Key Points
Activating STAT3 mutations are common in ENKTLs and associated with high CD30 expression.
ENKTLs with deleterious BCOR mutations overexpress MYC, and this BCOR-MYC is associated with decreased overall survival.
Abstract
Extranodal NK/T-cell lymphoma, nasal type (ENKTL) is an Epstein-Barr virus-positive, aggressive lymphoma with a heterogeneous cell of origin and variable clinical course. Several clinical prognostic indices have been proposed for ENKTL; however, there are few pathological biomarkers. This multi-institutional study sought to identify histologically assessable prognostic factors. We investigated mutation profiles by targeted next-generation sequencing (NGS) and immunohistochemical assessments of expression of MYC, Tyr705-phosphorylated (p-)STAT3, and CD30 in 71 ENKTL samples. The median age of the patients was 66 years (range, 6-100). The most frequent mutations were in STAT3 (27%), JAK3 (4%), KMT2D (19%), TP53 (13%), BCOR (10%), and DDX3X (7%). Immunohistochemistry (IHC) revealed that ENKTLs with STAT3 mutations exhibited higher expression of pSTAT3 and CD30. BCOR mutations were associated with increased MYC expression. Univariate analysis in the entire cohort showed that stage (II, III, or IV), BCOR mutations, TP53 mutations, and high MYC expression (defined as ≥40% positive neoplastic cells) were associated with reduced overall survival (OS). Multivariate modeling identified stage (II, III, or IV) and high MYC expression as independent adverse prognostic factors. In a subgroup analysis of patients treated with anthracycline (AC)-free chemotherapy and/or radiotherapy (RT) with curative intent, BCOR but not high MYC expression was an independent adverse prognostic factor. In conclusion, activating STAT3 mutations are common in ENKTLs and are associated with increased CD30 expression. MYC overexpression is, at least in part, associated with deleterious BCOR mutations, and this BCOR–MYC linkage may have prognostic significance, underscoring the potential utility of IHC for MYC in risk stratification of patients with ENKTL.
Introduction
Extranodal NK/T-cell lymphoma, nasal type (ENKTL) is an aggressive Epstein-Barr virus (EBV)-associated lymphoma of either cytotoxic T-cell or NK-cell origin.1 ENKTL predominantly arises in the upper aerodigestive tract, such as the nasal cavity. Other less frequent primary sites include the skin, gastrointestinal tract, soft tissue, and testes. ENKTL is a rare disease accounting for approximately 10% of T/NK-cell lymphomas2; however, its frequency varies geographically; it is relatively common in Asian or Pacific Islander, Hispanic White, and American Indian or Alaskan Native, and rarer in non-Hispanic White and Black populations.3
The clinical presentation of ENKTL is typically aggressive. Localized ENKTL is generally treated with concurrent chemoradiotherapy, such as radiotherapy (RT) with dexamethasone, etoposide, ifosfamide, and carboplatin (RT-DeVIC), with a 5-year overall survival (OS) rate of approximately 70%.4,5 In contrast, the survival of patients with advanced ENKTL is poor, and despite intensive multiagent chemotherapy containing l-asparaginase, the 5-year OS rate is approximately 25%.4,5 Nonetheless, clinical heterogeneity of ENKTL has been noted. For example, ENKTLs involving the nonupper aerodigestive tract region are more frequently disseminated, with a shorter 5-year OS than those involving the upper aerodigestive tract.6 Given such clinical heterogeneity, there have been multiple clinical prognostic or predictive models proposed, incorporating age, serum lactate dehydrogenase (LDH) concentration, performance status, stage, and nonnasal disease.5,7,8
Reflecting its diverse clinical courses, ENKTL molecular heterogeneity has been recognized. As ENKTL, by definition, involves neoplastic proliferation of NK or T cells, T-cell receptor (TCR) gene rearrangement and protein expression of TCR are detected in the subset of ENKTLs derived from T cells.9-11 Next-generation sequencing (NGS) profiling has identified commonly mutated genes, including TP53, DDX3X, STAT3, JAK3, MGA, and BCOR.12-18 A recent genomic and transcriptomic study proposed molecular subclassifications into 3 subtypes: (1) TSIM, characterized by alterations in tumor suppressors such as TP53 and immune modulators including JAK/STAT pathway genes and PD-L1/2 amplification; (2) HEA, characterized by aberrant histone acetylation driven by mutations in HDAC9, EP300, and ARID1A; and (3) MB, enriched for MYC-associated aberrations such as MGA mutation and loss of heterozygosity (LOH) at the BRDT locus.13 Interestingly, MB ENKTLs had significantly worse OS and progression-free survival (PFS) than those of the TSIM or HEA subtypes.
Despite these advances in understanding the clinical heterogeneity and molecular pathogenesis of ENKTL, there currently are few well-established pathological biomarkers.19 Because ENKTL typically presents with extranodal lesions, biopsy specimens are often small, yielding limited amounts of DNA and RNA, so extensive molecular analysis is often challenging. Therefore, there remains a need for a histological prognostic biomarker that reflects molecular abnormalities. In this multi-institution study, we performed mutational and immunohistochemical characterization of 71 ENKTLs, identifying prognostically relevant BCOR–MYC axis.
Materials and methods
Case selection and data acquisition
This retrospective study was approved by the institutional review board of the University of Yamanashi Hospital (approval number 2195) and other related hospitals. It was performed with the Declaration of Helsinki. We retrieved ENKTL cases diagnosed at the University of Yamanashi, Aichi Medical University Hospital, Mayo Clinic, and Tokai University Hospital from 2000 to 2020, for a total of 71 cases. These included cases were nearly diagnosed consecutively, but a small number of cases were excluded because of insufficient specimens for additional pathological and molecular analyses. Cytologic features of 1 ENKTL involving the uterine cervix have been previously reported.20 All ENKTL cases were histopathologically diagnosed on the basis of expression of T/NK-cell lineage markers, positivity for EBV-encoded small RNA, and at least focal expression of cytotoxic molecules such as granzyme B. No primary nodal EBV-positive T/NK-cell lymphomas were included. The following clinical information was retrieved from the medical records: age, sex, B symptoms, serum LDH concentration, stage, presence of nasal lesions, regional and distant lymph node involvement, NK/T-cell lymphoma prognostic index (NKPI),8 prognostic index of NK-cell lymphoma (PINK),7 and treatment. The median follow-up period was 12.5 months
Immunohistochemistry (IHC)
We measured the expression of 3 biomarkers, CD30, pSTAT3 (Tyr705-phosphorylated STAT3), and MYC, in ENKTLs (n = 71). Standard, indirect IHC was performed with antibodies specific for CD30 (clone 1G12, ready to use, Nichirei; Tokyo, Japan), pSTAT3 (clone D3A7, dilution 1:400; Cell Signaling Technology, Danvers, MA), and MYC (clone Y69, dilution 1:200, Abcam; Cambridge, UK). Membranous reactivity for CD30 and nuclear reactivity for pSTAT3 and MYC, at any intensity, were scored as positive on the single-cell level, then the sample was scored as the percentage of overall neoplastic cells in a sample with 10% increments. Expression was scored as high if positivity was ≥30% for CD30, ≥30% for pSTAT3, and ≥40% for MYC, as reported previously.21,22
NGS
For 67 ENKTL samples, we performed hybridization capture-based NGS, targeting 29 genes frequently mutated in T/NK-cell lymphomas: ARID1A, ASXL3, BCOR, CDKN2A, DDX3X, DNMT3A, ECSIT, EP300, FAT4, HDAC9, IDH1, IDH2, JAK1, JAK2, JAK3, KMT2C, KMT2D, MGA, MSN, NRAS, STAT1, STAT3, STAT5A, STAT5B, STAT6, TET1, TET2, TET3, and TP53. Briefly, DNA was extracted using the AllPrep DNA/RNA FFPE Tissue Kit (QIAGEN, Hilden, Germany) and QIAcube (QIAGEN). DNA was sheared into 200- to 300-bp fragments with an M220 focused ultrasonicator (Covaris, Woburn, MA) and repaired with NEBNext FFPE DNA Repair Mix (M6630; New England Biolabs, Ipswich, MA). We prepared libraries using the xGen Prism DNA Library Prep Kit and enriched for full coding sequences of the 29 genes using xGen Predesigned Gene Capture Pools and the xGen Hybridization and Wash Kit (Integrated DNA Technologies, Coralville, IA). Sequencing was performed with an Illumina NovaSeq 6000 instrument. Reads were trimmed by fastp version 0.20 using default parameters.23 We generated unmapped BAM files using FastqToSam (Picard Tools version 2.21.4, https://broadinstitute.github.io/picard). Reads were deduplicated with their start–stop positions, and unique molecular identifiers were created according to the manufacturer’s protocol. The resulting BAM files mapped to hg19 were imported into CLC Genomics Workbench version 21 (https://digitalinsights.qiagen.com/) and locally realigned. Putative somatic variants were called using 5 filters: (1) read length ≥20; (2) sequencing coverage (depth) ≥50; (3) variant read count ≥5; (4) allele frequency ≥0.05; and (5) relative read direction filter with a significance of 0.01. Variants were annotated using Annovar.24 SNPs, insertions, and deletions (indels) reported in the ExAC/gnomAD25 and ToMMo26 databases with allele frequencies >0.001 were excluded. We also required variants to have a frequency within the ENKTL dataset of <30% for single-nucleotide variants and <5% for indels to exclude obvious alignment artifacts. Finally, variants meeting ≥1 of the following 3 criteria were scored as significant: (1) start loss, stop gain, or frameshift/nonframeshift indel; (2) designated as pathogenic/likely pathogenic in ClinVar27; (3) present ≥2 times in the COSMIC database v94 (https://cancer.sanger.ac.uk/cosmic). All reported variants were validated by manual inspection on the Integrative Genomics Viewer28 and visualized by the cBioPortal OncoPrinter.29,30 NGS is deposited in the Japanese Genotype–Phenotype Archive (accession number: JGAS000548).
Statistical analysis
The Fisher exact test was used to analyze ordinal or nominal categorical variables. The Wilcoxon rank sum test was used to analyze continuous numerical data. The log-rank test was used to compare OS, as determined using the Kaplan-Meier method. Univariate and multivariate Cox proportional hazards models were used to identify specific variables associated with OS. For the multivariate analysis, variables with a P < .10 in the univariate analyses were included. All statistical analyses were performed using JMP Pro 16.2.0 (SAS Institute, Cary, NC). Graphs were generated using GraphPad Prism 8 (GraphPad Software LLC, San Diego, CA).
Results
Clinicopathologic characteristics
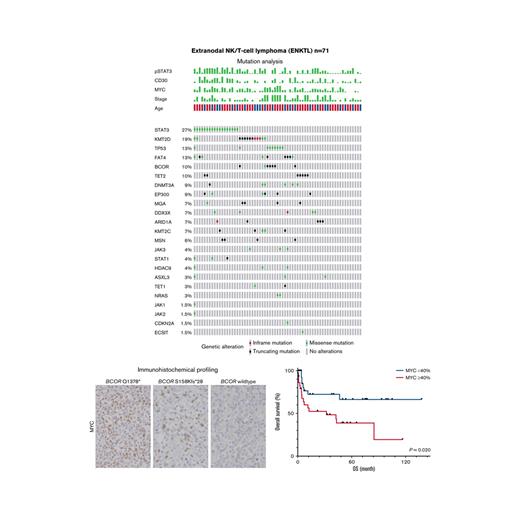
Clinical data for the 71 patients with ENKTL are summarized in Table 1. Their median age was 66 years (range, 6-100), with a slight male predominance (58%). Nasal lesions were identified in 70% (45 of 64) of cases. Advanced stages (III or IV) were noted in 30% (18 of 59). Group 3/4 NKPI and high-risk PINK were observed in 45% (26 of 58) and 17% (10 of 58) of the patients, respectively. Primarily because of poor performance status, 13% (8 of 61) of the patients received no treatment. For patients with stage I or II disease and available treatment history, 71% (25 of 35) of the patients were treated with concurrent RT-DeVIC. Among patients with stage III or IV disease who received multiagent chemotherapy, 40% (6 of 15) were treated with dexamethasone, methotrexate, ifosfamide, l-asparaginase, and etoposide (SMILE). Over a median follow-up period of 12.5 months, the median OS was 85 months, and 5-year OS was 52% (supplemental Figure 1 in the data supplement).
Clinicopathologic characteristics of patients with ENKTL
| Characteristic . | n = 71 . |
|---|---|
| Age, median (range), y | 66 (6-100) |
| Sex, male/female, n (%) | 41 (58)/30 (42) |
| B symptoms, yes/no, n (%) | 21 (36)/37 (64) |
| LDH above the UNL, yes/no, n (%) | 38 (64)/21 (36) |
| Stage, n (%) | |
| I | 29 (49) |
| II | 12 (20) |
| III | 3 (5) |
| IV | 15 (25) |
| Nasal lesion, yes/no, n (%) | 45 (70)/19(30) |
| Regional LN involvement, yes/no, n (%) | 20 (33)/41 (67) |
| Distant LN involvement, yes/no, n (%) | 7 (11)/54 (89) |
| NKPI group, n (%) | |
| Group 1 | 10 (17) |
| Group 2 | 22 (38) |
| Group 3 | 10 (17) |
| Group 4 | 16 (28) |
| PINK risk, n (%) | |
| Low | 21 (36) |
| Intermediate | 27 (47) |
| High | 10 (17) |
| Treatment, n (%) | |
| AC-free CTX and/or RT | 45 (74) |
| Other CTX | 8 (13) |
| No (including BSC) | 8 (13) |
| Characteristic . | n = 71 . |
|---|---|
| Age, median (range), y | 66 (6-100) |
| Sex, male/female, n (%) | 41 (58)/30 (42) |
| B symptoms, yes/no, n (%) | 21 (36)/37 (64) |
| LDH above the UNL, yes/no, n (%) | 38 (64)/21 (36) |
| Stage, n (%) | |
| I | 29 (49) |
| II | 12 (20) |
| III | 3 (5) |
| IV | 15 (25) |
| Nasal lesion, yes/no, n (%) | 45 (70)/19(30) |
| Regional LN involvement, yes/no, n (%) | 20 (33)/41 (67) |
| Distant LN involvement, yes/no, n (%) | 7 (11)/54 (89) |
| NKPI group, n (%) | |
| Group 1 | 10 (17) |
| Group 2 | 22 (38) |
| Group 3 | 10 (17) |
| Group 4 | 16 (28) |
| PINK risk, n (%) | |
| Low | 21 (36) |
| Intermediate | 27 (47) |
| High | 10 (17) |
| Treatment, n (%) | |
| AC-free CTX and/or RT | 45 (74) |
| Other CTX | 8 (13) |
| No (including BSC) | 8 (13) |
AC, anthracycline; BSC, best supportive care; CTX, chemotherapy; LN, lymph node; RT, radiotherapy; UNL, upper normal limit.
IHC revealed that the median positivity (range) for pSTAT3, CD30, and MYC was 50% (range, 0%-100%), 20% (range, 0%-90%), and 30% (range, 0%-30%), respectively. High expression of pSTAT3, CD30, and MYC (≥30%, ≥30%, and ≥40%, respectively) was observed in 65% (46 of 71), 42% (30 of 71), and 48% (34 of 71) of ENKTLs, respectively.
Mutation profiling
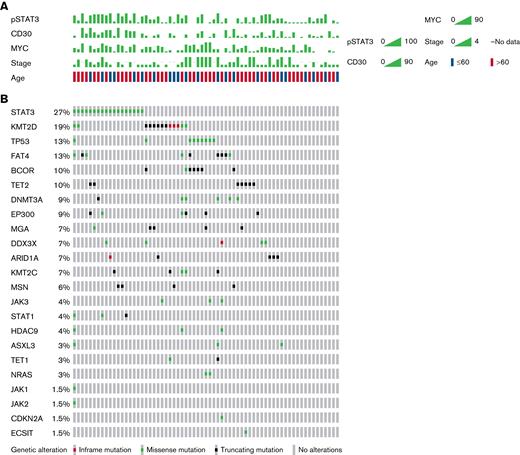
Targeted NGS for 29 genes was performed for 67 out of 71 ENKTLs examined; mutation profiles are shown in Figure 1 and listed in supplemental Table 1. A total of 125 variants were identified in 79% (53 of 67) of ENKTLs, whereas the remaining 21% (14 of 67) had no mutations in the 29 genes. The most frequently mutated gene was STAT3 in 27% of the patients (n = 18). Other JAK/STAT pathway genes were mutated at lower frequencies: JAK3 (4%, n = 3), STAT1 (4%, n = 3), JAK1 (1.5%, n = 1), and JAK2 (1.5%, n = 1). Consistent with earlier studies,13,15,18,31TP53 mutations were relatively common, in 13% (n = 9) of cases. Recurrent mutations involved epigenetic modifier genes, including KMT2D (19%, n = 13), TET2 (10%, n = 7), DNMT3A (9%, n = 6), EP300 (9%, n = 6), ARID1A (7%, n = 5), KMT2C (7%, n = 5), and HDAC9 (4%, n = 3). Other frequently mutated genes were FAT4 (13%, n = 9), BCOR (10%, n = 7), MGA (7%, n = 5), and DDX3X (7%, n = 5). Activating NRAS missense mutations (p.G13D and p.Q61K) were found in 3% (n = 2) of ENKTLs.
ENKTL mutation profiles from targeted NGS. (A) Clinical and IHC characteristics. (B) Mutation profiles of 29 examined genes.
ENKTL mutation profiles from targeted NGS. (A) Clinical and IHC characteristics. (B) Mutation profiles of 29 examined genes.
Activating STAT3 mutations are associated with high pSTAT3 and CD30 expression
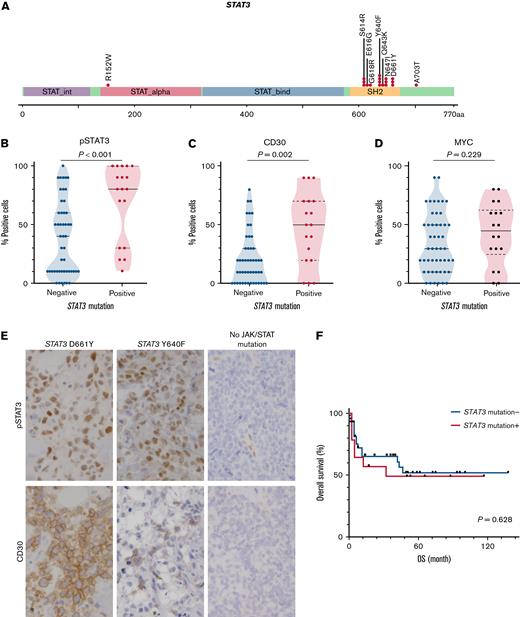
Next, we investigated the clinicopathological significance of STAT3 mutations in ENKTL, as it was the most frequently mutated gene in our ENKTL cohort. All STAT3 mutations identified were missense mutations, with all but 2 (16 of 18) in the SH2 domain of the STAT3 gene (Figure 2A). All STAT3 mutations, including 2 substitutions in the non-SH2 domain (p.R152W and p.A703T), have been reported to be activating, gain-of-function mutations associated with T/NK-cell malignancies or familial lymphoproliferation and early onset solid-organ autoimmunity.32,33 Consistent with their identification as activating mutations, ENKTLs with STAT3 mutations exhibited significantly higher pSTAT3 expression by IHC than those without (median positivity 80% vs 40%; Wilcoxon test P < .001) (Figure 2B,E). Furthermore, CD30 was expressed at higher concentrations in ENKTLs with STAT3 mutations than in those without (median positivity, 50% vs 20%; Wilcoxon test P = .002) (Figure 2C,E). In contrast, there was no significant association between STAT3 mutation status and MYC expression (median positivity, 45% vs 30%; Wilcoxon test P = .229) (Figure 2D). Survival analysis showed no significant difference in OS for patients with or without STAT3 mutations (log-rank test P = .628) (Figure 2F).
STAT3 mutations in ENKTLs. (A) Mutation distribution. (B-D) Semiquantitative measurements of pSTAT3, CD30, and MYC expression as a function of STAT3 mutation status. (E) Representative IHC images for pSTAT3 and CD30 by STAT3 mutation status. Original magnification ×400. (F) Kaplan-Meier plot of OS as a function of STAT3 mutation status.
STAT3 mutations in ENKTLs. (A) Mutation distribution. (B-D) Semiquantitative measurements of pSTAT3, CD30, and MYC expression as a function of STAT3 mutation status. (E) Representative IHC images for pSTAT3 and CD30 by STAT3 mutation status. Original magnification ×400. (F) Kaplan-Meier plot of OS as a function of STAT3 mutation status.
We have identified a few mutations in JAK/STAT pathway genes other than STAT3, including JAK3 (4%, n = 3), STAT1 (4%, n = 3), JAK1 (1.5%, n = 1), and JAK2 (1.5%, n = 1). Two JAK3 variants, p.A573V (n = 1) and p.M511I (n = 1), were activating and identified in ENKTLs with wild-type STAT3. The functionality of the other variants was of uncertain significance and mostly overlapped with STAT3 mutations (Figure 1B). Of note, after inclusion of non-STAT3 mutations, ENKTLs with a JAK/STAT gene mutation exhibited significantly higher expression of pSTAT3 (median positivity, 80% vs 40%; Wilcoxon test P = .013) and CD30 (median positivity, 40% vs 20%; Wilcoxon test P = .037) (supplemental Figure 2).
BCOR mutations are associated with high MYC expression and decreased OS
We focused on mutations in the gene encoding the BCL6 corepressor BCOR and their clinicopathological significance in ENKTLs. BCOR mutations were identified in 10% (7 of 67) of ENKTLs (Figure 1B); all but 1 missense mutation (p.V594I) were deleterious frameshift or nonsense mutations (Figure 3A). All ENKTLs with BCOR mutations tested negative for STAT3 mutations. In contrast, there was a tendency for cooccurrence between BCOR and TP53 mutations; 71% (5 of 7) of BCOR-mutated ENKTLs had TP53 mutations, and 55% (5 of 9) of TP53-mutated cases had BCOR mutations (P < .001; q-value = 0.062). While BCOR mutation status was not significantly associated with pSTAT3 and CD30 expression levels by IHC (Figure 3B-C), ENKTLs with BCOR mutations showed significantly higher MYC expression than those without BCOR mutations (median positivity 70% vs 30%; Wilcoxon test P = .009) (Figure 3D-E). Of note, patients with BCOR mutations (n = 7) had a significantly shorter survival than those without BCOR mutations (n = 52) (median OS, 5.9 months vs undefined; log-rank test P < .001) (Figure 3F).
BCOR mutations in ENKTLs. (A) Mutation distribution. (B-D) Semiquantitative measurements of pSTAT3, CD30, and MYC expression as a function of BCOR mutation status. (E) Representative IHC images for MYC by BCOR mutation status. Original magnification ×400. (F) Kaplan-Meier plot of OS as a function of BCOR mutation status.
BCOR mutations in ENKTLs. (A) Mutation distribution. (B-D) Semiquantitative measurements of pSTAT3, CD30, and MYC expression as a function of BCOR mutation status. (E) Representative IHC images for MYC by BCOR mutation status. Original magnification ×400. (F) Kaplan-Meier plot of OS as a function of BCOR mutation status.
High MYC expression and BCOR mutation are adverse prognostic factors in ENKTL
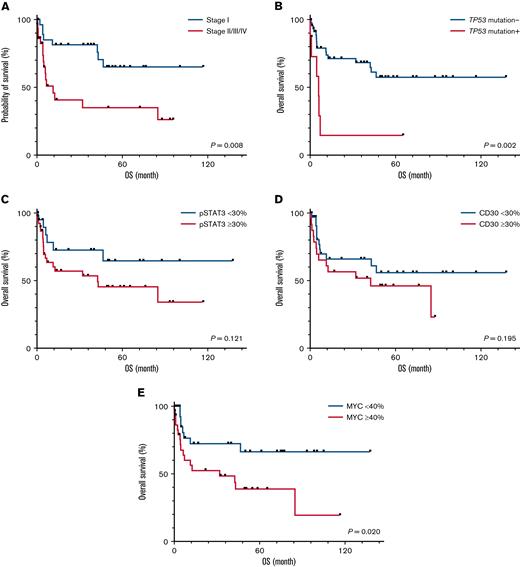
We examined OS with respect to known prognostic factors, genetic alterations, and IHC markers (Figure 4). Patients with stage II/III/IV disease (n = 30) had a significantly shorter OS than those with stage I disease (n = 29) (median OS, 11.4 months vs undefined; log-rank test, P = .008) (Figure 4A). In addition to the above-mentioned BCOR mutations, ENKTLs with TP53 mutations (n = 8) had significantly decreased OS than those without (n = 51) (median OS, 5.9 months vs undefined; log-rank test P = .002) (Figure 4B). Among the 3 IHC markers examined, ENKTLs with high MYC expression defined by positivity ≥40% (n = 31) showed inferior OS compared with those with low MYC expression (median OS 32.1 months vs undefined; log-rank test P = .020) (Figure 4E). No statistically significant association was observed between OS and pSTAT3 (P = .121) or CD30 (P = .195) expression (Figure 4C-D).
OS of patients with ENKTL. Kaplan-Meier plots of OS as a function of (A) stage, (B) TP53 mutation, (C) pSTAT3, (D) CD30, and (E) MYC expression.
OS of patients with ENKTL. Kaplan-Meier plots of OS as a function of (A) stage, (B) TP53 mutation, (C) pSTAT3, (D) CD30, and (E) MYC expression.
The univariate and multivariate analyses in the entire cohort are summarized in Table 2. In the univariate analyses, we identified that stage II, III, or IV disease (for OS: hazard ratio [HR], 3.07; 95% confidence interval [CI], 1.30-7.26; P = .011), BCOR mutation (HR, 6.66; 95% CI, 2.25-19.69; P < .001), TP53 mutation (HR, 4.06; 95% CI, 1.55-10.59; P = .004), and high MYC expression (HR, 2.62; 95% CI, 1.13-6.09; P = .025) were associated with an increased risk of death (Table 2). In a multivariate analysis, we found that stage II, III, or IV disease (HR, 3.07; 95% CI, 1.09-8.67; P = .034) and high MYC expression (HR, 3.92; 95% CI, 1.28-12.02; P = .017) were independently associated with decreased survival (Table 2). In contrast, anthracycline (AC)-free chemotherapy and/or RT were associated with decreased risk of death compared with no treatment/best supportive care (HR, 0.22; 95% CI, 0.06-0.88; P = .032).
Univariate and multivariate analyses of factors associated with OS of overall patients with ENKTL
| Variable . | Univariate . | Multivariate . | ||||
|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |
| Age > 60 y | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 0.70 | 0.32-1.57 | .392 | — | — | — |
| B symptoms | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 1.29 | 0.55-2.98 | .558 | — | — | — |
| Stage | ||||||
| I | 1 | — | — | 1 | — | — |
| II, III, or IV | 3.07 | 1.30-7.26 | .011 | 3.07 | 1.09-8.67 | .034 |
| Serum LDH > ULN | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 2.00 | 0.79-5.11 | .146 | — | — | — |
| Nasal lesion | ||||||
| Present | 1 | — | — | — | — | — |
| Absent | 0.93 | 0.40-2.16 | .863 | — | — | — |
| Regional LN involvement | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 1.24 | 0.54-2.83 | .605 | — | — | — |
| Distant LN involvement | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 1.04 | 0.31-3.50 | .952 | — | — | — |
| Treatment | ||||||
| AC–free CTX and/or RT | 0.29 | 0.08-1.01 | .051 | 0.22 | 0.06-0.88 | .032 |
| Other CTX | 0.47 | 0.10-2.14 | .331 | 0.45 | 0.07-2.97 | .404 |
| No (including BSC) | 1 | — | — | 1 | — | — |
| STAT3 mutation | ||||||
| Negative | 1 | — | — | — | — | — |
| Positive | 1.25 | 0.51-3.06 | .630 | — | — | — |
| BCOR mutation | ||||||
| Negative | 1 | — | — | 1 | — | — |
| Positive | 6.66 | 2.25-19.69 | <.001 | 5.11 | 0.56-46.97 | .149 |
| TP53 mutation | ||||||
| Negative | 1 | — | — | 1 | — | — |
| Positive | 4.06 | 1.55-10.59 | .004 | 0.89 | 0.13-6.17 | .902 |
| pSTAT3 ≥ 30% | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 2.04 | 0.81-5.11 | .129 | — | — | — |
| CD30 ≥ 30% | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 1.67 | 0.76-3.67 | .200 | — | — | — |
| MYC ≥ 40% | ||||||
| No | 1 | — | — | 1 | — | — |
| Yes | 2.62 | 1.13-6.09 | .025 | 3.92 | 1.28-12.02 | .017 |
| Variable . | Univariate . | Multivariate . | ||||
|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |
| Age > 60 y | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 0.70 | 0.32-1.57 | .392 | — | — | — |
| B symptoms | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 1.29 | 0.55-2.98 | .558 | — | — | — |
| Stage | ||||||
| I | 1 | — | — | 1 | — | — |
| II, III, or IV | 3.07 | 1.30-7.26 | .011 | 3.07 | 1.09-8.67 | .034 |
| Serum LDH > ULN | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 2.00 | 0.79-5.11 | .146 | — | — | — |
| Nasal lesion | ||||||
| Present | 1 | — | — | — | — | — |
| Absent | 0.93 | 0.40-2.16 | .863 | — | — | — |
| Regional LN involvement | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 1.24 | 0.54-2.83 | .605 | — | — | — |
| Distant LN involvement | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 1.04 | 0.31-3.50 | .952 | — | — | — |
| Treatment | ||||||
| AC–free CTX and/or RT | 0.29 | 0.08-1.01 | .051 | 0.22 | 0.06-0.88 | .032 |
| Other CTX | 0.47 | 0.10-2.14 | .331 | 0.45 | 0.07-2.97 | .404 |
| No (including BSC) | 1 | — | — | 1 | — | — |
| STAT3 mutation | ||||||
| Negative | 1 | — | — | — | — | — |
| Positive | 1.25 | 0.51-3.06 | .630 | — | — | — |
| BCOR mutation | ||||||
| Negative | 1 | — | — | 1 | — | — |
| Positive | 6.66 | 2.25-19.69 | <.001 | 5.11 | 0.56-46.97 | .149 |
| TP53 mutation | ||||||
| Negative | 1 | — | — | 1 | — | — |
| Positive | 4.06 | 1.55-10.59 | .004 | 0.89 | 0.13-6.17 | .902 |
| pSTAT3 ≥ 30% | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 2.04 | 0.81-5.11 | .129 | — | — | — |
| CD30 ≥ 30% | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 1.67 | 0.76-3.67 | .200 | — | — | — |
| MYC ≥ 40% | ||||||
| No | 1 | — | — | 1 | — | — |
| Yes | 2.62 | 1.13-6.09 | .025 | 3.92 | 1.28-12.02 | .017 |
BSC, best supportive care; LN, lymph node; RT, radiotherapy; UNL, upper normal limit.
We then performed a subgroup analysis on patients treated with AC-free chemotherapy and/or RT with curative intent (Table 3). Careful interpretation of the results is needed because of the limited number of patients in the subgroup (n = 45). In a univariate analysis, stage II, III, or IV disease (HR, 2.92; 95% CI, 1.09-7.81; P = .033) and BCOR mutation (HR, 9.71; 95% CI, 2.57-36.72; P < .001) were associated with increased risk of death (Table 3). In the multivariate analysis, only BCOR mutations remained significant (HR, 6.41; 95% CI, 1.33-30.86; P = .021).
Univariate and multivariate analyses of factors associated with OS of patients with ENKTL treated with curative intent
| Variable . | Univariate . | Multivariate . | ||||
|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |
| Age > 60 y | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 0.45 | 0.18-1.16 | .099 | 0.46 | 0.16-1.32 | .149 |
| B symptoms | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 1.20 | 0.43-3.3 | .729 | — | — | — |
| Stage | ||||||
| I | 1 | — | — | 1 | — | — |
| II, III, or IV | 2.92 | 1.09-7.81 | .033 | 1.77 | 0.55-5.73 | .337 |
| Serum LDH > ULN | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 2.09 | 0.67-6.53 | .204 | — | — | — |
| Regional LN involvement | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 1.33 | 0.5-3.59 | .567 | — | — | — |
| Nasal lesion | ||||||
| Present | 1 | — | — | — | — | — |
| Absent | 0.46 | 0.18-1.20 | .112 | — | — | — |
| Distant LN involvement | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 1.51 | 0.34-6.65 | .584 | — | — | — |
| STAT3 mutation | ||||||
| Negative | 1 | — | — | — | — | — |
| Positive | 0.75 | 0.24-2.33 | .617 | — | — | — |
| BCOR mutation | ||||||
| Negative | 1 | — | — | 1 | — | — |
| Positive | 9.71 | 2.57-36.72 | <.001 | 6.41 | 1.33-30.86 | .021 |
| TP53 mutation | ||||||
| Negative | 1 | — | — | — | — | — |
| Positive | 2.69 | 0.75-9.62 | .128 | — | — | — |
| pSTAT3 ≥ 30% | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 2.05 | 0.67-6.26 | .206 | — | — | — |
| CD30 ≥ 30% | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 1.64 | 0.65-4.16 | .299 | — | — | — |
| MYC ≥ 40% | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 1.96 | 0.73-5.26 | .179 | — | — | — |
| Variable . | Univariate . | Multivariate . | ||||
|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |
| Age > 60 y | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 0.45 | 0.18-1.16 | .099 | 0.46 | 0.16-1.32 | .149 |
| B symptoms | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 1.20 | 0.43-3.3 | .729 | — | — | — |
| Stage | ||||||
| I | 1 | — | — | 1 | — | — |
| II, III, or IV | 2.92 | 1.09-7.81 | .033 | 1.77 | 0.55-5.73 | .337 |
| Serum LDH > ULN | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 2.09 | 0.67-6.53 | .204 | — | — | — |
| Regional LN involvement | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 1.33 | 0.5-3.59 | .567 | — | — | — |
| Nasal lesion | ||||||
| Present | 1 | — | — | — | — | — |
| Absent | 0.46 | 0.18-1.20 | .112 | — | — | — |
| Distant LN involvement | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 1.51 | 0.34-6.65 | .584 | — | — | — |
| STAT3 mutation | ||||||
| Negative | 1 | — | — | — | — | — |
| Positive | 0.75 | 0.24-2.33 | .617 | — | — | — |
| BCOR mutation | ||||||
| Negative | 1 | — | — | 1 | — | — |
| Positive | 9.71 | 2.57-36.72 | <.001 | 6.41 | 1.33-30.86 | .021 |
| TP53 mutation | ||||||
| Negative | 1 | — | — | — | — | — |
| Positive | 2.69 | 0.75-9.62 | .128 | — | — | — |
| pSTAT3 ≥ 30% | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 2.05 | 0.67-6.26 | .206 | — | — | — |
| CD30 ≥ 30% | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 1.64 | 0.65-4.16 | .299 | — | — | — |
| MYC ≥ 40% | ||||||
| No | 1 | — | — | — | — | — |
| Yes | 1.96 | 0.73-5.26 | .179 | — | — | — |
BSC, best supportive care; CTX, chemotherapy; LN, lymph node; RT, radiotherapy; UNL, upper normal limit.
Discussion
In the present study, we investigated the mutational profile and protein expression of 71 ENKTLs using targeted NGS and IHC, revealing multiple genotype–immunophenotype associations with prognostic impacts. We confirmed that mutations that activate the JAK/STAT pathway are common in ENKTL. IHC for pSTAT3 revealed that 65% of the ENKTLs had JAK/STAT pathway activation, partly explained by activating mutations in JAK/STAT pathway genes, including STAT3, consistent with earlier reports on T/NK-cell malignancies.31,34,35 The identification of frequent JAK/STAT mutations and active signaling offers insight into therapeutic approaches. A recent phase 2 study demonstrated that the JAK1/2 inhibitor ruxolitinib is clinically efficacious across multiple T-cell lymphoma subtypes, particularly for T-cell lymphomas with JAK/STAT mutations or active signaling defined by pSTAT3 IHC positivity ≥30%,22 although this study enrolled no patients with ENKTL. Given the high frequency of JAK/STAT activation in ENKTLs and the promising efficacy of ruxolitinib in non-ENKTL T-cell lymphomas, although the controversial risk of secondary lymphoproliferative disorders should be carefully considered,36-38 our data suggest that at least a subset of patients with ENKTL would benefit from this precise therapeutic approach to JAK/STAT inhibition. pSTAT3 was also positive in a considerable number of ENKTLs without JAK/STAT gene mutations, suggesting that there are additional underlying molecular mechanisms leading to pathway activation. Because our targeted NGS panel did not include genes encoding JAK/STAT pathway suppressors, such as SOCS1 and SOCS3, loss-of-function mutations in them would provide a potential explanation,39,40 although a previous whole-exome sequencing study on ENKTL reported no SOCS mutations.13
ENKTLs frequently exhibit increased CD30 expression41-43; using a cutoff of 30%, in our cohort, 42% were CD30+. There was no significant difference in OS between CD30+ and CD30− ENKTLs. The prognostic impact of CD30 expression in ENKTL is controversial; one explanation could be the different cutoffs of CD30 IHC positivity used.42 A systematic review and meta-analysis of 10 retrospective cohort studies reported that CD30 expression is associated with better OS in patients with ENKTL.44 To our knowledge, ours is the first study to demonstrate an association between STAT3 mutation and CD30 expression in ENKTL. High CD30 expression correlated with STAT3 mutation, implying underlying molecular interactions between the activated JAK/STAT pathway and the TNFRSF8 gene that encodes CD30. Supporting this hypothesis, a study using CRISPR library screening found that TNFRSF8 is a transcriptional target of STAT3 and that its transcription is significantly downregulated in STAT3-depleted anaplastic large cell lymphoma cell lines.45 The CD30-directed antibody–drug conjugate brentuximab vedotin (BV) has been introduced for the treatment of peripheral T-cell lymphomas46 and classic Hodgkin lymphomas47 expressing CD30, although the proportion of CD30+ tumor cells is not necessarily correlated with BV efficacy. However, there have been a limited number of studies on the efficacy of BV in ENKTLs, with varying overall response rates.48-51 Therefore, it is pivotal to establish a predictive biomarker to identify patients with ENKTL who can respond best to BV. Collectively, our data suggest that CD30-directed therapy could be a therapeutic option for ENKTL, especially those harboring activating STAT3 mutations.
We demonstrated that MYC overexpression is an adverse prognostic factor for patients with ENKTL. A limited number of reports have suggested the prognostic significance of high MYC expression. Huang and colleagues demonstrated that MYC expression in 53 ENKTLs, defining positivity as ≥5%, is associated with unfavorable OS and decreased PFS.52 Wang and colleagues performed IHC for MYC in 53 ENKTLs, defining positivity as ≥20%, demonstrating an association with decreased PFS and OS.53 A study by Chen and colleagues of 54 patients with ENKTL found that OS was significantly shorter in the MYC+ group (defined as positivity ≥40%).54 Although they used different cutoff values, all showed high MYC expression to be an independent prognostic factor by multivariate analyses. Our finding further confirms the utility of MYC IHC for risk stratification. Because IHC for MYC is widely used for the risk stratification of B-cell malignancies, especially diffuse large B-cell lymphoma,55 the addition of MYC to a diagnostic IHC panel for ENKTL would be simple to implement and of significant prognostic value. Taken together, although the cutoff value needs to be validated, MYC overexpression determined by IHC is an adverse prognostic factor useful for stratifying the survival of patients with ENKTL. The precise molecular mechanism(s) underlying this association is currently unknown. An in vivo study showed that MYC overexpression upregulates transcription of the long noncoding RNA SNHG12, decreasing sensitivity to cisplatin56; however, further studies are required to elucidate the functional consequences of MYC overexpression in ENKTL.
The molecular mechanism underlying MYC overexpression in ENKTL is interesting since, unlike B-cell lymphomas, gene rearrangements involving MYC are absent or very rare, although increased MYC copy number is present in a subset of ENKTLs.54 In the present study, ENKTLs with high MYC expression were enriched in those with mutations in the BCOR gene, encoding the BCL6 corepressor BCOR, which have been reported in T-cell– and NK-cell–derived malignancies with varying frequencies.12,13,15-18 In ENKTL, the frequency of BCOR mutations varies from 0% to 32%.12,13,15-18 Consistent with the observed association between high MYC expression and BCOR mutation in the current ENKTL series, an earlier study demonstrated that BCOR represses MYC transcription in T cells.57 Mice missing Bcor exon 4, thus lacking the BCL6-binding domain of BCOR, showed a strong propensity to develop acute T-cell lymphoblastic leukemia (T-ALL); interestingly, Myc was highly upregulated in the T-ALL induced in the Bcor-deleted mice. Therefore, human ENKTLs with deleterious BCOR mutations likely represent an aggressive subgroup of ENKTL, which is presumably driven by aberrant MYC activation by abrogation of BCOR. In contrast, Xiong and colleagues identified a clinically aggressive ENKTL MB subtype, characterized by tumor suppressor gene MGA mutations, 1p22.1/BRDT LOH, and overexpression of MYC.13 Like BCOR, MGA is a tumor suppressor that inhibits MYC-dependent tumor progression.58 It is of interest whether the MB subtype reported by Xiong and colleagues overlaps with the MYC-high ENKTLs in our series. We observed no significant association between MGA mutations (identified only in 7% of cases) and MYC expression (data not shown), although we did not test for 1p22.1/BRDT LOH. Another potential mechanism leading to high MYC expression is the activation of the JAK/STAT pathway: MYC is a downstream target of STAT3, and Song and colleagues reported that Ba/F3 cells forced to express mutant STAT3 show higher mRNA expression levels of MYC.59 In our ENKTL samples, however, there was no significant association between STAT3 or JAK/STAT pathway gene mutations and MYC protein expression levels by IHC (Figure 2D; supplemental Figure 2). Collectively, there seem to be multiple molecular alterations capable of driving MYC overexpression and ENKTL aggressiveness, including BCOR and MGA mutations.
The present study has several limitations. ENKTL is uncommon, even in East Asian populations. Our multi-institutional study investigated the OS of 63 patients with ENKTLs incorporating clinicopathological, IHC, and genetic characteristics; however, the relatively small number of patients included was the primary limitation of our study. Another potential limitation is treatment heterogeneity. Because our case series dated from 2000, not all patients with advanced-stage ENKTL underwent current standard AC-free multiagent chemotherapies, such as SMILE.60 MYC overexpression was an independent adverse prognostic factor in the entire cohort; however, it was not significant in the subgroup treated with AC-free chemotherapy and/or RT for curative intent. Therefore, our heterogeneous patient population requires further validation using a more clinically and temporally uniform ENKTL cohort.
Conclusions
Using targeted NGS and IHC profiling, we showed that activation of the JAK/STAT pathway, partly induced by gain-of-function STAT3 mutations, is common in ENKTLs and associated with high CD30 expression. We also identified the prognostically relevant BCOR-MYC association. ENKTLs with high MYC expression are enriched in those with BCOR mutations and exhibit shorter OS, highlighting the potential utility of IHC for MYC for risk stratification. Further multiomics studies incorporating transcriptomic and epigenomic analyses should be conducted in larger ENKTL cohorts to delineate the functional consequences of MYC overexpression.
Acknowledgments
The authors thank Wakaba Iha for technical support.
This work was supported by a grant from the Kurozumi Medical Foundation and Japan Society for the Promotion of Science Kakenhi grant 21K06883, both to N.O. The sponsors of this study are public and nonprofit organizations that support science in general. Neither had any role in gathering, analyzing, or interpreting the data.
Authorship
Contribution: N.O., K. Mochizuki, and T.K. conceived and designed the study; N.O. performed experiments, analyzed data, and wrote the paper; T.S. supervised the NGS mutational analysis; K. Miyake, N.O., A.S., M.M., I.K., K.K., A.L.F., and N.N. reviewed the cases and collected clinical information; N.O. designed the research and drafted the paper; and A.S. and A.L.F. helped in writing the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Naoki Oishi, Department of Pathology, University of Yamanashi, 1110 Shimokato, Chuo-shi, Yamanashi-ken 409-3898, Japan; e-mail: nohishi@yamanashi.ac.jp.
References
Author notes
Next-generation sequencing is deposited in the Japanese Genotype–Phenotype Archive (accession number: JGAS000548). For original data, please contact nohishi@yamanashi.ac.jp.
The full-text version of this article contains a data supplement.