Key Points
IRF8 is a transcriptional regulator of CD37 expression in DLBCL, which may have implications for anti-CD37 therapies.
Patients with poor prognostic CD37-negative DLBCL show significantly lower IRF8 expression compared with patients with CD37-positive DLBCL.
Abstract
Diffuse large B-cell lymphoma (DLBCL) represents the most common form of non-Hodgkin lymphoma (NHL) that is still incurable in a large fraction of patients. Tetraspanin CD37 is highly expressed on mature B lymphocytes, and multiple CD37-targeting therapies are under clinical development for NHL. However, CD37 expression is nondetectable in ∼50% of DLBCL patients, which correlates with inferior treatment outcome, but the underlying mechanisms for differential CD37 expression in DLBCL are still unknown. Here, we investigated the regulation of the CD37 gene in human DLBCL at the (epi-)genetic and transcriptional level. No differences were observed in DNA methylation within the CD37 promoter region between CD37-positive and CD37-negative primary DLBCL patient samples. On the contrary, CD37-negative DLBCL cells specifically lacked CD37 promoter activity, suggesting differential regulation of CD37 gene expression. Using an unbiased quantitative proteomic approach, we identified transcription factor IRF8 to be significantly higher expressed in nuclear extracts of CD37-positive as compared with CD37-negative DLBCL. Direct binding of IRF8 to the CD37 promoter region was confirmed by DNA pulldown assay combined with mass spectrometry and targeted chromatin immunoprecipitation (ChIP). Functional analysis indicated that IRF8 overexpression enhanced CD37 protein expression, while CRISPR/Cas9 knockout of IRF8 decreased CD37 levels in DLBCL cell lines. Immunohistochemical analysis in a large cohort of primary DLBCL (n = 206) revealed a significant correlation of IRF8 expression with detectable CD37 levels. Together, this study provides new insight into the molecular mechanisms underlying differential CD37 expression in human DLBCL and reveals IRF8 as a transcriptional regulator of CD37 in B-cell lymphoma.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common type of non-Hodgkin lymphoma (NHL). It accounts for approximately one-third of all NHL diagnoses, and a large fraction of DLBCL patients develop relapsed/refractory disease.1,2 Tetraspanin CD37 is a 4-transmembrane protein exclusively expressed on hematopoietic cells, with the highest levels detected in the B-cell lineage from pre-B to mature B cells, but is absent on plasma cells.3,4 CD37 recently regained attention as a promising therapeutic target for the treatment of mature B-cell malignancies,5 and several CD37-targeting therapies are currently under clinical investigation.6,7 Although many B-cell malignancies express CD37 on the cell surface,4 we discovered that approximately 50% of DLBCL shows undetectable CD37 protein expression by immunohistochemistry (IHC), which is correlated with inferior overall and progression-free survival of these patients.8,9
Inactivating CD37 gene mutations have been reported in ∼20% of DLBCL presenting at immune-privileged sites.10,11 However, CD37 mutations are rarely observed in nonimmune-privileged DLBCL,10,12,13 and cannot provide an explanation for the undetectable CD37 expression in a significant fraction of DLBCL. Thus, we hypothesized that other mechanisms like epigenetic or transcriptional gene regulation might underlie the differential expression of CD37. Aberrant DNA methylation patterns are known to occur in DLBCL.14,15 Moreover, it is well-established that altered expression of transcriptional regulators is common in B-cell NHL (B-NHL), including gene rearrangements of MYC and BCL6.16 Other transcription factors, such as FOXO1, can modulate the expression of cell surface protein CD20, thereby affecting therapy response and prognosis in B-NHL.17
Here, we performed an unbiased quantitative proteomic approach of CD37-positive and CD37-negative DLBCL cells and identified transcription factor interferon regulatory factor 8 (IRF8) as a regulator of CD37 expression in DLBCL. IRF8, also known as interferon consensus sequence binding protein (ICSBP), is part of the IRF family of transcription factors that bind to specific DNA sequences named interferon-stimulated response elements (ISRE). IRF8 plays an important role in immune cell development and innate and adaptive immune responses,18 including the promotion of the development of pro- to pre-B cells,19 as well as the germinal center response.20,21 Altogether, we provide new insight into the molecular mechanisms underlying differential CD37 expression in human DLBCL.
Methods
Collection of primary DLBCL patient samples
Primary human DLBCL samples diagnosed between 2000 and 2015 were retrieved from the archives of the Department of Pathology at Radboudumc and CWZ Hospital (Nijmegen, The Netherlands) and Rijnstate Hospital (Arnhem, The Netherlands), and collected in accordance with the Declaration of Helsinki.
Culture and transfection of DLBCL cell lines
DLBCL cell lines were cultured in RPMI-1640 containing 10% fetal bovine serum, 1% antibiotics/antimycotics, and 1% Ultraglutamine and maintained at 37°C, 5% CO2. BJAB cells were transfected using the Neon Transfection System (Invitrogen) according to the manufacturer’s protocol. Other cell lines were transfected using SF Cell Line 4D-Nucleofector X Kit L (Lonza) and the AMAXA Nucleofector biosystem (OCI-Ly1: program DN-100; OCI-Ly8 and SU-DHL-10: program DN-103; SU-DHL-6 and OCI-Ly19: program CV-104; Lonza).
Flow cytometry
Membrane expression of CD37 and CD19 was determined using CD37-FITC (clone M-B371; Biolegend), CD37-APC (clone MB-1; eBioscience), or CD19-APC (clone HIB19, eBioscience) antibodies. Total IRF8 expression was determined with anti-IRF8-PE (clone U31-644; BD Pharmingen) using Transcription Factor Buffer Set (BD Pharmingen) according to the manufacturer’s protocol. All fluorescence intensities were measured on a BD FACSLyric or BD FACSVerse Flow cytometer (BD Biosciences), and data were analyzed using FlowJo X software.
Transient reporter assay
Plasmid psGFP2-C1 (Addgene Plasmid #22881) was digested with AseI and AgeI restriction enzymes (New England BioLabs) to remove the cytomegalovirus (CMV) enhancer and promoter sequence. A region of approximately 2 kb upstream of the CD37 transcription start site (chr19:49 836,812-49 838,766 [GRCh37/hg19]) was amplified from genomic DNA of SU-DHL-5 cells (supplemental Table 2) and ligated into digested psGFP2-C1 to replace the CMV enhancer and promoter region. Cell lines were cotransfected with CMV promoter-GFP (green fluorescent protein) or CD37 promoter-GFP construct and pmScarlet-C1 (Addgene plasmid #85042) as the transfection loading control. In case of cotransfection of the CD37 promoter-GFP with PU.1 and/or IRF2 (both in pCDNA3.1, Genscript), pIRF670-N1 plasmid22 was used as a loading control. If required, pCDNA3.1 empty vector was added to obtain equal amounts of total plasmid DNA. Fluorescent protein expression was analyzed by flow cytometry 24 hours after transfection. Viable cells were gated on positive expression of Scarlet and/or GFP, and the percentage of GFP-expressing cells within this population was determined.
Nuclear proteome
DNA pulldown assay
Oligo baits for DNA pulldown assay were ordered from IDT, with the forward strand containing a 5′ biotin moiety (supplemental Table 3). DNA affinity purifications and on-bead trypsin digestion were performed as described previously.26 Tryptic peptides were first desalted on C18 Stage Tips (without acidification) as described previously.25 On-Stage Tip dimethyl labeling was performed by applying 300 μL of labeling reagent (0.2% CH2O [light] or CD2O [medium]) 2 mg/mL sodium cyanoborohydride, 10 mM NaH2PO4, 35 mM Na2HPO4) to the Stage Tip. Stage Tips were centrifuged for 10 minutes at 2200 xg to pass the labeling reagent through, followed by a wash with 100 μL Buffer A (0.1% formic acid). Samples were analyzed using mass spectrometry.
Mass spectrometry analysis
Samples were eluted from the Stage Tips with 30 μL of Buffer B (80% acetonitrile and 0.1% formic acid). Light and medium labeled DNA pulldown pairs were combined into the same tube. Acetonitrile was evaporated by SpeedVac centrifugation at room temperature, after which Buffer A (0.1% formic acid) was added to a total volume of 12 μL. Half of the sample was applied to reverse phase chromatography using an Easy-nLC 1000 with a ∼30 cm C18 column coupled online to a Q Exactive HF-X Hybrid Quadrupole-Orbitrap mass spectrometer (ThermoFisher). For nuclear proteome samples, a 120-minute gradient, and for DNA pulldown samples, a 60-minute gradient of Buffer B (80% acetonitrile and 0.1% formic acid) was applied. Peptides with a charge state between 2 and 6 were selected for fragmentation with a dynamic exclusion list of 30 peptides for 45 seconds.
Mass spectrometry data analysis
Raw data were analyzed using MaxQuant27 version 1.5.1.0 (DNA pulldowns) and version 1.6.1.0 (nuclear proteome) with a database coding for all human proteins (downloaded June 2017). For DNA pulldown data, requantify was enabled. For the proteome data, match between runs, LFQ, and iBAQ options were enabled. Data were then further processed and filtered using Perseus.28 Protein intensities were log10 transformed, and proteins were filtered for common contaminants, reverse hits, having at least 1 unique peptide, and LFQ nuclear proteome data: having at least 3 valid values in 1 group of triplicates. Missing LFQ values were imputed using default settings, and a Student t test was performed to identify outliers. For the DNA pulldowns, outliers were determined using boxplot statistics.
ChIP assay
Cells were crosslinked with formaldehyde and subsequently resuspended in ice-cold lysis buffer (50 mM HEPES-KOH [pH 7.6]; 140 mM NaCl; 1mM EDTA [pH 8.0]; 1% Triton X-100; 0.1% sodium deoxycholate; Complete Protease Inhibitor Cocktail [Roche]). After lysis, chromatin extracts were sonicated with the Bioruptor PICO (Diagenode). Fragmented chromatin extracts were subjected to ChIP using Protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology) conjugated to anti-IRF8 (E-9 X, Santa Cruz Biotechnology). Controls were subjected to Protein A/G beads only. Following the pulldown, DNA was decrosslinked and purified. Isolated DNA fragments were amplified by quantitative polymerase chain reaction (qPCR) using 2 primer sets to analyze CD37 upstream loci (supplemental Table 4). CD74 and MB primer sets were used as positive and negative controls, respectively (supplemental Table 4). Ct values were used to calculate primer binding efficiency in anti-IRF8 ChIP samples relative to the input chromatin samples, shown as the percentage of input.
Generation of IRF8 plasmid constructs and knockout (KO) cell lines
IRF8 wild-type (WT) sequence was amplified from cDNA of cell line OCI-Ly8 (supplemental Table 5), digested using EcoRI-HF and SalI-HF (New England BioLabs), and subsequently ligated into pIRES2-EGFP plasmid to create an IRF8 expression plasmid. A guide RNA (gRNA) pair targeting the first coding exon of IRF8 was designed using CRISPOR29 (supplemental Table 6), annealed, and cloned into the pX330-U6-Chimeric_BB-CBh-hSpCas9 (Zhang,30 Addgene plasmid #42230) that was linearized using BbsI-HF (New England BioLabs). To obtain IRF8 KO cells, cell lines were transfected with 2 μg of IRF8gRNA-pX330 plasmid or empty pX330 vector (control) and 0.5 μg of pSGFP2-C1, and GFP-positive cells were sorted on a FACS Aria (BD Biosciences) 24 hours later to obtain a polyclonal KO cell population. Additional IRF8 KO cells in BJAB and OCI-Ly8 cells were generated using 3 new gRNAs designed using the CRISPOR database (supplemental Table 6; set #2). Oligos for these gRNAs were annealed and ligated into the pSpCas9(BB)-2A-Puro (PX459) V2.0 (Addgene Plasmid #62988)31 using the BbsI site. The efficiency of these gRNAs was determined using a T7 endonuclease assay. For KO experiments, cells were nucleofected with the different gRNA constructs. To select for cells that expressed Cas9, puromycin was added to the BJAB and OCI-Ly8 cells at a concentration of respectively 2 and 1 µg/mL, dead cells were removed using Ficoll separation 24 hours after nucleofection, and cells were analyzed for IRF8 and CD37 expression.
IHC
IHC staining on lymphoma tissues was performed on formalin-fixed, paraffin-embedded tissue microarrays following the manufacturer’s instructions using mouse anti-CD37 antibody (clone 2B8; ThermoScientific and Novus Biologicals) or mouse anti-ICSBP (IRF8, clone E-9; Santa Cruz Biotechnology) followed by hematoxylin counterstaining.
Results
Mutation and DNA methylation analysis of the CD37 promoter region
Previously we reported that ∼50% of primary DLBCL show undetectable CD37 expression by IHC, which is associated with inferior treatment outcome after R-CHOP (a combination therapy of anti-CD20 antibody rituximab, cyclophosphamide, hydroxydaunorubicin, vincristine [oncovin], and prednisone) treatment in both germinal center B cell-like (GCB) and non-GCB DLBCL.8,9 We analyzed a cohort of CD37-negative DLBCL (n = 12) that displayed no genetic alterations within the CD37 coding sequence,10 for the presence of potential mutations in the CD37 promoter sequence. This region of approximately 1.7 kb upstream of the CD37 transcription start site was selected based on epigenetic profiling data from ENCODE in a malignant B-cell line at this locus (supplemental Figure 1).32 No mutations were detected in this locus in any of the CD37-negative DLBCL cases, indicating that alternative mechanisms are responsible for the attenuated CD37 expression in these tumors.
Next, we determined the methylation status in the upstream regulatory locus of CD37, as well as the 3′ untranslated region (UTR), in DLBCL cell lines and primary human DLBCL tumor samples using bisulfite sequencing PCR (BSP) (supplemental Figures 1 and 2; supplemental Tables 8-13). BSP analysis was performed for 2 loci in the CD37 promoter region and 1 locus in the 3′ UTR, which contains a higher density of CpG pairs than the promoter, which lacks CpG islands. Little differences in methylation levels were observed between CD37-negative and CD37-positive DLBCL cell lines. This was confirmed in primary DLBCL samples, where similar methylation levels between CD37-negative and CD37-positive samples were detected (supplemental Figure 2). Human embryonic kidney cells that do not express CD37 were included as a control in these experiments, showing a high level of methylation in the promoter region (supplemental Figure 2). Together, these data indicate no significant role for aberrant DNA methylation at these 3 loci in the regulation of CD37 expression in human DLBCL.
Identification of IRF8 as candidate transcriptional regulator of CD37 expression
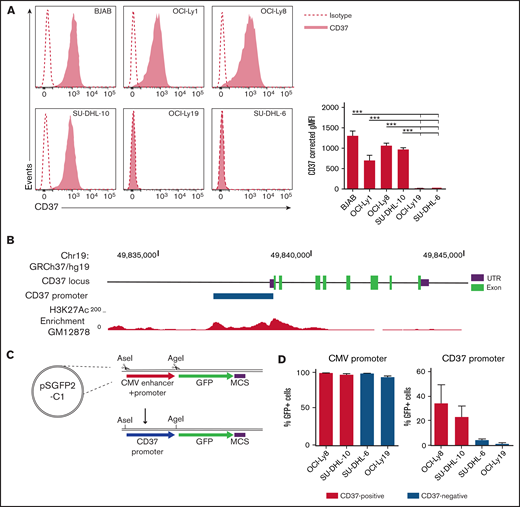
While no mutations or differential DNA methylation were detected in the CD37 promoter region in CD37-negative DLBCL cells, lymphoma cells with reduced CD37 expression may still show diminished promoter activity. To study differential gene regulation, human DLBCL cell lines were first analyzed for membrane expression of CD37 and the B-cell marker CD19 as control. DLBCL lines OCI-Ly19 and SU-DHL-6 were CD37-negative, and BJAB, OCI-Ly1, OCI-Ly8, and SU-DHL-10 were CD37-positive (Figure 1A). CD19 was expressed on all DLBCL cell lines as expected (supplemental Figure 3). In line with CD37 membrane expression, CD37 mRNA expression was strongly reduced in CD37-negative cell lines (supplemental Figure 4). Sequence analysis of the CD37 coding region in these CD37-negative cell lines showed no mutations. To investigate the CD37 promoter activity in the different DLBCL cell lines, we exchanged the CMV promoter and enhancer region of a GFP-reporter plasmid for the ∼2 kb upstream sequence of the CD37 gene (Figure 1B-C). DLBCL cell lines were transfected with CD37 promoter-GFP or the original CMV promoter plasmid as control. CD37-negative DLBCL cell lines showed lower GFP expression than CD37-positive DLBCL cell lines (Figure 1D), suggesting that transcriptional activators or repressors involved in the regulation of CD37 gene transcription are differentially expressed in CD37-negative DLBCL lines.
CD37-negative DLBCL cell lines lack CD37 promoter activity. (A) Representative histograms (left) and quantification (right) of flow cytometry analysis of CD37 membrane expression on human DLBCL cell lines. Corrected geometric mean fluorescence intensity (gMFI): gMFI corrected for gMFI of isotype control. One-way ANOVA analysis was followed by Tukey's multiple comparisons test. ***P < .001 represents the significance of each cell line compared with OCI-Ly19 and SU-DHL-6 separately. Data represent mean + SEM of 3 independent experiments. (B) Schematic representation of the CD37 gene locus and upstream sequence. The blue bar indicates the locus of the CD37 promoter as used in (C) and (D), based on the H3K27Ac enrichment in the lymphoblastoid GM12878 cell line (as obtained via the University of California, Santa Cruz Genome Browser, data from the Bernstein Laboratory at the Broad Institute32 ). UTR, untranslated region. (C) Schematic representation of the GFP-reporter plasmid. GFP, green fluorescent protein; MCS, multiple cloning site. (D) Percentage of GFP-positive cells in CD37-positive and CD37-negative DLBCL cell lines transfected with CMV promoter-GFP or CD37 promoter-GFP construct. Data represent mean + SEM of 3 independent experiments.
CD37-negative DLBCL cell lines lack CD37 promoter activity. (A) Representative histograms (left) and quantification (right) of flow cytometry analysis of CD37 membrane expression on human DLBCL cell lines. Corrected geometric mean fluorescence intensity (gMFI): gMFI corrected for gMFI of isotype control. One-way ANOVA analysis was followed by Tukey's multiple comparisons test. ***P < .001 represents the significance of each cell line compared with OCI-Ly19 and SU-DHL-6 separately. Data represent mean + SEM of 3 independent experiments. (B) Schematic representation of the CD37 gene locus and upstream sequence. The blue bar indicates the locus of the CD37 promoter as used in (C) and (D), based on the H3K27Ac enrichment in the lymphoblastoid GM12878 cell line (as obtained via the University of California, Santa Cruz Genome Browser, data from the Bernstein Laboratory at the Broad Institute32 ). UTR, untranslated region. (C) Schematic representation of the GFP-reporter plasmid. GFP, green fluorescent protein; MCS, multiple cloning site. (D) Percentage of GFP-positive cells in CD37-positive and CD37-negative DLBCL cell lines transfected with CMV promoter-GFP or CD37 promoter-GFP construct. Data represent mean + SEM of 3 independent experiments.
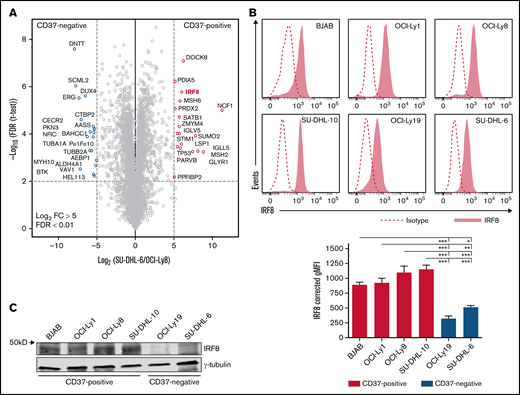
In order to identify these components, the nuclear proteome of CD37-positive (OCI-Ly8) and CD37-negative (SU-DHL-6) DLBCL cell lines were investigated for differentially expressed candidate transcriptional regulators using an unbiased quantitative proteomic approach. Transcription factors p53 and IRF8 were both higher expressed in CD37-positive DLBCL compared with CD37-negative DLBCL, with IRF8 being the most significantly upregulated transcription factor, showing about 60 times higher expression in OCI-Ly8 (Figure 2A; supplemental Table 14; for a summary of hits see supplemental Table 15). To confirm this finding, protein expression of IRF8 was analyzed with both flow cytometry and western blotting in multiple DLBCL lines (Figure 2B-C). Significantly lower IRF8 expression was observed in CD37-negative DLBCL lines (SU-DHL-6 and OCI-Ly19) as compared with CD37-positive cell lines (BJAB, OCI-Ly1, OCI-Ly8, and SU-DHL-10). Moreover, analysis of publically available data from the Cancer Dependency Portal (DepMap)33,34 showed that expression levels of IRF8 protein positively correlated with CD37 mRNA levels in NHL cell lines (supplemental Figure 5). Together these data indicate that IRF8 is a candidate regulator of CD37 expression in human DLBCL.
Identification of IRF8 in CD37-positive DLBCL cell lines. (A) Volcano plot of the label-free quantification (LFQ) of protein abundance in the nuclear protein fractions of CD37-negative (SU-DHL-6) vs CD37-positive (OCI-Ly8) DLBCL cells. Gene names are indicated. The volcano plot presents data of n = 3 technical replicates for both SU-DHL-6 and OCI-Ly8 extract. (B) Representative histograms (top) and quantification (bottom) of flow cytometry analysis of IRF8 expression in human DLBCL cell lines. Corrected gMFI: gMFI corrected for gMFI of isotype control. One-way ANOVA analysis was followed by Tukey's multiple comparisons test. ***P < .001, **P < .01, *P < .05. Data represent mean + SEM of 3 independent experiments. (C) Representative western blot stained for IRF8 and γ-tubulin as loading control in indicated CD37-positive and CD37-negative DLBCL cell lines.
Identification of IRF8 in CD37-positive DLBCL cell lines. (A) Volcano plot of the label-free quantification (LFQ) of protein abundance in the nuclear protein fractions of CD37-negative (SU-DHL-6) vs CD37-positive (OCI-Ly8) DLBCL cells. Gene names are indicated. The volcano plot presents data of n = 3 technical replicates for both SU-DHL-6 and OCI-Ly8 extract. (B) Representative histograms (top) and quantification (bottom) of flow cytometry analysis of IRF8 expression in human DLBCL cell lines. Corrected gMFI: gMFI corrected for gMFI of isotype control. One-way ANOVA analysis was followed by Tukey's multiple comparisons test. ***P < .001, **P < .01, *P < .05. Data represent mean + SEM of 3 independent experiments. (C) Representative western blot stained for IRF8 and γ-tubulin as loading control in indicated CD37-positive and CD37-negative DLBCL cell lines.
IRF8 binds to the upstream regulatory region of the CD37 gene and induces its expression
Next, we assessed whether IRF8 was able to interact with the CD37 promoter region in DLBCL. IRF8 is known to bind several conserved DNA motifs, including ISREs and ETS-IRF composite elements (EICE).35,36 We identified an EICE motif within the 2 kb upstream sequence of the CD37 gene (supplemental Figure 6). WT and mutated DNA oligo baits of this potential binding site (Figure 3A) were incubated with nuclear extracts of CD37/IRF8-positive OCI-Ly8 cells. Oligo bait interactors were subsequently analyzed by liquid chromatography-mass spectrometry using dimethyl labeling for quantification. Analysis of the interacting proteins showed significant binding of multiple factors, including IRF8, to the WT sequence over the mutated sequence (Figure 3B). In addition, the WT oligo bait also demonstrated binding of IRF2, another related family member that may act in a complex with IRF8.
IRF8 binds to the upstream regulatory region of the human CD37 gene. (A) Schematic representation of the CD37 gene locus and upstream sequence. The blue bar indicates the locus of the CD37 promoter, as shown in Figure 1. The red bar indicates the sequence locus of the DNA oligo bait. Orange boxes show amplified loci in qPCR preceded by immunoprecipitation using an anti-IRF8 antibody (ChIP-seq). (B) Scatterplot showing the results of a DNA pulldown experiment with CD37 oligo bait, analyzed by liquid chromatography-mass spectrometry using dimethyl labeling for quantification. The x-axis shows the ratio of proteins labeled as mutant bait: heavy/WT CD37 bait: light, whereas the y-axis shows the label-swap experiment (WT CD37 bait: heavy/mutant bait: light). Proteins labeled in blue are specific interactors of the mutated control sequence, and proteins labeled in red are specific interactors of the WT sequence. (C) ChIP using anti-IRF8 antibody or control, followed by qPCR analysis in 2 CD37-positive (BJAB [left], OCI-Ly8 [right]) and 2 CD37-negative (OCI-Ly19 [left], SU-DHL-6 [right]) DLBCL cell lines. Two primer sets were used to analyze the CD37 upstream locus, as indicated in (A). CD74 was used as a positive control and MB as a negative control. Differences were determined using a 2-way ANOVA followed by Tukey's multiple comparisons test. *P < .05, ***P < .001. Data represent mean + SEM of 3 independent experiments.
IRF8 binds to the upstream regulatory region of the human CD37 gene. (A) Schematic representation of the CD37 gene locus and upstream sequence. The blue bar indicates the locus of the CD37 promoter, as shown in Figure 1. The red bar indicates the sequence locus of the DNA oligo bait. Orange boxes show amplified loci in qPCR preceded by immunoprecipitation using an anti-IRF8 antibody (ChIP-seq). (B) Scatterplot showing the results of a DNA pulldown experiment with CD37 oligo bait, analyzed by liquid chromatography-mass spectrometry using dimethyl labeling for quantification. The x-axis shows the ratio of proteins labeled as mutant bait: heavy/WT CD37 bait: light, whereas the y-axis shows the label-swap experiment (WT CD37 bait: heavy/mutant bait: light). Proteins labeled in blue are specific interactors of the mutated control sequence, and proteins labeled in red are specific interactors of the WT sequence. (C) ChIP using anti-IRF8 antibody or control, followed by qPCR analysis in 2 CD37-positive (BJAB [left], OCI-Ly8 [right]) and 2 CD37-negative (OCI-Ly19 [left], SU-DHL-6 [right]) DLBCL cell lines. Two primer sets were used to analyze the CD37 upstream locus, as indicated in (A). CD74 was used as a positive control and MB as a negative control. Differences were determined using a 2-way ANOVA followed by Tukey's multiple comparisons test. *P < .05, ***P < .001. Data represent mean + SEM of 3 independent experiments.
To further confirm the binding of IRF8 to the CD37 promoter region in CD37-positive DLBCL, we selected 2 loci for targeted ChIP analysis. One of these contained the EICE motif to which IRF8 binding was detected in the DNA pulldown assay (supplemental Figure 6). This element contains both an IRF8 binding motif and one for ETS proteins, including transcription factor PU.1, a known partner of IRF8.18,21,35,36 We observed higher PU.1 expression in CD37-positive DLBCL cell lines compared with CD37-negative DLBCL (supplemental Figure 7A). CD37-promoter activity was induced upon combined PU.1/IRF2 expression in CD37-negative DLBCL, but not in CD37-positive DLBCL (supplemental Figure 7B). The second ChIP locus that was selected also harbored a binding motif for PU.1.
ChIP-qPCR analysis with anti-IRF8 antibody showed enrichment of IRF8 to these 2 loci (Figure 3A,C). Specificity for IRF8 binding was confirmed by significant binding to the known IRF8 target gene CD74,35 while there was no recruitment of IRF8 in the negative controls (ie, to the myoglobin [MB] locus and in the CD37-negative DLBCL cell lines that express significantly less IRF8 than the CD37-positive cell lines) (Figure 3C). Together, these data confirm that IRF8 can directly bind to the promoter region of the CD37 gene in DLBCL.
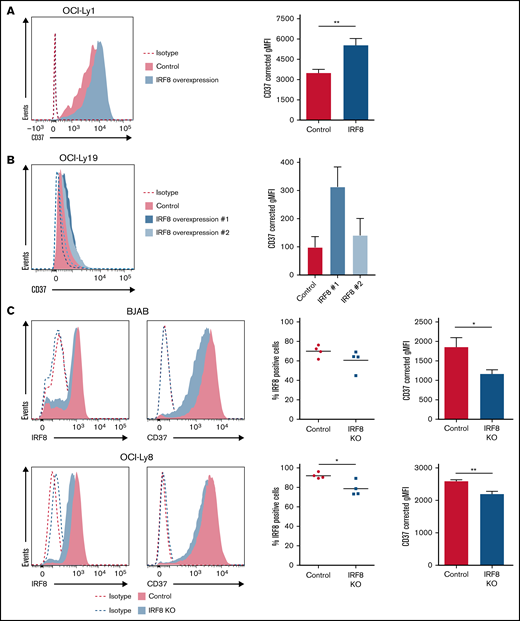
To determine whether the expression of IRF8 directly affects CD37 expression levels, both IRF8 overexpression and IRF8 KO experiments were performed in DLBCL cell lines. Transient overexpression of IRF8 in OCI-Ly1 cells resulted in a significant upregulation of CD37 protein expression at the cell surface (Figure 4A). Moreover, 2 independent pools of CD37-negative OCI-Ly19 cells with stable overexpression of IRF8 were created (supplemental Figure 8), resulting in increased CD37 and PU.1 expression compared with the nontransduced control cells (Figure 4B; supplemental Figure 8). Additionally, the importance of IRF8 in regulating CD37 expression was validated in 2 IRF8 KO DLBCL cell lines (Figure 4C). Analysis of CD37 membrane expression in these IRF8 KO DLBCL cells showed a significant decrease (15% to 36%) in CD37 levels by flow cytometry (Figure 4C). Independently generated IRF8 KO DLBCL cells using different CD37 guide RNAs showed a similar trend (supplemental Figure 9). The moderate decrease in CD37 expression by IRF8 CRISPR KO in both DLBCL cell lines is likely caused by the limitation to generate a complete IRF8KO, which was incompatible with the viability of the cells. Together these results demonstrate that IRF8 can directly bind to the upstream promoter region of the CD37 gene and affects its expression in DLBCL cells.
IRF8 directly affects the expression of CD37 in DLBCL cell lines. (A) Representative histogram (left) and quantification (right) of flow cytometry analysis of CD37 membrane expression on GFP-positive OCI-Ly1 cells transfected with empty IRES-EGFP (control) or IRF8-IRES-EGFP expression plasmid (IRF8). **P < .01, paired t test. Data represent mean + SEM of 4 independent experiments. (B) Representative histogram (left) and quantification (right) of flow cytometry analysis of CD37 membrane expression on 2 independent batches of OCI-Ly19 cells that stably overexpress IRF8. Data represent mean + SEM of 3 independent experiments. (C) Representative histograms (left) and quantification (right) of flow cytometry analysis of BJAB (top) and OCI-Ly8 (bottom) control and IRF8 KO cells. The percentage of IRF8-positive cells was determined as the percentage of cells with IRF8 signal above isotype level: *P < .05, unpaired t test. Quantification of CD37 expression: *P < .05, **P < .01, unpaired t test. Data represent mean + SEM of 4 independent experiments. Corrected gMFI: gMFI corrected for gMFI of isotype control.
IRF8 directly affects the expression of CD37 in DLBCL cell lines. (A) Representative histogram (left) and quantification (right) of flow cytometry analysis of CD37 membrane expression on GFP-positive OCI-Ly1 cells transfected with empty IRES-EGFP (control) or IRF8-IRES-EGFP expression plasmid (IRF8). **P < .01, paired t test. Data represent mean + SEM of 4 independent experiments. (B) Representative histogram (left) and quantification (right) of flow cytometry analysis of CD37 membrane expression on 2 independent batches of OCI-Ly19 cells that stably overexpress IRF8. Data represent mean + SEM of 3 independent experiments. (C) Representative histograms (left) and quantification (right) of flow cytometry analysis of BJAB (top) and OCI-Ly8 (bottom) control and IRF8 KO cells. The percentage of IRF8-positive cells was determined as the percentage of cells with IRF8 signal above isotype level: *P < .05, unpaired t test. Quantification of CD37 expression: *P < .05, **P < .01, unpaired t test. Data represent mean + SEM of 4 independent experiments. Corrected gMFI: gMFI corrected for gMFI of isotype control.
IRF8 expression correlates with CD37 mRNA and protein levels in primary DLBCL
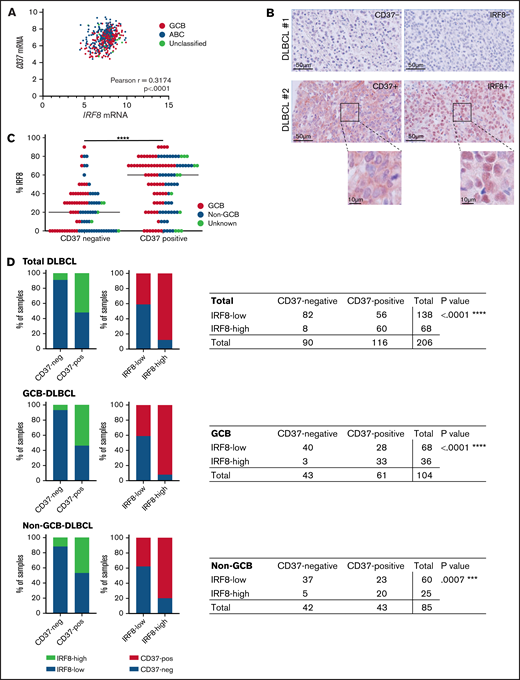
To assess the role of IRF8 in regulating CD37 expression in primary human DLBCL, data on mRNA expression of IRF8 and CD37 were obtained from gene expression profiling previously performed on tumor samples (n = 498) of the International DLBCL R-CHOP Consortium Program.9 The prognostic association of CD37 expression was also evident in this cohort (supplemental Figure 10A), and correlation analysis showed a positive linear relationship between IRF8 and CD37 mRNA levels (Pearson correlation r = 0.3174; P < .0001) (Figure 5A) in both GCB and ABC subtype (supplemental Figure 10B). The correlation between IRF8 and CD37 mRNA expression was not affected by IRF8 mutations as analyzed in a publicly available RNA-Seq database (supplemental Figure 11). Protein expression of IRF8 and CD37 in DLBCL tissues was determined by IHC analysis in an independent DLBCL cohort (n = 206) (Figure 5B) consisting of both GCB and non-GCB subtypes (supplemental Figure 12). IRF8 was expressed in a significantly lower fraction of tumor cells in CD37-negative DLBCL than CD37-positive DLBCL (P = .0001) (Figure 5C). In line with this, absence of CD37 expression highly correlated with low IRF8 expression in total DLBCL, as well as in GCB and non-GCB tumors separately (protein expression by IHC <60%; P < .0001, P < .0001, and P = .0007, respectively) (Figure 5D). IRF8 expression was low in 138 out of 206 DLBCL samples (67%) and 82 out of 90 CD37-negative DLBCL samples (91%). Taken together, these data reveal that IRF8 expression positively correlates with CD37 expression in human primary DLBCL.
IRF8 expression correlates with CD37 expression in primary human DLBCL. (A) Analysis of IRF8 and CD37 mRNA levels in primary DLBCL (n = 498). Linear regression R2= 0.1007. Pearson correlation r = 0.3174; 95% CI, 0.2361-0.3943; ****P < .0001. (B) CD37 and IRF8 IHC staining (red) of 2 representative DLBCL biopsies: CD37/IRF8 double-negative (top) and CD37/IRF8 double-positive (bottom) human DLBCL. Cell nuclei were counterstained with hematoxylin (blue). (C) IHC staining of CD37 and IRF8 was scored in 206 primary DLBCL samples. The percentage indicates the percentage of IRF8-positive tumor cells per sample. Each dot represents 1 tumor sample. GCB/non-GCB status was known for 189 samples and is indicated (red: GCB; blue: non-GCB). The bar indicates the median percent IRF8 value per group. χ-square test of the absolute number of total DLBCL samples per group showed a significant association between the percentage of IRF8 and CD37 expression. ****P < .0001. (D) Samples were determined CD37-negative and IRF8-low when scoring was <10% and <60%, respectively. Data about GCB (middle) and non-GCB (bottom) status were available for 189 out of the 206 analyzed patients. P value shows the statistical significance obtained using Fisher’s exact test. ****P < .0001 (total DLBCL), ****P < .0001 (GCB), and ***P = .0007 (non-GCB).
IRF8 expression correlates with CD37 expression in primary human DLBCL. (A) Analysis of IRF8 and CD37 mRNA levels in primary DLBCL (n = 498). Linear regression R2= 0.1007. Pearson correlation r = 0.3174; 95% CI, 0.2361-0.3943; ****P < .0001. (B) CD37 and IRF8 IHC staining (red) of 2 representative DLBCL biopsies: CD37/IRF8 double-negative (top) and CD37/IRF8 double-positive (bottom) human DLBCL. Cell nuclei were counterstained with hematoxylin (blue). (C) IHC staining of CD37 and IRF8 was scored in 206 primary DLBCL samples. The percentage indicates the percentage of IRF8-positive tumor cells per sample. Each dot represents 1 tumor sample. GCB/non-GCB status was known for 189 samples and is indicated (red: GCB; blue: non-GCB). The bar indicates the median percent IRF8 value per group. χ-square test of the absolute number of total DLBCL samples per group showed a significant association between the percentage of IRF8 and CD37 expression. ****P < .0001. (D) Samples were determined CD37-negative and IRF8-low when scoring was <10% and <60%, respectively. Data about GCB (middle) and non-GCB (bottom) status were available for 189 out of the 206 analyzed patients. P value shows the statistical significance obtained using Fisher’s exact test. ****P < .0001 (total DLBCL), ****P < .0001 (GCB), and ***P = .0007 (non-GCB).
Discussion
Tetraspanin proteins have been linked to cancer progression and patient outcome.37 In particular, the expression of tetraspanin CD37 is significantly attenuated in ∼50% of DLBCL, which correlates with inferior survival of these DLBCL patients.8,9 However, the underlying cause for differential CD37 expression levels in DLBCL is largely unknown. Here, we investigated diverse molecular mechanisms that may contribute to the regulation of CD37 in human DLBCL, where we identified IRF8 as a transcriptional regulator of CD37 expression.
We detected a significantly higher expression of transcription factor IRF8 in CD37-positive DLBCL compared with CD37-negative DLBCL cell lines. In line with our findings, the expression pattern of IRF8 during normal B-cell development is similar to that of CD37, with high IRF8 expression observed in GC B cells and low levels in plasma cells.20 IRF8 can function as a transcriptional activator or repressor in B cells.35 We observed that overexpression of IRF8 induced CD37 expression in DLBCL cells, while IRF8 KO decreased CD37 expression, implying IRF8 as a transcriptional activator of CD37 gene expression. Our findings are in accordance with an IRF8 transcriptional profiling study, in which binding of IRF8 to the CD37 gene in GC B cells was observed.35
Moreover, IRF8 expression positively correlated with CD37 expression in primary human DLBCL tissue samples. IRF8 expression was low as determined by IHC in 138 out of 206 DLBCL samples (67%), which correlated with attenuated CD37 expression. The role of IRF8 in DLBCL pathogenesis has not been fully elucidated. One study showed that loss of IRF8 in DLBCL cells reduced their growth in vitro and in vivo, suggesting an oncogenic role.38 In line with this, we observed the complete IRF8 KO in DLBCL cell lines was difficult to generate. Moreover, IRF8 was detected as a novel fusion partner of IGH in DLBCL,39,40 and this fusion resulted in higher IRF8 expression levels.39 In a mouse model of the IGH-IRF8 fusion, this deregulation resulted in lower survival of these mice compared with the WT control; however, no increased incidence of B-cell lymphoma was observed.41 IRF8 is shown to be recurrently mutated in comprehensive whole-genome studies of DLBCL.12,13,42 In addition, aberrant DNA methylation of the IRF8 promoter region has been observed in multiple myeloma and B-cell acute lymphoblastic leukemia.43,44 Our studies did not show a relation between IRF8 mutations and CD37 expression; thus, how IRF8 expression is differentially regulated in CD37-negative DLBCL is subject for future studies.
We observed no significant differences in DNA methylation in the CD37 promoter and 3′UTR gene region in CD37-positive compared with CD37-negative DLBCL, although we cannot exclude that methylation in other CpG-rich regions of the CD37 gene regulates CD37 expression. Alterations in DNA methylation of tetraspanin genes in other hematological cancers have been reported in multiple myeloma (hypermethylation of tetraspanin genes CD9, CD81, and CD82)45,46 and mantle cell lymphoma (CD37 hypomethylation).47
It is expected that supplemental (co)factors are involved in the regulation of CD37 expression in DLBCL. We confirmed direct binding of IRF8 to 2 sequences that included an EICE or a PU.1 binding motif in the CD37 locus in CD37-positive DLBCL cells. Next to IRF8, we observed the binding of IRF2 and ELF1 to the CD37 EICE sequence upon incubation with DNA oligo baits. IRF2 also belongs to the IRF family of transcription factors and is able to heterodimerize with IRF8.48 Like PU.1, ELF1 is a member of the ETS family.49 Both transcription factors have also been indicated to be required for normal B-cell development.50,51 Furthermore, the EICE is a well-known binding motif for IRF8-PU.1, as well as IRF4-PU.1 complexes.18,35,36 The expression of IRF4 is, like IRF8, restricted to hematopoietic cells and implicated in the development of dendritic cells and B cells.18,19,52 Our initial data shows a potential role for PU.1 and/or IRF2 in the activation of CD37 expression; however, the exact role of IRF2, IRF4, PU.1, and ELF1 in this regulation, and whether or not this is in conjunction with IRF8, remains to be elucidated.
Although our study was focused on positive regulators of CD37 expression, it is possible that negative regulators also contribute to the regulation of its expression. For example, higher expression of SCML2 was observed in CD37-negative cells, which is a member of the Polycomb group proteins and contributes to target gene repression.53 Whether SCML2 or chromatin remodeling by Polycomb group complexes are involved in the silencing of the CD37 locus requires further investigation.
CD37 is a prognostic factor for R-CHOP-treated DLBCL independent of TP53 mutation and the international prognostic index as shown in a large international DLBCL R-CHOP Consortium Program.9 CD37 facilitates the formation of tetraspanin nanodomains in the cell membrane and regulates intracellular signaling pathways through lateral interactions with partner proteins.37,54 It has no known ligand, although crossligation of CD37 can induce apoptosis.55 Multiple antibody-based CD37-targeting therapies are under (pre)clinical development for B-NHL.6,7 Current targeted immunotherapies have shown the pivotal importance of target expression, exemplified by the downregulation of CD20 membrane expression as a proposed mechanism of rituximab resistance.56 In this context, it is of high interest to induce CD37 membrane expression, and it remains to be investigated whether this can be achieved by manipulating IRF8 levels or activity in DLBCL. Taken together, we have shown that IRF8 is a transcriptional activator of CD37 gene expression in human DLBCL. Full elucidation of the regulation of CD37 expression can provide new therapeutic strategies for the large population of patients with poor prognostic CD37-negative DLBCL.
Acknowledgments
The authors thank Christiaan J. Stavast and Stefan J. Erkeland (Department of Immunology, Erasmus MC Rotterdam) and Femke Doubrava-Simmer (Department of Pathology, Radboudumc, Nijmegen) for their valuable help with the DNA methylation studies; Nelleke Spruijt (Department of Molecular Biology, Radboud Institute for Molecular Life Sciences, Nijmegen) for her valuable help with the mass spectrometry; and Alie van der Schaaf and Erik Jansen (Department of Tumor Immunology, Radboudumc, Nijmegen) for their contribution to the IHC and (q)PCR analyses.
S.E. is supported by a Radboudumc grant; A.B.v.S. is supported by the Netherlands Organization for Scientific Research NWO Gravitation Programme 2013 grant (ICI 000-23), the Dutch Cancer Society (KWF) (11618/2018 and 12949/2020), and was awarded a European Research Council Consolidator Grant (Secret Surface, 724281); B.S. is a recipient of a Dutch Cancer Society grant (11137/2017); and the Vermeulen laboratory is part of the Oncode Institute, which is partly funded by the Dutch Cancer Society.
Authorship
Contribution: S.E., B.S., and A.B.v.S. designed the study and wrote the manuscript; S.E. and M.t.B. performed flow cytometry transfection experiments and western blotting; S.E. performed sequencing and statistical analyses; L.J. performed exon sequencing analyses and ChIP experiments; S.E., A.N.K., R.M.S., and D.d.W. performed targeted DNA methylation analysis; M.t.B., A.N.K., R.M.S., and D.d.W. cloned plasmids; P.M.H. performed DepMap analysis; M.v.d.B., C.H., B.H., E.v.d.S., D.-J.v.S., J.v.H., and E.H. collected DLBCL tissues and corresponding pathological information; C.M.d.W. and A.N.K. stained tissue microarray sections that were scored by M.v.d.B.; C.M.d.W. and M.t.B. performed qPCR experiments; S.E. and M.P.B. prepared nuclear lysates and M.P.B. performed the DNA pulldown; P.W.T.C.J. performed mass spectrometry analyses that were supervised by M.V.; Z.Y.X.-M. and K.H.Y. provided gene expression profiling data and survival analysis; S.B. and C.K. performed IRF8 mutation analysis on RNA-Seq dataset; S.E., B.S., and A.B.v.S. supervised the work; and all authors read and approved the final manuscript.
Conflict-of-interest disclosures: The authors declare no competing financial interests.
Correspondence: Annemiek B. van Spriel, Department of Tumor Immunology, Radboud Institute for Molecular Life Sciences, Radboud University Medical Center, GeertGrooteplein-Zuid 26-28, 6525 GA Nijmegen, The Netherlands; e-mail: annemiek.vanspriel@radboudumc.nl.
References
Author notes
M.t.B. and L.J. contributed equally to this study.
All data generated or analyzed during this study are included in this published article and its supplemental information files. For additional original data not included in the supplemental information files, please contact the corresponding author.
The full-text version of this article contains a data supplement.

![IRF8 binds to the upstream regulatory region of the human CD37 gene. (A) Schematic representation of the CD37 gene locus and upstream sequence. The blue bar indicates the locus of the CD37 promoter, as shown in Figure 1. The red bar indicates the sequence locus of the DNA oligo bait. Orange boxes show amplified loci in qPCR preceded by immunoprecipitation using an anti-IRF8 antibody (ChIP-seq). (B) Scatterplot showing the results of a DNA pulldown experiment with CD37 oligo bait, analyzed by liquid chromatography-mass spectrometry using dimethyl labeling for quantification. The x-axis shows the ratio of proteins labeled as mutant bait: heavy/WT CD37 bait: light, whereas the y-axis shows the label-swap experiment (WT CD37 bait: heavy/mutant bait: light). Proteins labeled in blue are specific interactors of the mutated control sequence, and proteins labeled in red are specific interactors of the WT sequence. (C) ChIP using anti-IRF8 antibody or control, followed by qPCR analysis in 2 CD37-positive (BJAB [left], OCI-Ly8 [right]) and 2 CD37-negative (OCI-Ly19 [left], SU-DHL-6 [right]) DLBCL cell lines. Two primer sets were used to analyze the CD37 upstream locus, as indicated in (A). CD74 was used as a positive control and MB as a negative control. Differences were determined using a 2-way ANOVA followed by Tukey's multiple comparisons test. *P < .05, ***P < .001. Data represent mean + SEM of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/6/7/10.1182_bloodadvances.2021004366/5/m_advancesadv2021004366f3.png?Expires=1769083504&Signature=GIgbyN8csD1JkKgpOVqFEaMBqdfHsorokjQ6g~oInYsUA8taHcSzeL0p6ZqxvtfH4BsYhT47PlTMz5lDiaRNrHfCXDhoC~z-Pv2~eTwKurYhP07r9n5CeURHEZdgygKFoA3LJWa2DaFVStTJwSDKhrKPtMl~gv0muRn6OmtA56uHjumC3igU9lsLlYfTSnJYb9SuR6mlaFHACIYrRRwHcM~Mbi83UnXdmKGwGuGbFw9srq5Q~lLYC9F3Iv-zydrL0VXAcqUd-MYaaxCf9Xj19rFql5pL5U~mLL3U8K90jBFc5vhlmBMN-lAfeesiQ7vxV-DmLeJ9iM1BOa8JoeFP9A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)