Key Points
BRAFV600E alleles are mainly found in myeloid cells at diagnosis of multisystem LCH.
After more than 2 years, T cells account for 85% of persistent BRAFV600E mutation detected in peripheral blood mononuclear cells.
Abstract
Most children with high-risk Langerhans cell histiocytosis (LCH) have BRAFV600E mutation. BRAFV600E alleles are detectable in myeloid mononuclear cells at diagnosis but it is not known if the cellular distribution of mutation evolves over time. Here, the profiles of 16 patients with high-risk disease were analyzed. Two received conventional salvage chemotherapy, 4 patients on inhibitors were tracked at intervals of 3 to 6 years, and 10 patients, also given inhibitors, were analyzed more than 2 years after diagnosis. In contrast to the patients responding to salvage chemotherapy who completely cleared BRAFV600E within 6 months, children who received inhibitors maintained high BRAFV600E alleles in their blood. At diagnosis, mutation was detected predominantly in monocytes and myeloid dendritic cells. With time, mutation switched to the T-cell compartment, which accounted for most of the mutational burden in peripheral blood mononuclear cells, more than 2 years from diagnosis (median, 85.4%; range, 44.5%-100%). The highest level of mutation occurred in naïve CD4+ T cells (median, 51.2%; range, 3.8%-93.5%). This study reveals an unexpected lineage switch of BRAFV600E mutation in high-risk LCH, which may influence monitoring strategies for the potential withdrawal of inhibitor treatment and has new implications for the pathogenesis of neurodegeneration, which occurred in 4 patients.
Introduction
Langerhans cell histiocytosis (LCH) is a histiocytic neoplasm associated with inappropriate MAPK activation, caused by somatic mutation or fusion of BRAF or other MAPK pathway genes.1-3 LCH cells accumulate, forming destructive lesions and impairing organ function. Multisystem LCH (MS-LCH) with risk organ involvement affects the bone marrow, liver, and spleen leading to death in up to 10% of cases.4 When BRAFV600E is present in patients with high-risk disease, mutation is detectable in cellular DNA isolated from monocytes and myeloid dendritic cells (DCs).5-10
BRAF and MEK inhibitors can induce dramatic clinical improvement in patients with MS-LCH with risk organ involvement who fail salvage chemotherapy.11-16 However, many studies have shown that 1% to 10% of BRAFV600E alleles persist in peripheral blood mononuclear cells (PBMCs) and cell-free DNA, and that treatment withdrawal often results in rapid relapse.11-16 Because monocytes and myeloid DCs are potential precursors of LCH cells, this has led to the assumption that mutation persists mainly in the myeloid cell compartment. In this study, we performed longitudinal and cross-sectional analyses of patients with high-risk LCH from 3 centers and found that the persistence of mutation in peripheral blood was associated with switching of the reservoir from myeloid cells to T cells.
Methods
Patients and samples
Samples were obtained from UK patients joining the LCHIV Biology Study sponsored by Newcastle upon Tyne Hospitals NHS Foundation Trust (Newcastle upon Tyne, United Kingdom) with informed consent according to research ethics committee–approved study protocols. Blood samples from Cincinnati, Ohio, and Houston, Texas, were collected from patients with LCH under protocols approved by the institutional review boards of Cincinnati Children’s Hospital Medical Center and Baylor College of Medicine, respectively.
Full clinical details are given in the supplemental material and supplemental Table 1.
Flow cytometry
Absolute quantitation was performed using TruCount tubes (Becton Dickinson Biosciences), acquired on an LSRFortessa X-20 (Becton Dickinson Biosciences), and analyzed with FlowJo version 10.7.1 (Tree Star). Mononuclear cells were sorted using a BD FACSAria Fusion System (Becton Dickinson Biosciences). Antibodies and gating are given in supplemental Table 2 and supplemental Figure 1.
Measurement of BRAFV600E
DNA was extracted using QIAamp Micro/Mini and Circulating Nucleated Acid Kits (Qiagen). BRAF mutation was measured with mutation allele assay, BRAF_476_mu 4465804 Hs00000111_mu and gene reference assay, BRAF_rf 4465807 Hs00000172_rf (Life Technologies) on QuantStudio 3 Real-Time PCR System (ThermoFisher Scientific) with 50 cycles. Ctmutant and Ctreference wild-type values were manually inspected and compared with negative controls as described in the supplemental information and supplemental Figure 2. The fraction of mutated alleles was calculated from 1/2[Ct(reference wild-type) − Ct(mutant)]. The fraction of mutated cells is double this value, assuming heterozygous mutation. The absolute number of mutated cells per μL is the product of the absolute cell count and the fraction of mutated cells. The percentage contribution to mutation burden is the proportion contributed by each cell type of the sum of all of the mutated alleles measured. Graphs and statistics were done with Prism version 5.0 (GraphPad Software Inc).
Results and discussion
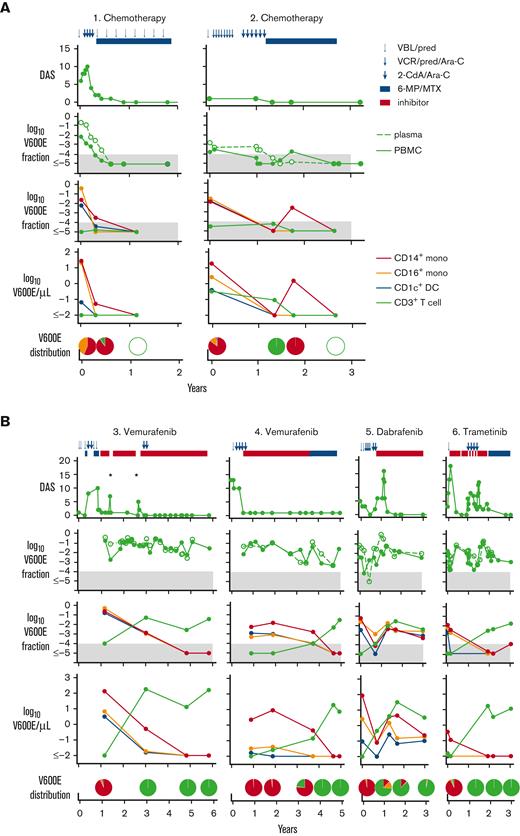
The kinetics of BRAFV600E mutation were observed in 2 patients undergoing salvage chemotherapy with cladribine and cytarabine. The level of BRAFV600E declined in parallel with clinical improvement, until it was undetectable (Figure 1A). During treatment, mutation was slightly higher in plasma cell-free DNA than in PBMC (supplemental Figure 3A). Although these patients remained in clinical remission with undetectable mutation more than 3 years later, 4 patients who failed salvage chemotherapy and were treated with MAPK inhibitors rapidly improved but maintained a high level of mutant alleles at similar levels in PBMC and plasma cell-free DNA (Figure 1B; supplemental Figure 3B).
Clinical progress and distribution of BRAFV600E alleles in PBMC and sorted cell fractions. (A) Patients 1 and 2 treated with conventional chemotherapy. (B) Patients 3 to 6 treated with inhibitors. Pie charts depict the distribution of mutated cells as a proportion of all mutated cells detected in blood. Each chart is aligned as closely as possible to the corresponding time point. Blue symbols at the top indicate chemotherapy cycles; red bar indicates MAPK inhibitor. Clinical scores and mutation analyses over time are shown in rows (top to bottom). Filled gray areas indicate negative results below 0.0001. Disease Activity Score (DAS) was measured according to Donadieu et al17; BRAFV600E mutation fraction measured in total PBMC (filled symbols and lines) and plasma cell-free DNA (open symbols and broken line); BRAFV600E mutation fraction measured in sorted subsets; and mutated cells per μL in sorted subsets. Asterisks indicate rapid disease reactivation when treatment was suspended. Red line indicates classical CD14+ monocytes; orange line indicates CD16+ nonclassical monocytes; blue line indicates CD1c+ myeloid DC; and green line indicates CD3+ T cells. VBL/pred, vinblastine and prednisone; VCR/pred/Ara-C, vincristine, prednisone, and cytarabine; 2CdA/Ara-C, cladribine and cytrarabine; 6-MP/MTX: 6-mercaptopurine and methotrexate maintenance.
Clinical progress and distribution of BRAFV600E alleles in PBMC and sorted cell fractions. (A) Patients 1 and 2 treated with conventional chemotherapy. (B) Patients 3 to 6 treated with inhibitors. Pie charts depict the distribution of mutated cells as a proportion of all mutated cells detected in blood. Each chart is aligned as closely as possible to the corresponding time point. Blue symbols at the top indicate chemotherapy cycles; red bar indicates MAPK inhibitor. Clinical scores and mutation analyses over time are shown in rows (top to bottom). Filled gray areas indicate negative results below 0.0001. Disease Activity Score (DAS) was measured according to Donadieu et al17; BRAFV600E mutation fraction measured in total PBMC (filled symbols and lines) and plasma cell-free DNA (open symbols and broken line); BRAFV600E mutation fraction measured in sorted subsets; and mutated cells per μL in sorted subsets. Asterisks indicate rapid disease reactivation when treatment was suspended. Red line indicates classical CD14+ monocytes; orange line indicates CD16+ nonclassical monocytes; blue line indicates CD1c+ myeloid DC; and green line indicates CD3+ T cells. VBL/pred, vinblastine and prednisone; VCR/pred/Ara-C, vincristine, prednisone, and cytarabine; 2CdA/Ara-C, cladribine and cytrarabine; 6-MP/MTX: 6-mercaptopurine and methotrexate maintenance.
As previously reported, mutation was mainly present in CD14+ classical monocytes, CD16+ nonclassical monocytes, and CD1c+ myeloid DCs at diagnosis.5-10 Lymphoid cells including CD3+ T cells, CD19+ B cells, and CD56+ natural killer cells were either an order of magnitude lower or negative at this point. However, with time, we observed an increase in both the mutated allele fraction and the absolute number of mutated alleles per μL, attributable to T cells. At 1.5 to 4.5 years after diagnosis, the mutation burden of T cells exceeded that of myeloid cells (Figure 1B).
Early attempts to withdraw inhibitor therapy were not successful in patient 3. However, patients 4 and 6 were successfully moved to oral maintenance with 6-mercaptopurine and methotrexate, without systemic relapse at the point at which BRAFV600E had switched completely to T cells (Figure 1B). Patient 6 had also received high-dose chemotherapy during treatment with trametinib, a strategy also reported by others in association with clearance of mutation.18 However, in our patients, BRAFV600E persisted in T cells, but systemic disease did not recur. Further studies will be required to determine whether these observations are causally related.
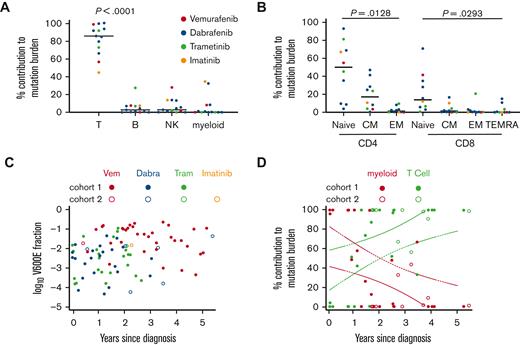
Seeking to validate the observation that mutation switched from myeloid cells to T cells and to exclude the contribution of other lymphoid cells that had not been directly measured in the first cohort, we recruited a second cohort of 10 patients. A single blood sample was obtained at least 2 years after diagnosis and between 6 months and 5 years on inhibitor therapy. This confirmed that a median of 85.4% of mutated alleles were present in T cells, compared with 2.7% in B cells, 2.8% in natural killer cells, and 0.3% in myeloid cells (Figure 2A). Granulocytes were consistently negative in our hands at all time points.7BRAFV600E was most abundant in naïve CD4+ and CD8+ cells, suggesting that enrichment in the T-cell lineage was not simply because of the expansion of memory clones (Figure 2B). Combining the data from all patients in both cohorts, we observed that the level of mutation remained roughly constant in PBMC, but with time, switched from myeloid to T cells (Figure 2C-D).
Comparison and quantitation of alleles in lymphoid subsets. (A) The percentage contribution of each PBMC subset to the total mutation burden (obtained by summing the parts) among CD3+ T cells, CD19+ B cells, and myeloid cells (monocytes and DCs). (B) The percentage contribution of each T-cell subset to the mutation burden present in T cells among naïve, central memory (CM), effector memory (EM), and effector memory cells re-expressing CD45RA (TEMRA). Supplemental Figure 1 shows the sorting strategy. In panels A and B, the graphs show data from 1 late time point of all patients with a sufficient material available (n = 13; analysis of variance). (C) Summary of the mutant allele fraction in PBMC in all samples as a function of time on treatment. (D) Summary of the percentage contribution from T cells (green) and all myeloid cells (red) to the total mutation burden of these subsets as a function of time since diagnosis (n = 33). Broken lines depict the 95% confidence intervals for a simple linear regression. Dabra, dabrafenib; NK, natural killer; Tram, trametinib; Vem, vemurafenib.
Comparison and quantitation of alleles in lymphoid subsets. (A) The percentage contribution of each PBMC subset to the total mutation burden (obtained by summing the parts) among CD3+ T cells, CD19+ B cells, and myeloid cells (monocytes and DCs). (B) The percentage contribution of each T-cell subset to the mutation burden present in T cells among naïve, central memory (CM), effector memory (EM), and effector memory cells re-expressing CD45RA (TEMRA). Supplemental Figure 1 shows the sorting strategy. In panels A and B, the graphs show data from 1 late time point of all patients with a sufficient material available (n = 13; analysis of variance). (C) Summary of the mutant allele fraction in PBMC in all samples as a function of time on treatment. (D) Summary of the percentage contribution from T cells (green) and all myeloid cells (red) to the total mutation burden of these subsets as a function of time since diagnosis (n = 33). Broken lines depict the 95% confidence intervals for a simple linear regression. Dabra, dabrafenib; NK, natural killer; Tram, trametinib; Vem, vemurafenib.
Most reports agree that BRAFV600E is found primarily in myeloid cells at diagnosis of MS-LCH.5-10,13 Mutation has previously been reported exclusively in T cells of 1 patient treated with inhibitors who had undergone stem cell transplantation.13 Our data now show that T cells become the dominant reservoir in all patients with high-risk LCH with persisting mutation. Although most of the patients in this study had been maintained on MAPK inhibitor therapy for several years, 1 received imatinib and 2 were diagnosed more than 4 years earlier, before being switched to MAPK inhibitors for only 6 to 12 months before sampling. Although it is conceivable that MAPK inhibitors might exert a greater selective pressure on myeloid-primed progenitors allowing lymphoid lineages to grow out, inhibitor treatment may have simply allowed us to observe a natural time-dependent process in patients with high-risk disease.
Although we noted that 2 patients who received chemotherapy with MAPK inhibitors were able to withdraw from inhibitors, the confinement of mutation to T cells may not correlate with safe treatment cessation in all patients. Vigilance for late effects must also be maintained. Patient 4 developed neurodegeneration 1 year after vemurafenib was withdrawn (supplemental Figure 5), and 3 patients in cohort 2 developed neurodegeneration just before starting inhibitor therapy. In all, their T cells accounted for >90% of BRAFV600E, at the onset of symptoms. Mutation has previously been reported in the T cells of conventionally treated patients who developed neurodegeneration19 and are known to access the central nervous system in other forms of neuropathology.20 Given the similar latency of lineage switching and the onset of neurodegeneration, it will be of great interest to determine whether there is a causal link with T cells carrying activated BRAF.
Acknowledgments
The authors thank the patients and families who participated in this study.
This work was supported by funding from the Cancer Research UK biomarker project (grant C30484/A21025), Histio UK, Histiocytosis Association, and Bright Red.
This work would not have been possible without the inspiration and enthusiasm of our late friend and colleague, Johann Visser.
Authorship
Contribution: P.M. designed and performed experiments, interpreted data, and wrote the manuscript; S.B., A. Kumar, and C.A. provided samples and clinical data and interpreted data; O.S., J.N., D.Y., R.G., A.P., J. Pears, G.M., B.M., and J.H. provided samples and clinical data; A.N., S.R., and J.V. provided samples; K.M. provided and interpreted clinical data; A. Kamal, C.R.-G., and M. Minkov provided clinical data; J.L., M. Madhkali, J.M., and P.S. processed samples; S.P. coordinated sample collection and processing; J.D. and V.B. interpreted data; J. Picarsic provided samples and interpreted data; and M.C. designed experiments, interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: A. Kumar is a consultant for Sobi and SpringWorks Therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Matthew Collin, Translational and Clinical Research Institute, Newcastle University, Framlington Place, Newcastle upon Tyne NE2 4HH, United Kingdom; e-mail: matthew.collin@newcastle.ac.uk.
References
Author notes
Data are available on request from the corresponding author, Matthew Collin (matthew.collin@newcastle.ac.uk).
The full-text version of this article contains a data supplement.