TO THE EDITOR:
Somatic genome abnormality is a hallmark of B-cell acute lymphoblastic leukemia (B-ALL) with implications in disease diagnosis and risk stratification.1 Similarly, germline genetic variants in several genes such as TP53, ETV6, PAX5, or IKZF1 have also been associated with the biology of B-ALL and its prognosis.2-5 Transcription factor 3 (TCF3) gene fusion with PBX1 or HLF1 are common somatic alterations in B-ALL but the contribution of germline TCF3 variants to these cancers is not clearly understood.1TCF3 encodes for the E protein E2A, a member of the basic helix–loop–helix (b-HLH) transcription factor family.6 Alternative splicing gives rise to 2 distinct isoforms, E47 and E12, that only differ in exon 18 related to the b-HLH domain.6,7 These 2 isoforms differentially contribute to the E2A transcriptional network, which initiates a series of events during normal hematopoiesis, especially B-cell development.6-8 The loss of TCF3-E47 leads to differentiation arrest at the pre–pro-B stage, whereas TCF3-E12 is dispensable during early B-cell development.8,9 E12 and E47 are both required for VJ rearrangement but with distinctive target genes.7,8 Case reports of homozygous or heterozygous TCF3 germline variants have linked them to congenital immunodeficiency, B- and T-cell Lymphoma.7,9-14 Here we present a comprehensive screening and characterization of TCF3 germline variants in pediatric B-ALL.
A total of 4183 patients enrolled in Children’s Oncology Group (COG) trials AALL0232, P9904/5/6, and AALL0331 and St. Jude Total Therapy XIII and XV clinical trials15-20 for newly diagnosed B-ALL were included for TCF3-targeted sequencing. The study was approved by institutional review boards at St. Jude Children’s Research Hospital, COG member institutions, and Tokyo Medical and Dental University. Informed consent was obtained from parents, guardians, or patients.
TCF3-targeted sequencing was performed as described previously.2-4,21 In brief, genomic DNA was extracted from bone marrow or peripheral blood samples obtained during remission. Illumina dual-indexed libraries were generated from patient germline DNA and pooled in sets of 96 before hybridization with customized Roche NimbleGene SeqCap EZ probes (Roche, Roche NimbleGen, Madison, WI) to capture TCF3 genomic region. Quantitative PCR was used to define the appropriate titer of the capture product necessary to efficiently populate an Illumina HiSeq 2000 flowcell for paired-end 2 × 100 bp sequencing.
For the luciferase assay, full-length coding sequence of TCF3 (E12 and E47) was amplified from cDNA of the B-ALL cell line REH, subcloned into MSCV-IRES-GFP vector, and TCF3 variants were introduced by site-directed mutagenesis. MSCV-TCF3(WT or mutants)–IRES-GFP or mock vector were cotransfected with pGL4-(μE5-μE2)×4 and pRL-SV40 into HEK 293T cells. After 24 hours, luciferase activity was assessed with the Dual-Luciferase Reporter Assay System (Promega, Madison, WI).
For the statistical analyses, rare deleterious TCF3 germline variants in B-ALL were evaluated using CoCoRV (consistent summary counts based rare variant burden test),22 which calculates final statistics based on Cochran–Mantel-Haenszel exact test stratified by ethnicities. Characteristics for 10 B-ALL patients with TCF3 germline variant were compared with 3799 patients with B-ALL without these variants. The analyses were restricted to AALL0232 (n = 2180) and COG P9904/5/6 (n = 1619) clinical trials because of their large sample sizes. Fisher t-test or nonparametric Wilcoxon rank-sum test was used to assess statistical significance.
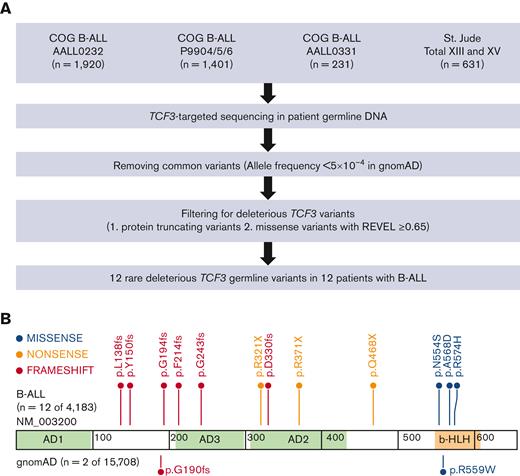
To evaluate the pattern and prevalence of TCF3 germline variants in pediatric B-ALL, we performed targeted sequencing of TCF3 coding regions in 4183 children with newly diagnosed B-ALL from COG and St. Jude frontline trials. Rare deleterious TCF3 variants were identified based on 2 criteria: (1) a population allele frequency ≤5 × 10-4 in the general population derived from the gnomAD data set and (2) protein truncating or missense variants with a REVEL score ≥0.65, implemented using the CoCoRV analysis pipeline22 (Figure 1A). We also examined rare variant-based burden by comparing B-ALL cases and gnomAD noncancer cohort as controls (n = 15 708) stratified by population groups.23 We focused on the E12 isoform (NM_003200) because it is the predominant TCF3 transcript expressed in B-ALL (supplemental Figure 1). In total, 12 unique rare and deleterious TCF3 coding variants were identified in 12 patients with B-ALL, significantly more frequent than in non-ALL control cases (P = 1.24 × 10−5; odds ratio 19.9; supplemental Tables 1-2). The allele fraction of each variant in each sample was confirmed to be ∼50%, consistent with a heterozygous genotype. None of the 12 patients with TCF3 risk variants had a pathogenic variant in the known ALL predisposing genes, which included PAX5, TP53, IKZF1, RUNX1, or ETV6. For 4 patients, we examined whole-exome sequencing or RNA sequencing data and ruled out additional somatic mutations on the wildtype TCF3 allele. The exact penetrance of these TCF3 variants on ALL remains unclear owing to the lack of family history data for the index cases.
TCF3 germline variants in patients with pediatric B-ALL identified by targeted sequencing. (A) CONSORT diagram of COG and St. Jude patients included in this study. (B) Protein domain plot of TCF3 E12 isoform (NM_003200): Activation Domain (AD) 1 to 3 and b-HLH domain. The upper panel shows rare deleterious TCF3 E12 germline variants in patients with B-ALL, and the lower panel shows TCF3 E12 variants in the gnomAD control participants. Each dot represents 1 case.
TCF3 germline variants in patients with pediatric B-ALL identified by targeted sequencing. (A) CONSORT diagram of COG and St. Jude patients included in this study. (B) Protein domain plot of TCF3 E12 isoform (NM_003200): Activation Domain (AD) 1 to 3 and b-HLH domain. The upper panel shows rare deleterious TCF3 E12 germline variants in patients with B-ALL, and the lower panel shows TCF3 E12 variants in the gnomAD control participants. Each dot represents 1 case.
Of these 12 TCF3 variants, 9 led to protein truncation with the loss of the b-HLH domain (Figure 1B). The remaining 3 missense variants clustered within exon 18a, which encodes the b-HLH domain that is responsible for TCF3 dimerization and DNA binding.6,24 This differs from previously published pathogenic TCF3 variants in Burkitt Lymphoma, which exclusively affects exon 18b of the E47 isoform.24 E12 and E47 differentially regulate TCF3 target gene expression7,8 and thus have a likely distinct impact on oncogenic transformation. However, the mechanism by which TCF3 E12 variants contribute to B-ALL development remains unclear.
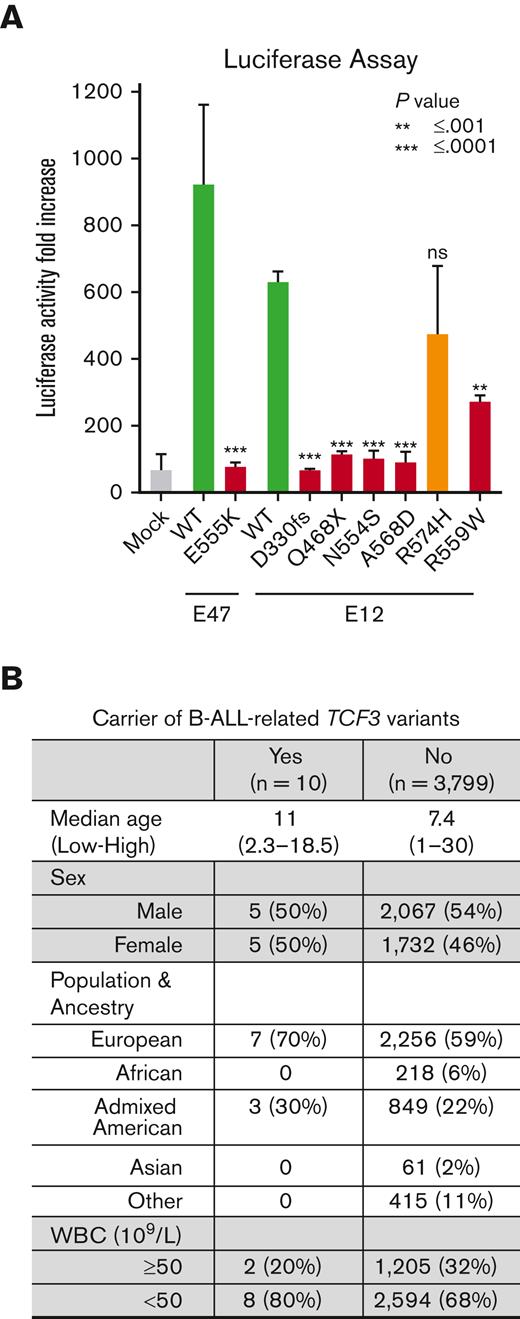
To functionally evaluate B-ALL–related TCF3 variants, we examined transcription factor activity using luciferase reporter assay in HEK 293T cells. TCF3 E47 p.E555K is a known loss-of-function variant9,13 and was included as a positive control. Two of the 3 B-ALL–related missense variants in the E12 isoform (p.N554S and p.A568D) showed a remarkable decrease in transcription factor activity (P < .0001). The missense variant E12 p.R559W, identified in a single non-ALL gnomAD case, was also confirmed as a loss-of-function variant (Figure 2A). These results are consistent with the notion that genetic variation within the b-HLH domain impairs TCF3 dimerization and DNA binding.13,25 However, the E12 p.R574H variant showed WT-like activity and was therefore considered of uncertain significance. Protein truncating variants were predicted to result in the loss of transcriptional activity and this was validated for 2 representative variants (p.D330fs and p.Q468X; Figure 2A). It should be noted that reporter gene assays have inherent limitations, and more sophisticated functional characterization is needed in future studies to fully understand the transcriptional consequences of ALL-related TCF3 variants.
Functional characterization of TCF3 variants and their association with clinical features of pediatric B-ALL. (A) Relative μE5/μE2 promoter activity in luciferase reporter assay using HEK293T cells ectopically expressing TCF3–E12 WT or TCF3-E12 variants. TCF3-E47 p.E555K was included as a positive control. The results are represented as the average ± standard deviation of 6 independent experiments. (B) Characteristics of 10 patients with TCF3 germline variant compared with 3799 patients with B-ALL treated in COG P9904/5/6 and COG AALL0332 clinical trials. WBC, white blood count.
Functional characterization of TCF3 variants and their association with clinical features of pediatric B-ALL. (A) Relative μE5/μE2 promoter activity in luciferase reporter assay using HEK293T cells ectopically expressing TCF3–E12 WT or TCF3-E12 variants. TCF3-E47 p.E555K was included as a positive control. The results are represented as the average ± standard deviation of 6 independent experiments. (B) Characteristics of 10 patients with TCF3 germline variant compared with 3799 patients with B-ALL treated in COG P9904/5/6 and COG AALL0332 clinical trials. WBC, white blood count.
Focusing on patients treated in COG clinical trials, we compared 10 patients with B-ALL with rare deleterious TCF3 germline variants with 3799 patients with B-ALL without these variants. We did not observe any difference in age, sex, population, ancestry, or white blood count at diagnosis (Figure 2B). In addition, TCF3 variant status was not associated with treatment response, that is either end-of-induction minimal residual disease or event-free survival in this cohort (supplemental Table 3). None of the 10 patients with the TCF3 germline variant had somatic fusion genes involving TCF3 (ie, TCF3-PBX1 or TCF3-HLF1).
In summary, we systematically examined germline genetic variants in the TCF3 gene in patients with B-ALL, reporting rare deleterious variants with a cumulative frequency of 0.29%. Two of the 12 variants (p.N554S and p.R574H) are listed in the ClinVar database as variants of uncertain significance. B-ALL–related variants directly affect the TCF3 b-HLH domain, which is critical for TCF3 transcription factor activity and involved in regulating B- and T-cell homeostasis.8,13,14,25 We hypothesize that these variants alter B-cell maturation which may increase the risk for preleukemic clone emergence. Future studies are warranted to fully characterize the mechanism of B-ALL development in children carrying TCF3 risk variants.
Acknowledgments: This work was supported by grants-in-aid for scientific research (grant P50GM115279, R01CA241452, P30CA21765, 19H03614) and The Japan Agency for Medical Research and Development (grant 20ck0106467h0002). COG clinical trials were supported by the National Institutes of Health (grant U10 CA98543, U10 CA98413, U10CA180886, and U10 CA180899). S.P.H. is the Jeffrey E. Perelman Distinguished Chair in the Department of Pediatrics at the Children’s Hospital of Philadelphia.
Contribution: M.T. and J.J.Y. initiated and led the project; C.H.P., S.P.H., M.L.L., and J.J.Y. designed the study; M.D., D.T.T., E.A.R., E.L., P.L.M., W.P.B., S.P.H., C.H.P., and M.L.L. contributed to the data collection; W.C., G.W., W.Y. and Z.L. analyzed genomic and patient data; S.M., Y.N., and M.T. performed luciferase assay; C.E., J.J.Y., and M.T. interpreted the data; C.E. and J.J.Y. wrote the manuscript; and all the authors reviewed and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Masatoshi Takagi, Department of Pediatrics, Tokyo Medical and Dental University, 1-5-45 Yushima, Bunkyo-ku, Tokyo, 113-8510, Japan; e-mail: m.takagi.ped@tmd.ac.jp; and Jun J. Yang, St. Jude Children’s Research Hospital, Pharmaceutical Sciences, 262 Danny Thomas Place, Memphis, TN 38105; e-mail: jun.yang@stjude.org.
References
Author notes
Sequencing data reported in this article have been deposited can in the European Genome-phenome Archive (accession numbers EGAS00001001952 and EGAS00001003266).
Data are available on request from the corresponding author, Jun J. Yang (jun.yang@stjude.org).
The full-text version of this article contains a data supplement.
C.E., W.C., and S.M. contributed equally to this study.