Key Points
Both patients showed sequential exaggerated TPO, platelet gene, and platelet count cycling, as well as trilineage gene fluctuations.
Both had somatic GOF STAT3 mutations and clonal T cells; 1 had a novel germline LOF MPL mutation correlating with cycling parameters.
Abstract
Cyclic thrombocytopenia (CTP) is a rare disease of periodic platelet count oscillations. The pathogenesis of CTP remains elusive. To study the underlying pathophysiology and genetic and cellular associations with CTP, we applied systems biology approaches to 2 patients with stable platelet cycling and reciprocal thrombopoietin (TPO) cycling at multiple time points through 2 cycles. Blood transcriptome analysis revealed cycling of platelet-specific genes, which are in parallel with and precede platelet count oscillation, indicating that cyclical platelet production leads platelet count cycling in both patients. Additionally, neutrophil and erythrocyte-specific genes also showed fluctuations correlating with platelet count changes, consistent with TPO effects on hematopoietic progenitors. Moreover, we found novel genetic associations with CTP. One patient had a novel germline heterozygous loss-of-function (LOF) thrombopoietin receptor (MPL) c.1210G>A mutation, and both had pathogenic somatic gain-of-function (GOF) variants in signal transducer and activator of transcription 3 (STAT3). In addition, both patients had clonal T-cell populations that remained stable throughout platelet count cycles. These mutations and clonal T cells may potentially involve in the pathogenic baseline in these patients, rendering exaggerated persistent thrombopoiesis oscillations of their intrinsic rhythm upon homeostatic perturbations. This work provides new insights into the pathophysiology of CTP and possible therapies.
Introduction
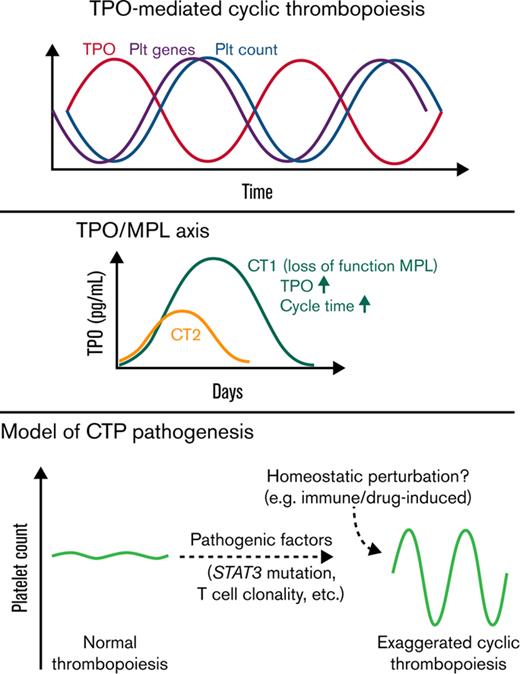
The production of megakaryocytes and platelets in the bone marrow (BM) is strictly regulated by the balance of thrombopoietin (TPO) and platelet mass to maintain platelet homeostasis. Patients with cyclic thrombocytopenia (CTP) have substantial periodic platelet oscillations with amplitudes ranging from thrombocytopenic to normal/thrombocytosis levels. Patients with CTP typically have a cycling time of 3 to 5 weeks.1,2 Various potential etiological associations including cyclical megakaryopoiesis, menstrual cycles, cyclical autoimmune platelet destruction, and T-cell clonality have been observed, yet the pathogenesis remain elusive.1-3
The oscillatory nature of hematopoiesis in general has been recognized and mathematically modeled as a nonlinear dynamical system with feedback loops involving time delays.4 After perturbations, such systems entail oscillations with a constant period of about twice the time delay, and certain conditions introducing moderate thrombocytopenic baselines may increase the “gain” of the system and result in a stable and exaggerated oscillatory stage. Morley, Mackey, and others postulated that CTP could be a consequence of errors in or perturbation of thrombopoiesis.5-7 Biological findings correlating with these concepts are needed to improve specific modeling for CTP.7 We hypothesized that inherited or acquired genetic mutations affecting thrombopoiesis could create a vulnerability of this system to transition to a CTP phenotype and that patterns of gene expression during cycling could shed light on the regulation of this process.
Methods
The study was approved by the Stanford University Institutional Review Board and conducted in accordance with the Declaration of Helsinki. Two patients enrolled with informed consent. Twenty-four consecutive blood samples from patient 1 (CT1) and 15 from patient 2 (CT2) were collected every 3 to 4 days.
MPL was Sanger sequenced. MPL c.1210G>A mutation was constructed. Wild-type (WT) or the mutant MPL cDNA was stably transfected into Ba/F3 cells for functional analyses.
The Stanford Tumor Actionable Mutation Panel for Hematopoietic and Lymphoid Neoplasms, or Heme-STAMP in short, is a targeted next-generation sequencing (NGS)-based panel. Heme-STAMP was performed on whole blood DNA to screen for hematolymphoid neoplasm–associated mutations in 164 genes.
Peripheral blood mononuclear cell DNA were used for NGS-based T- and B-cell receptor clonality profiling (LymphoTrack, Invivoscribe).
Blood RNAs were extracted after red blood cell lysis and sequenced for longitudinal transcriptome analysis using SAMseq algorithm to investigate platelet count–associated gene expressions.
Details of these methods are provided in supplemental Methods.
Results and discussion
CT1, a 58-year-old male, exhibited platelet count oscillation (range, 1-409 × 109/L; mean, ∼128 × 109/L) and an inverse plasma TPO oscillation (6-2745 pg/mL) with a prolonged cycling period of 39 days.8 CT2, an 86-year-old male, also exhibited reciprocal platelet count (range, 7-142 × 109/L; mean, ∼65 × 109/L) and TPO oscillations (12-405 pg/mL) over a period of 26 days (Figure 1A). Neither patient had known comorbidities or medications known to affect platelet number or function.
Oscillations of platelet count, plasma TPO, and trilineage-specific genes. (A) Reciprocal platelet count and plasma TPO oscillations in CT1 and CT2. CT2 received a platelet transfusion on day 2 (green arrow). TPO levels were measured in platelet-poor plasma by Quantikine ELISA (R&D Systems). (B) Venn diagram of the number of genes quantitatively associated with platelet count changes in both patients and gene list of the 3 lineage-specific groups. (C) Heatmap of the 151 platelet-specific (Plt-specific), the 19 erythrocyte-specific (Ery-specific), and the 7 immature neutrophil-specific (Neut-specific) genes displaying their longitudinal expression patterns over 2 platelet cycles for each patient. Using Cluster 3.0, the expression levels of individual genes were mean-centered across all time points and log-transferred for heatmaps, which were visualized using Java TreeView (Version 1.1.6r4) and color-scaled into yellow (high expression) to blue (low expression) or gray (below detection limit). (D) Quantification of the heatmap by showing the median levels of each gene group at every time point to illustrate their longitudinal patterns. (E) The median expression levels (in transcripts per million [TPM]) of the 34 platelet-specific genes that showed the highest fold changes in relation with platelet count and TPO oscillations. These platelet-specific genes are in parallel with and leading platelet count by 3 to 4 days in both patients. Together, they mirror the TPO oscillations. ELISA, enzyme-linked immunosorbent assay.
Oscillations of platelet count, plasma TPO, and trilineage-specific genes. (A) Reciprocal platelet count and plasma TPO oscillations in CT1 and CT2. CT2 received a platelet transfusion on day 2 (green arrow). TPO levels were measured in platelet-poor plasma by Quantikine ELISA (R&D Systems). (B) Venn diagram of the number of genes quantitatively associated with platelet count changes in both patients and gene list of the 3 lineage-specific groups. (C) Heatmap of the 151 platelet-specific (Plt-specific), the 19 erythrocyte-specific (Ery-specific), and the 7 immature neutrophil-specific (Neut-specific) genes displaying their longitudinal expression patterns over 2 platelet cycles for each patient. Using Cluster 3.0, the expression levels of individual genes were mean-centered across all time points and log-transferred for heatmaps, which were visualized using Java TreeView (Version 1.1.6r4) and color-scaled into yellow (high expression) to blue (low expression) or gray (below detection limit). (D) Quantification of the heatmap by showing the median levels of each gene group at every time point to illustrate their longitudinal patterns. (E) The median expression levels (in transcripts per million [TPM]) of the 34 platelet-specific genes that showed the highest fold changes in relation with platelet count and TPO oscillations. These platelet-specific genes are in parallel with and leading platelet count by 3 to 4 days in both patients. Together, they mirror the TPO oscillations. ELISA, enzyme-linked immunosorbent assay.
Whole blood transcriptome analyses identified 236 genes that were quantitatively correlated with platelet count in both patients. These genes can be parsed into 3 groups based on their expression patterns (Figure 1B; supplemental Table 1). The largest group consisted of 151 platelet-specific genes that oscillated preceding platelet count fluctuations by 3 to 4 days (Figure 1B-D). Thirty-four of these 151 genes (eg, MYL9, TUBB1, ITGA2B) demonstrated over 50-fold and 10-fold expression changes during platelet count cycles in CT1 and CT2, respectively (Figure 1B). As platelet RNAs are inherited from megakaryocytes and are positively correlated with the number of newly formed platelets,9 the expression patterns suggest cyclical platelet production underlie platelet count oscillations in both patients (Figure 1E).
Both patients also shared fluctuations of immature neutrophil-specific10,11 (eg, AZU1, ELANE, LCN2) and erythrocyte-specific (eg, ALAS2, EBP42, HBM) genes, with cycling patterns preceding platelet-specific genes (Figure 1B-D). Gene ontology analysis showed enrichment of neutrophil-specific and erythrocyte-specific biological processes in these platelet count–associated genes (supplemental Table 2). In contrast to gene expression patterns, only neutrophil counts in CT1 showed a cycling trend, all within normal range (supplemental Figure 1). This may be due to dampening (buffering) effects of marrow neutrophil release control and long lifespan of erythrocytes that absorbed the mild production fluctuations.4
The TPO and trilineage gene expression patterns are consistent with TPO-mediated oscillations4 in both patients. For CT2, the 14d interval between TPO and platelet count oscillations resembles the normal time delay (about 12 days) of TPO effect on platelet count in healthy people,12 whereas CT1 displayed a prolonged time delay of 21d. Moreover, compared with CT2 and prior CTP cases,3,13-16 CT1 had much higher TPO peak levels, similar to those observed in congenital amegakaryocytic thrombocytopenia.17 These prompted us to sequence the TPO receptor, MPL, in CT1, which revealed a novel germline c.1210G>A heterozygous mutation, resulting in a p.Gly404Arg (G404R) substitution in a highly conserved site (Figure 2A-B; supplemental Figure 2). No MPL mutation was detected in CT2.
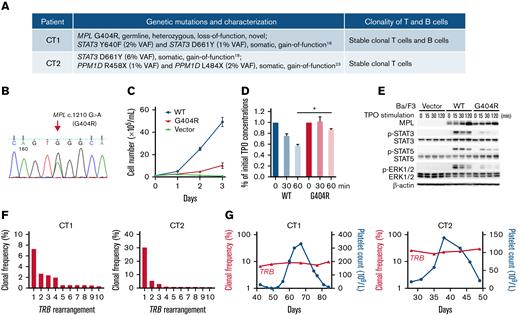
Genetic alterations and clonal lymphocytes patterns in these patients. (A) A list of the pathogenic blood cell mutations and clonality of T and B cells detected in each patient. (B) Sanger sequencing data of the MPL c.1210G>A (G404R) heterozygous mutation in CT1 using both blood and hair DNA samples. (C) The MPL G404R mutation failed to support TPO-stimulated growth of Ba/F3 cells. Ba/F3 cells expressing WT MPL, mutant MPL, or vector were incubated in medium without IL-3 but containing 10 ng/mL TPO. Cell numbers were counted daily using hemocytometer and displayed as mean plus or minus SD. (D) The mutant MPL is impaired in TPO uptake, a process involving TPO-MPL binding at the cell surface and its subsequent internalization. Ba/F3 cells expressing WT or mutant MPL were seeded in 6-well plates containing equal amounts of TPO with 5 × 106 cells per well. TPO uptake was monitored by measuring the remaining TPO concentration in cell supernatant after designated incubation time. After 60 minutes, the remaining TPO in WT cells was significantly lower than that of mutant cells (P = .0108) based on paired Student t test. Mean plus or minus SD of 2 separate experiments is displayed. (E) Mutant MPL leads do reduced JAK-STAT pathway activation upon TPO stimulation. Compared with WT MPL (WT), Ba/F3 cells expressing the mutant MPL (G404R) had reduced phosphorylation of JAK-STAT pathway proteins such as p-STAT3, p-STAT5, and p-ERK1/2 after 15 to 120 minutes of TPO stimulation as revealed by western blot. Ba/F3 cells transfected with the expression vector (Vector) was a negative control. β-Actin was a loading control for western blot. (F) Clonality of T cells in CT1 and CT2. Histograms show the frequencies of the top 10 clones of T-cell receptor β locus (TRB) in a sample at platelet count descending phase. Both CT1 and CT2 had clonal TRB. (G) The top clones of TRB in 6 sequential samples in CT1 and 5 in CT2 were identical. Their clonal frequencies were plotted and linked to show their stable longitudinal patterns over a platelet count cycle. IL-3, interleukin-3; SD, standard deviation; VAF, variant allele frequency.
Genetic alterations and clonal lymphocytes patterns in these patients. (A) A list of the pathogenic blood cell mutations and clonality of T and B cells detected in each patient. (B) Sanger sequencing data of the MPL c.1210G>A (G404R) heterozygous mutation in CT1 using both blood and hair DNA samples. (C) The MPL G404R mutation failed to support TPO-stimulated growth of Ba/F3 cells. Ba/F3 cells expressing WT MPL, mutant MPL, or vector were incubated in medium without IL-3 but containing 10 ng/mL TPO. Cell numbers were counted daily using hemocytometer and displayed as mean plus or minus SD. (D) The mutant MPL is impaired in TPO uptake, a process involving TPO-MPL binding at the cell surface and its subsequent internalization. Ba/F3 cells expressing WT or mutant MPL were seeded in 6-well plates containing equal amounts of TPO with 5 × 106 cells per well. TPO uptake was monitored by measuring the remaining TPO concentration in cell supernatant after designated incubation time. After 60 minutes, the remaining TPO in WT cells was significantly lower than that of mutant cells (P = .0108) based on paired Student t test. Mean plus or minus SD of 2 separate experiments is displayed. (E) Mutant MPL leads do reduced JAK-STAT pathway activation upon TPO stimulation. Compared with WT MPL (WT), Ba/F3 cells expressing the mutant MPL (G404R) had reduced phosphorylation of JAK-STAT pathway proteins such as p-STAT3, p-STAT5, and p-ERK1/2 after 15 to 120 minutes of TPO stimulation as revealed by western blot. Ba/F3 cells transfected with the expression vector (Vector) was a negative control. β-Actin was a loading control for western blot. (F) Clonality of T cells in CT1 and CT2. Histograms show the frequencies of the top 10 clones of T-cell receptor β locus (TRB) in a sample at platelet count descending phase. Both CT1 and CT2 had clonal TRB. (G) The top clones of TRB in 6 sequential samples in CT1 and 5 in CT2 were identical. Their clonal frequencies were plotted and linked to show their stable longitudinal patterns over a platelet count cycle. IL-3, interleukin-3; SD, standard deviation; VAF, variant allele frequency.
In vitro functional analysis demonstrated that MPL G404R is a loss-of-function mutation with deficiencies in TPO-stimulated growth, TPO uptake, and TPO pathway activation (Figure 2C-E; supplemental Figure 2). Thus, this mutation may explain the unusually high TPO peaks without rebound thrombocytosis, and it may also slow the TPO-mediated thrombopoiesis, explaining the prolonged cycle period in CT1. Family studies demonstrated a paternal origin of the mutant allele. The father exhibited nonperiodic fluctuations in platelet count (166-223 × 109/L) during an 80-day period. Therefore, this MPL mutation is insufficient to cause CTP.
Heme-STAMP testing of peripheral blood revealed somatic STAT3 variants in both patients: CT1 had STAT3 Y640F (2% VAF) and STAT3 D661Y (1%VAF); CT2 had STAT3 D661Y (6% VAF) (Figure 2A). These STAT3 variants are pathogenic gain-of-function mutations first identified in T-cell large granular lymphocyte (T-LGL) leukemia and are associated with immune-mediated cytopenias, including neutropenia or pure red cell aplasia, even when present at subclonal (low VAF) levels.18-21 Background LGL populations were observed in both CT1 and CT2 (supplemental Figure 3), and T-LGL lymphoproliferation has been reported in 2 patients with CTP.3,22
NGS profiling demonstrated clonal T cells in both patients (Figure 2F). This is consistent with a recent report where clonal T-cell receptor rearrangement was found in 6 out of 8 patients with CTP, including 1 who later developed T-LGL leukemia.3 The clonal populations did not show cyclical variation in either patient (Figure 2G).
Additional findings were identified in each patient. CT1 had a monoclonal B-cell population by flow cytometry and B-cell receptor clonality profiling (Figure 2A; supplemental Figure 4). CT2 had pathogenic gain-of-function mutations in the last exon of PPM1D (R458X, 1% VAF; L484X, 2% VAF), which impair the DNA damage response.23
In summary, our study revealed novel genetic and cellular associations in 2 patients with CTP, who displayed exaggerated oscillations with their intrinsic periodic rhythms. The fact that pathogenic somatic gain-of-function STAT3 mutations and stable clonal T-cell populations are identified in both patients and are associated with autoimmunity and cytopenias suggest their involvement in the pathogenic baseline of CTP in these patients. In line with the previous model,4 we hypothesize that these factors could impair thrombopoiesis, increasing gain of the system, leading to overcorrection and persistence of cycling in the setting of perturbations eliciting a thrombopoietic response. In support of this concept, a CTP phenotype linked to hydroxyurea therapy has been observed in patients with JAK2 V617F–positive polycythemia vera.24,25
The true incidence of underlying somatic and germline genetic alterations in CTP such as seen here in STAT3 and MPL will need to be explored in additional patients. Identifying these mutations in patients who require therapy could represent a potential opportunity for individualized, novel targeted therapeutic approaches.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (NIH National Cancer Institute) (2P01CA04960529A1) and an award from the Department of Pathology, Stanford University (Pathology in Precision Health Research Award).
Authorship
Contribution: H.Z., M. Chien, Y.H., W.S., R.S.B., C.H., P.H., D.X., N.C.W., A.G.T., J.J., W.H.W, Z.C., and P.A. performed experiments; H.Z., Y.H., B.M.Z., X.G., L.L.T., D.J., M.S.K., and M. Craig analyzed the data; H.Z., J.R.G., and J.L.Z. designed the study; H.Z., J.B.B., and J.L.Z wrote the paper; and all authors provided critical reviews and edits.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: James L. Zehnder, Department of Pathology, Stanford University School of Medicine, 300 Pasteur Dr, Lane 235, Stanford, CA 94305; e-mail: zehnder@stanford.edu.
References
Author notes
Thirty-eight RNA sequencing data of sequential blood samples of both patients are available at the Gene Expression Omnibus (GEO) under accession number GSE179076.
Please contact the corresponding author for other data sharing, James L. Zehnder (zehnder@stanford.edu).
The full-text version of this article contains a data supplement.

![Oscillations of platelet count, plasma TPO, and trilineage-specific genes. (A) Reciprocal platelet count and plasma TPO oscillations in CT1 and CT2. CT2 received a platelet transfusion on day 2 (green arrow). TPO levels were measured in platelet-poor plasma by Quantikine ELISA (R&D Systems). (B) Venn diagram of the number of genes quantitatively associated with platelet count changes in both patients and gene list of the 3 lineage-specific groups. (C) Heatmap of the 151 platelet-specific (Plt-specific), the 19 erythrocyte-specific (Ery-specific), and the 7 immature neutrophil-specific (Neut-specific) genes displaying their longitudinal expression patterns over 2 platelet cycles for each patient. Using Cluster 3.0, the expression levels of individual genes were mean-centered across all time points and log-transferred for heatmaps, which were visualized using Java TreeView (Version 1.1.6r4) and color-scaled into yellow (high expression) to blue (low expression) or gray (below detection limit). (D) Quantification of the heatmap by showing the median levels of each gene group at every time point to illustrate their longitudinal patterns. (E) The median expression levels (in transcripts per million [TPM]) of the 34 platelet-specific genes that showed the highest fold changes in relation with platelet count and TPO oscillations. These platelet-specific genes are in parallel with and leading platelet count by 3 to 4 days in both patients. Together, they mirror the TPO oscillations. ELISA, enzyme-linked immunosorbent assay.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/1/10.1182_bloodadvances.2021006701/4/m_blooda_adv-2021-006701-gr1.jpeg?Expires=1769089088&Signature=zyfQRYu8il1KK3U7n06pc6768~WRStB~uJM-4GF~pjF1~sIqfOB1L-P2L4hRfevWmzxHHEcuvC50Osxpwu6B8pJqIwLjIa~VZ0QhMP~mej2dABRqaf-JAhjlpKoFshZR7dVB0iqZnDfa7Va3LiAF2YRxuhVCD4K0~kRniw3qeCOVOABxGOGT7Dlfh5KbX-8dyoU7ZNcBsNUfo17yVkhnyETh5Lsf5Tl0AfhmRbbBvsxETNtaIm8-ItOPsdScKcs4jIfJwOkhBqm~hrL23SO~zFY8zsSUbo408SN5~28k-FNxpml6In2iFFnReoDN9iLfI~TcoRDoxSq1tHstuFQdBg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)