TO THE EDITOR:
The immunoglobulin glycoprotein VI (GPVI) is a major signaling receptor on platelets for collagen, fibrin, and fibrinogen.1 There have been 3 reports of inheritable mutations in the GP6 gene, 2 describing single individuals with compound heterozygous mutations,2,3 whereas the third reports a homozygous mutation originally described in 5 individuals in Chile.4 This homozygous mutation (c.711dup [GP6InsA/InsA]) leads to the insertion of a premature stop codon in exon 6 (p.Val238SerfsTer5), preventing the surface expression of GPVI. The identification and wider screening of this founder GP6 mutation provides an opportunity to assess the role of GPVI in cardiovascular health, because this unique group of patients can offer novel insight into its etiology and role in platelet biology.
Since the original report, a total of 11 patients with the homozygous mutation have been identified, all of Chilean nationality and residing in distant locations, with no relatedness and nonconsanguineous parents (Figure 1). We estimated that 2.9% of Chileans are heterozygous for the GP6 mutation in a genetic screening of ∼1200 individuals representative of the population,5 suggesting more than 4000 individuals in Chile with the homozygous mutation, which is twice the number of patients with hemophilia.
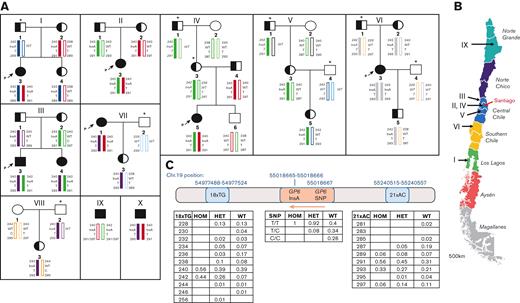
The genetic etiology of the founder GP6 mutation found in Chilean patients. (A) Pedigrees showing a pattern of inheritance and different haplotypes surrounding the GP6InsA mutation site in identified Chilean families. (i-viii), Pedigrees including 6 probands (marked by a P and an arrow) and 2 heterozygous cases, wherein which the DNA of family members was also available for sequencing. For relatives who were not sequenced, we inferred the most likely haplotype based on sequencing results from samples provided; these individuals are marked with an asterisk (∗) and their inferred haplotype, assuming a normal pattern of inheritance and no recombination events, is in italics. (ix-x), Haplotypes of 2 individuals with homozygous mutation in which no DNA of family members was available. Color coding has been used to show inheritance patterns of alleles (block color alleles carry the GP6InsA haplotype). Squares indicate males, and circles indicate females. Blank shapes indicate GP6WT/WT status, half-filled shapes indicate GP6WT/InsA status and filled shapes indicate GP6InsA/InsA status. (B) Map of Chile indicating geographical sites of origin where patients have been diagnosed (arrowed). Santiago, Chile’s capital city, is indicated in red. Map adapted from www.touropia.com with permission. (C) The position of the microsatellite markers (18xTG and 21xAC), the GP6 exon 6 mutation (clinVar ID number 1072837), and the GP6 common SNP rs1654416 on chromosome 19 are shown. Tables show the relative frequencies of fragment size for each microsatellite marker and SNP status, in the 3 genetic groups investigated. The orange arrow shows the direction of GP6 transcription. HOM = GP6InsA/InsA, HET = GP6WT/InsA, WT = GP6WT/WT. n = 9 for GP6InsA/InsA, 50 for GP6WT/InsA and 36 for GP6WT/WT.
The genetic etiology of the founder GP6 mutation found in Chilean patients. (A) Pedigrees showing a pattern of inheritance and different haplotypes surrounding the GP6InsA mutation site in identified Chilean families. (i-viii), Pedigrees including 6 probands (marked by a P and an arrow) and 2 heterozygous cases, wherein which the DNA of family members was also available for sequencing. For relatives who were not sequenced, we inferred the most likely haplotype based on sequencing results from samples provided; these individuals are marked with an asterisk (∗) and their inferred haplotype, assuming a normal pattern of inheritance and no recombination events, is in italics. (ix-x), Haplotypes of 2 individuals with homozygous mutation in which no DNA of family members was available. Color coding has been used to show inheritance patterns of alleles (block color alleles carry the GP6InsA haplotype). Squares indicate males, and circles indicate females. Blank shapes indicate GP6WT/WT status, half-filled shapes indicate GP6WT/InsA status and filled shapes indicate GP6InsA/InsA status. (B) Map of Chile indicating geographical sites of origin where patients have been diagnosed (arrowed). Santiago, Chile’s capital city, is indicated in red. Map adapted from www.touropia.com with permission. (C) The position of the microsatellite markers (18xTG and 21xAC), the GP6 exon 6 mutation (clinVar ID number 1072837), and the GP6 common SNP rs1654416 on chromosome 19 are shown. Tables show the relative frequencies of fragment size for each microsatellite marker and SNP status, in the 3 genetic groups investigated. The orange arrow shows the direction of GP6 transcription. HOM = GP6InsA/InsA, HET = GP6WT/InsA, WT = GP6WT/WT. n = 9 for GP6InsA/InsA, 50 for GP6WT/InsA and 36 for GP6WT/WT.
This study delineated the genetic etiology of the founder GP6 mutation by genotyping individuals with GP6InsA/InsA and their families, as well as heterozygotes and unrelated wild-type (WT) controls identified in the representative population of 1200 individuals.5 We performed sequencing of GP6 and genotyping of flanking polymorphic microsatellite markers (short tandem repeats)6,7 located close to GP6 to identify shared haplotypes. Blood and DNA samples were obtained after obtaining informed consent according to the ethical scientific committee at the Pontificia Universidad Catolica de Chile, as described.5 A total of 10 families and their relatives were investigated (families I-X, Figure 1A). Seven families had a single individual with the homozygous mutation and 1 family had 2 individuals with the homozygous mutation (Family III). Families V and VIII had only individuals with heterozygote mutation. The bleeding assessment tool score and symptoms for these individuals have been previously reported.5
Microsatellite markers flanking GP6 were genotyped and haplotypes were generated for the 10 families (Figure 1A). A total of 5 distinct individual haplotypes were identified in individuals with GP6InsA/InsA, defined by the minimal rates of recombination at each microsatellite marker studied (Table 1). The same alleles (orange and green) were found in families I, II, IV, VII, and X and families II, III, IV, and V, respectively. Families II and IV comprised unrelated individuals with GP6InsA/InsA who have inherited an identical haplotype, both of which originate from Santiago (Figure 1B, Table 1). Individual IX could have either a green or an orange haplotype, depending on which allele is inherited with the 291 marker, therefore also sharing one allele with the individuals in Santiago. In addition, the individuals with GP6InsA/InsA from families VII and X share identical haplotypes, also sharing one allele with the individuals in Santiago. Only 1 individual with the homozygous mutation inherited identical alleles from both the parents (family VI, 2 gray alleles). Interestingly, this is the only patient to share no common alleles with the patients in Santiago.
Summary of the 5 distinct haplotypes that were identified in the individuals with homozygous GP6InsA/InsAand the colors they are assigned inFigure 1A
| Haplotype color in Figure 1A . | STR 18xTG . | GP6 c.711dup mutation (InsA) . | GP6 SNP rs1654416 . | STR 21xAC . | Identified in families . |
|---|---|---|---|---|---|
| Orange | 240 | InsA | T | 291 | I, II, IV, VII, X |
| Purple | 240 | InsA | T | 293 | III, VII, VIII |
| Blue | 242 | InsA | T | 289 | I |
| Green | 242 | InsA | T | 291 | II, III, IV, V |
| Gray | 242 | InsA | T | 293 | VI |
| Haplotype color in Figure 1A . | STR 18xTG . | GP6 c.711dup mutation (InsA) . | GP6 SNP rs1654416 . | STR 21xAC . | Identified in families . |
|---|---|---|---|---|---|
| Orange | 240 | InsA | T | 291 | I, II, IV, VII, X |
| Purple | 240 | InsA | T | 293 | III, VII, VIII |
| Blue | 242 | InsA | T | 289 | I |
| Green | 242 | InsA | T | 291 | II, III, IV, V |
| Gray | 242 | InsA | T | 293 | VI |
Each allele is defined by the minimal rates of recombination at each microsatellite (STR) marker studied (18x TG and 21xAC). The families in which each of these haplotypes were found are listed.
STR, short tandem repeat.
Afterward, we analyzed the frequencies of microsatellite repeat alleles associated with GP6 in the families (Figure 1A) and compared them with the corresponding allele frequencies. We included additional unrelated individuals with the heterozygous mutation and control Chilean subjects identified from a previous study5 (Figure 1C). The total number of individuals who have been studied in this manuscript is therefore 9 with homozygous mutation (GP6InsA/InsA), 50 with heterozygous mutation (GP6WT/InsA), and 36 with WT (GP6WT/WT). The first marker, 18xTG, located approximately 41 kb upstream of GP6InsA, showed that there were only 2 alleles associated with GP6InsA/InsA (56% for one allele [240] and 44% for the other allele [242]). Similarly, all individuals with GP6InsA/InsA were T/T for single nucleotide polymorphisms (SNP), 2 base pairs away from the mutation insertion site (rs1654416) (allele frequency = 0.76125), which has previously been associated with sticky platelet syndrome.8
For the second microsatellite marker, 21xAC, located approximately 220 kb downstream of GP6, the individuals with GP6InsA/InsA showed slightly more variation, but 89% of haplotypes carried 1 of 2 alleles (56% for the one allele [291] and 33% for the other [293]). As expected, more variation was seen in the GP6WT/InsA and GP6WT/WT samples, with more allelic heterogeneity present for both. The allele 240 for marker 18xTG, which was predominant in GP6InsA/InsA samples, was also present in 39% of samples from the GP6WT/InsA and GP6WT/WT groups. Interestingly, allele 242, which was seen in 44% of GP6InsA/InsA samples, was seen in almost half of this frequency in GP6WT/InsA samples (26%), whereas it was only present in 7% of GP6WT/WT samples. The SNP haplotype seen in all GP6InsA/InsA samples was seen in 92% of GP6WT/InsA samples and 40% of GP6WT/WT samples.
Our study observed that individuals with GP6InsA/InsA had a bi-allelic distribution of the closest marker to GP6, 18xTG. If no recombination is assumed at this point since the GP6InsA mutation occurred, two founder events could have occurred for the 18xTG STR. However, based on the small shared region of identity between the different patients, we can conclude that the GP6InsA is likely to be an old mutation because young mutations would have a much larger shared region of identity. We cannot exclude that the mutation may be due to multiple de novo events, as the background haplotype was not that uncommon. Furthermore, although the microsatellite genotypes were not homozygous, they differ by only 2 bp, which may be the result of slippage, where the number of repeats increases or decreases between generations. The distal marker to GP6, 21 x AC, was homozygous for allele 291 for families II and IV, and heterozygous for allele 291 for families I, III, and V, suggesting a common yet inclusive genetic background. Overall, the GP6InsA mutation is likely to be old, due to the lack of shared and homozygous conserved haplotypes surrounding GP6, despite some families being closely geographically located within Chile.
Recent genetic analysis has revealed that the Chilean population is composed of 84% Amerindian mitochondrial DNA and 32% Amerindian Y chromosome,9 owing to limited immigration due to the Pacific Ocean in the west, the Andes mountains in the east, and the Atacama Desert in the north. With the arrival of the Spanish in 1541 (predominantly males), many of the 1 million indigenous inhabitants were killed (Instituto Nacional de Estadísticas, INE 200). Due to both its geography and history of colonization, Chile has favorable settings for the establishment of founder mutations following colonization by the Spanish, because population numbers remained low for several centuries.10 We have analyzed genetic data from 300 Spanish individuals with an unknown bleeding and/or platelet disorder and found no cases carrying the c.711dup mutation.11 Despite this, we cannot exclude that the GP6InsA variant was introduced to the Chilean population by the Spanish between the mid- sixteenth and nineteenth centuries, especially as all of the families have Spanish surnames; however, the apparent absence in Spain may suggest that the mutation arose in a Chilean male or female within this time period when the population numbers remained low.
With an estimated 4000 individuals with GP6InsA/InsA in Chile and so far only 11 identified, it is possible that GPVI loss is not always associated with bleeding, explaining why so few homozygotes have been identified in the clinic. Clinically, it is also possible that the GP6InsA mutation offers a modern-day protective benefit, reducing the risk of developing cardiovascular diseases.5 Because there is a clinical need to identify novel antithrombotic drugs that are not associated with bleeding, long-term monitoring of the cardiovascular health of these patients may shed more light on the targeting of GPVI as an antithrombotic. As it stands, young age and a low number of identified patients prevent more meaningful insight from being deduced from their GP6 status, other than their mild bleeding phenotypes.
Acknowledgments: The authors would like to acknowledge help from the DNA sequencing services by the School of Biosciences, University of Birmingham, United Kingdom. The work in the author’s laboratories is supported by the British Heart Foundation (FS/18/11/33443) (NVM), S.P.W. holds a British Heart Foundation Chair (CH03/003). A.D. is supported by a B.H.F Accelerator Award (AA/18/2/34218). D.M. is supported by Grant 1181681, ANID/FONDECYT Chile. J.R. research is supported by grants from Instituto de Salud Carlos III (ISCIII) & Feder (PI20/00926 and PMP21/00052) and the Spanish Society of Thrombosis and Haemostasis (SETH, Ayuda GEAPC).
Contribution: A.D., S.P.W., and N.V.M. designed the research; D.M. diagnosed the patients and provided samples and patient data; J.R. performed GP6 molecular analysis in Spanish patients; A.D. performed experiments and analyzed data; and all authors contributed to the initial drafts and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Neil Morgan, Institute of Cardiovascular Sciences, College of Medical and Dental Sciences, Edgbaston, University of Birmingham, Birmingham, United Kingdom; e-mail: n.v.morgan@bham.ac.uk.
References
Author notes
Contact the corresponding author for data sharing, Neil Morgan (n.v.morgan@bham.ac.uk).