specific antibodies directed against MICA in patients undergoing solid organ transplantation and survival has been described.63 Although the current study did not test antibodies, the findings are of interest in HCT, particularly with the new observations of MICA and relapse.
The effects of genetic variation in transplantation are not always manifested through donor mismatching. A patient’s MICB-52 may function similarly to the HLA-B leader, HLA-DRβ peptide-binding motifs, and HLA-DQ heterodimers in unrelated donor HCT.46,56,57 The MICB-Asn52Asp substitution is a conservative change, and its location is not likely to affect its structure or contact with NKG2D;24,25,27 however, the regulation of MICB is highly complex with transcriptional and posttranscriptional events that shape its overall expression and the availability of the ligand to interact with NKG2D.64,65 It is also possible that MICB-52Asp–associated effects could stem from a variant in strong positive linkage disequilibrium with the residue. Previous studies identified mismatching at other MICB residues to be risk factors.37,38 We found a suggestive association between MICB-57GluLys and outcome, but a firm assessment of the effects of mismatching at MICB-98 and -189 was hampered by the high frequencies of the major alleles.
The single missense change at NKG2D-72 was itself associated with relapse. The location of NKG2D-72 in the cytoplasmic domain is not predicted to directly alter ligand/receptor interactions.24,25,27 NKG2D-72 is a putative expression quantitative trait locus for NKG2D and NKG2C.26 The association of NKG2D-72 with relapse might reflect the amount of NKG2D that is available to engage with its ligands and/or broader involvement of other NKG2 resident genes in relapse. These intriguing mechanisms remain to be examined in future studies.
Although variation in each gene affects relapse and survival, the combination of the ligand with its receptor informs outcome beyond what any 1 factor contributes and reflects the important dual role of the ligand and the receptor in the biology of relapse. For NKG2D paired with MICB, the data suggest better survival for patients with MICB-52Asp when the donor is NKG2D-72Thr. Likewise, relapse may be lower when NKG2D-72Thr–positive donors are also MICA-STR–mismatched. The number of patients and donors were limited, and the findings remain to be validated when a larger clinical experience becomes available. The selection of donors who are both STR-mismatched and NKG2D-72Thr is an attractive strategy to supplement pre- and posttransplant therapies to lower the risk of relapse. Because MICA and NKG2D segregate independently, the probability of achieving both is potentially feasible, particularly when 55% of the haploidentical pairs in the current study were STR-mismatched. Although NKG2D-72Thr is a minor allele, it is found in 18% of US Black donors. Similar to the early HLA-DP experience,66 because NKG2D was not a criterion for donor selection, the frequency of STR-mismatched NKG2D-72Thr donors cannot be accurately predicted. Current next-generation sequencing platforms include MICA and MICB loci.17 Testing for NKG2D-72 is technically feasible and would permit the identification of STR-mismatched NKG2D-72Thr donors. When donors with 2 favorable features are not available, transplantation from either STR-mismatched or NKG2D-72Thr donors remains beneficial. Finally, we recently identified large differences in survival among US Black, Hispanic, Asian, and White patients undergoing unrelated donor HCT.67 Population differences in the frequencies of MICA, MICB, and NKG2D variants suggest that a more thorough understanding of the role of the NKG2D axis in relapse and survival in diverse populations will be important to effectively translate data for clinical decision-making.64,68
It is of interest that none of the polymorphisms tested for MICA, MICB, or NKG2D showed an association with acute or chronic GVHD. A major difference between the current study and previous analyses is the use of PTCy as GVHD prophylaxis. The direct cytotoxic effects of PTCy in the immediate posttransplantation period include not only the elimination of alloreactive T cells but also purge mature KIR-expressing NK cells infused in the allograft.44,45 Confirmation of the effects of NKG2D ligand/receptor gene variation in independent populations of patients undergoing haploidentical and unrelated donor transplantation with and without PTCy remain important objectives of future studies. Furthermore, relapse was studied in patients with malignant diagnoses, including leukemia, lymphoma, and myeloma; the effects of ligand/receptor variation might differ depending on the patient’s underlying diagnosis and remain an important question for future studies with a larger transplantation experience.
MICA and MICB have been defined in many disease models in which their function in the stress response and immune evasion are well-known. Together with recent findings of classical HLA in haploidentical transplantation,43 the results of the current study extend information on the constituents of the transplantation barrier. Although the current study was not designed to rank the classical HLA loci with MICA, MICB, and NKG2D, this remains a highly interesting question that will require a larger transplantation experience representative of (mis)matching at each locus and allele and sequence features for MICA, MICB, and NKG2D. Furthermore, given the striking differences in genotype frequencies in individuals of diverse backgrounds, future algorithms for risk assessment and donor selection may consider inclusion of population-informative risks. In transplantation, the effects of patient MICA and MICB on relapse align with their roles as ligands for the donor NKG2D receptor. The identification of specific functional features of MICA, MICB, and NKG2D underscores the importance of understanding ligand-receptor interactions and offers promising opportunities to translate predictive genetics to clinical care.
Acknowledgments
The authors thank Dawn Miller and Mark Gatterman for their outstanding technical assistance.
This study was supported by grants from the National Institutes of Health, (AI069197 [E.W.P., T.G., K.C.H., M.M., C.M., P.S., and S.R.S.]), (CA218285, CA231838, and CA100019 [E.W.P., T.G., M.M., P.S., and C.M.]), (CA015704 [T.G. and S.R.S.]), (5U24CA076518 [S.R.S.]), (HL069294 [S.R.S.]), and the US Office of Naval Research (N00014-20-1-2832 and N000014-21-1-2954 [S.R.S.]).
The funding agencies had no role in the study design, data collection and analysis, the decision to submit the manuscript for publication, or manuscript preparation.
Authorship
Contribution: E.W.P. designed the study; P.S. and T.G. performed statistical analysis; E.W.P. drafted the manuscript; and all authors assembled the data, critically reviewed, edited, and approved the final version of the manuscript.
Conflict-of-interest disclosure: E.W.P., M.M., K.C.H., T.G., C.M., S.R.S., and P.S. report grants from National Institutes of Health, during the conduct of the study. R.K.S. reports patent US20180371051. M.H. declares no competing financial interests.
Correspondence: Effie W. Petersdorf, University of Washington, Fred Hutchinson Cancer Center, 1100 Fairview Ave North, Seattle, WA 98109; e-mail: epetersd@fredhutch.org.
References
Author notes
Data are available on request from the corresponding author, Effie W. Petersdorf (epetersd@fredhutch.org).
Data sharing for research purposes may have partial restrictions, consistent with the informed consent of study participants from whom the data were obtained.
The full-text version of this article contains a data supplement.