Key Points
We detail at clinical, cytological, cytogenetic, and molecular levels 113 cases of MDS and MDS/MPN with del(11q), a rare recurrent event.
CADM1, a tumor suppressor gene identified initially in solid tumors, ATM, CBL, and KMT2A are deleted and/or mutated in del(11q).
Abstract
Myelodysplastic syndromes (MDS) represent a heterogeneous group of clonal hematopoietic stem cell disorders characterized by ineffective hematopoiesis leading to peripheral cytopenias and in a substantial proportion of cases to acute myeloid leukemia. The deletion of the long arm of chromosome 11, del(11q), is a rare but recurrent clonal event in MDS. Here, we detail the largest series of 113 cases of MDS and myelodysplastic syndromes/myeloproliferative neoplasms (MDS/MPN) harboring a del(11q) analyzed at clinical, cytological, cytogenetic, and molecular levels. Female predominance, a survival prognosis similar to other MDS, a low monocyte count, and dysmegakaryopoiesis were the specific clinical and cytological features of del(11q) MDS. In most cases, del(11q) was isolated, primary and interstitial encompassing the 11q22-23 region containing ATM, KMT2A, and CBL genes. The common deleted region at 11q23.2 is centered on an intergenic region between CADM1 (also known as Tumor Suppressor in Lung Cancer 1) and NXPE2. CADM1 was expressed in all myeloid cells analyzed in contrast to NXPE2. At the functional level, the deletion of Cadm1 in murine Lineage-Sca1+Kit+ cells modifies the lymphoid-to-myeloid ratio in bone marrow, although not altering their multilineage hematopoietic reconstitution potential after syngenic transplantation. Together with the frequent simultaneous deletions of KMT2A, ATM, and CBL and mutations of ASXL1, SF3B1, and CBL, we show that CADM1 may be important in the physiopathology of the del(11q) MDS, extending its role as tumor-suppressor gene from solid tumors to hematopoietic malignancies.
Introduction
Myelodysplastic syndromes (MDS) are defined according to the World Health Organization (WHO) classification by cytopenias, bone marrow (BM) dysplasia, and recurrent cytogenetic abnormalities.1,2 They represent a heterogeneous group of clonal hematopoietic stem cell disorders characterized by ineffective hematopoiesis leading to peripheral cytopenias and, in a substantial proportion of cases, to acute myeloid leukemia (AML).3,4 According to the revised International Prognostic Scoring System (IPSS-R), their prognosis is mainly based on the degree of cytopenias, the percentage of blasts in the BM, and the presence of karyotypic abnormalities.5
Recurrent driver chromosomal abnormalities and mutations are found in MDS.6 More specifically, copy-number abnormalities are a hallmark of MDS with complete or partial deletions of chromosomes 5 (-5/5q), 7 (-7/7q), 11 (11q), 12 (12p), 13 (13q), 17 (17p), 18 (18q), and 20 (20q). The most frequent among these chromosomal deletions are −7/7q (del(7q)) and −5/5q (del(5q)) deletions.6 MDS with isolated del(5q), recognized as a specific entity by the WHO classification, is associated with unique features, such as anemia; frequent, increased platelet counts; low blast percentage in bone marrow; the presence of characteristic small, nonlobated megakaryocytes; and a low rate of leukemic transformation.6 Del(5q) leads to haploinsufficiency of numerous genes, including RPS14, miR-145, and CSNK1A1 within the 5q32-33 critical deleted region underlying the necessary combination of various events in the development of MDS consequently to this deletion.6
Deletion of the long arm of chromosome 11 (del(11q)) is a rare clonal abnormality found in 0.6% to 3% of MDS and myelodysplastic/myeloproliferative neoplasms (MDS/MPN).2,7,8 According to the WHO 2008 classification, it represents a hallmark of MDS in a context of unexplained cytopenia.9 Since its first description in 1992,10 few studies have investigated this deletion in MDS.7,10-14 The first substantial study analyzed 19 cases,12 leading to assign a very good prognosis to del(11q) as a sole abnormality in the IPSS-R classification.5,13 The largest cohort published so far included 56 cases demonstrating an association of del(11q) with SF3B1 mutations without delineating a common deleted region (CDR).8
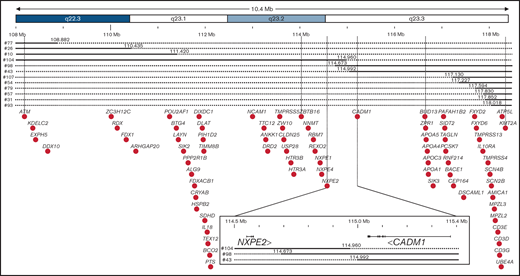
Here, we explore the clinical, cytological, cytogenetic, and molecular features of 113 MDS and MDS/MPN cases harboring a del(11q), representing the largest series of patients so far. Cytogenetic and molecular analyses showed that ATM and/or KMT2A were deleted in all tested cases, and that CBL was deleted or mutated in most of the cases. Our approach allowed to define a CDR located within the 11q23.2 band centered on an intergenic region between CADM1 and NXPE2, the former gene being the only 1 consistently expressed in myeloid cells. Finally, CADM1 deletion by genomic editing leads to hematopoietic features reminiscent of MDS. Therefore, del(11q) MDS as del(5q) MDS could be the result of multiple combined haploinsufficiencies of genes located in the 11q22-q23 region, such as ATM, CADM1, KMT2A, or CBL genes.
Methods
Patients and inclusion criteria
In this retrospective study, 113 patients (103 MDS and 10 MDS/MPN) were included over an 11-year period by 24 French and Belgian laboratories affiliated to the Groupe Francophone de Cytogénétique Hématologique. Inclusion criteria were the presence of a clonal del(11q) at karyotype from MDS or MDS/MPN patients defined according to the WHO 2016 classification and the availability of remaining cytogenetic pellet for fluorescence in situ hybridization (FISH) analysis (supplemental Table 1). The research was approved by the Toulouse University ethical committee and all human participants gave written informed consent. Karyotypes were established on a median of 20 metaphases per patient (range 5-33). All analyses were performed on the cytogenetic pellet used for diagnosis. Karyotypes were reviewed at 2 successive workshops and written according to International System for Human Cytogenomic Nomenclature 2016.15 Scoring of clonal chromosomal abnormalities was established according to IPSS-R.13,16 Morphological classification was done locally and validated centrally using the WHO 2016 classification.2 Cytopenias were defined according to WHO 2016.2 Each case was assigned a risk score according to the IPSS-R for MDS5 and therapy-related MDS (t-MDS).17 A cohort of 88 MDS without del(11q) enrolled by Groupe Français des Myélodysplasies clinical centers in the PHRC MDS-04 was included as control (Other MDS group, Table 1).
Summary of clinical and cytological data of del(11q) MDS and MDS/MPN compared with other MDS
| Characteristics . | del(11q) MDS and MDS/MPN . | del(11q) MDS . | Other MDS . | P value MDS . |
|---|---|---|---|---|
| Patients | 113 | 103 | 88 | |
| Age, years, median (range) | 74 (31-94) | 74 (31-94) | 71 (44-94) | 0.24 |
| Sex ratio | 0.74 | 0.74 | 1.26 | 0.020 |
| Blood count | ||||
| Hemoglobin (Hb), g/dL, median (range) | 9.9 (4.5-14.8) | 9.9 (5.5-14.8) | 9.4 (6.4-14.9) | 0.14 |
| Anemia (Hb <10 g/dL) | 52% | 52% | 58% | 0.43 |
| Mean corpuscular volume, fL, median (range) | 101 (77-139) | 101 (77-139) | 99.5 (76-129) | 0.035 |
| Platelets (plt), G/L, median (range) | 173 (17-1234) | 173 (17-1234) | 183 (5-494) | 0.71 |
| Thrombocytopenia (plt <100 G/L) | 32% | 32% | 27% | 0.44 |
| White blood count (WBC), G/L, median (range) | 4.0 (1.0-94) | 4.0 (1.0-18.4) | 4.2 (1.0-18.4) | 0.85 |
| Neutrophil Count (ANC), G/L median (range) | 2.3 (0.2-52.6) | 2.0 (0.2-13.4) | 2.2 (0.3-15.8) | 0.61 |
| Neutropenia (ANC <1.8 G/L) | 36% | 36% | 37% | 0.96 |
| Monocyte count, G/L, median (range) | 0.3 (0-15) | 0.2 (0-0.9) | 0.4 (0-0.9) | 0.0004 |
| Lymphocyte count, G/L, median (range) | 1.2 (0-5.0) | 1.1 (0.1-3.5) | 1.3 (0.3-3.7) | 0.57 |
| Bone marrow | ||||
| % BM blasts, median (range) | 4 (0-14) | 4 (0-13) | 3 (0-19) | 0.086 |
| Dyserythropoiesis | 73% | 75% | 71% | 0.58 |
| Dysgranulopoiesis | 51% | 51% | 69% | 0.025 |
| Dysmegakaryopoiesis | 78% | 80% | 57% | 0.001 |
| Characteristics . | del(11q) MDS and MDS/MPN . | del(11q) MDS . | Other MDS . | P value MDS . |
|---|---|---|---|---|
| Patients | 113 | 103 | 88 | |
| Age, years, median (range) | 74 (31-94) | 74 (31-94) | 71 (44-94) | 0.24 |
| Sex ratio | 0.74 | 0.74 | 1.26 | 0.020 |
| Blood count | ||||
| Hemoglobin (Hb), g/dL, median (range) | 9.9 (4.5-14.8) | 9.9 (5.5-14.8) | 9.4 (6.4-14.9) | 0.14 |
| Anemia (Hb <10 g/dL) | 52% | 52% | 58% | 0.43 |
| Mean corpuscular volume, fL, median (range) | 101 (77-139) | 101 (77-139) | 99.5 (76-129) | 0.035 |
| Platelets (plt), G/L, median (range) | 173 (17-1234) | 173 (17-1234) | 183 (5-494) | 0.71 |
| Thrombocytopenia (plt <100 G/L) | 32% | 32% | 27% | 0.44 |
| White blood count (WBC), G/L, median (range) | 4.0 (1.0-94) | 4.0 (1.0-18.4) | 4.2 (1.0-18.4) | 0.85 |
| Neutrophil Count (ANC), G/L median (range) | 2.3 (0.2-52.6) | 2.0 (0.2-13.4) | 2.2 (0.3-15.8) | 0.61 |
| Neutropenia (ANC <1.8 G/L) | 36% | 36% | 37% | 0.96 |
| Monocyte count, G/L, median (range) | 0.3 (0-15) | 0.2 (0-0.9) | 0.4 (0-0.9) | 0.0004 |
| Lymphocyte count, G/L, median (range) | 1.2 (0-5.0) | 1.1 (0.1-3.5) | 1.3 (0.3-3.7) | 0.57 |
| Bone marrow | ||||
| % BM blasts, median (range) | 4 (0-14) | 4 (0-13) | 3 (0-19) | 0.086 |
| Dyserythropoiesis | 73% | 75% | 71% | 0.58 |
| Dysgranulopoiesis | 51% | 51% | 69% | 0.025 |
| Dysmegakaryopoiesis | 78% | 80% | 57% | 0.001 |
FISH analyses
FISH analyses were performed for all patients, either locally or centrally, using the following chromosome 11 probes from centromere to 11q telomere (supplemental Figure 1A): D11Z1 (centromeric probe, Vysis), CCND1 (Vysis), ATM (Vysis), ZBTB16 (Kreatech), CADM1 (RP11-713B9), ZPR1 (RP11-87O6 and RP11-4N9), KMT2A (Vysis), CBL (RP11-1105H9 and RP11-122H4), MIR125B1 (RP11-382J20), FLI1 (RP11-432L13 and RP11-744N12), and D11S1037 (subtelomeric probe, Vysis).
Mutational screening
A specific CBL mutational screening was performed for 41 MDS with del(11q) targeting the CBL mutations hotspots (exons 7 to 9) by high resolution melting PCR (detailed in supplemental Methods). Positive signals were confirmed by Sanger sequencing.
Specific resequencing detailed in supplemental Methods was performed for 36 del(11q) samples by next generation sequencing of 28 genes (ASXL1, CBL, CSF3R, DNMT3A, ETV6, EZH2, GATA1, GATA2, IDH1, IDH2, JAK2, KDM6A, KIT, KRAS, NPM1, NRAS, PHF6, PTPN11, RAD21, RUNX1, SF3B1, SMC1A, SMC3, STAG2, TET2, TP53, U2AF1, and WT1). Additional resequencing for CSMD1, ETNK1, PRPF8, SRSF2, and ZRSR2 was performed for 18 del(11q) samples. Specific resequencing of the complete coding regions of CADM1 and NXPE2 was performed for 89 samples, 19 MDS with del(11q), 3 MDS, 2 MDS/MPN, and 60 AML without del(11q).
Genomic microarray analysis
Twelve samples with a deletion boundary located between ATM and KMT2A were analyzed for copy number changes at a very high resolution using Affymetrix Cytoscan HD arrays (Affymetrix) and detailed in supplemental Methods.
NXPE2 and CADM1 expression
cDNA was synthesized using 1 µg of ribonucleic acid (RNA) and the SuperScript Vilo kit (Invitrogen) according to the manufacturer’s instructions in a final volume of 25 µL. Quantitative PCR was performed as detailed in supplemental Methods in 21 MDS, 10 MDS/MPN and 55 AML samples (supplemental Table 2).
CADM1 genomic editing
Single-guide RNAs (sgRNAs) targeting the Cadm1 exon 2 were subcloned in pLKO5-sgRNA-EFS-GFP.18 Editing was performed using Rosa26-Cas9 mice. Lineage negative (Lin-) BM cells were purified using a magnetic microbeads Lineage Cell Depletion Kit (Miltenyi Biotech) from 6-8-week-old Rosa26-Cas9-expressing (CD45.2+) mice and analyzed using antibodies from Pharmingen (BD Biosciences) detailed in supplemental Methods.
Purified Lin-Sca1+kit+ (LSK) cells were plated in suspension in StemSpan medium (StemCell Technologies) with 50 ng/mL murine thrombopoietin (TPO) and 50 ng/mL murine stem cell factor (SCF). After 24 hours of prestimulation, LSK cells were transduced by lentiviruses at a multiplicity of infection of 10 (MOI10) and 2 μg/mL polybrene (Sigma Aldrich). Efficiency of the genomic editing was analyzed using next generation sequencing of the targeted Cadm1 exon 2 and the CRISPResso software.19
Clonogenic and transplantation assays
Transduced LSK cells were plated in triplicate in multipotential methylcellulose (MethoCult 3434, StemCell Technologies). Colonies were numbered and characterized 8 days later. Transduced LSK (CD45.2+) were transplanted into 6-8 week-old recipient mice (CD45.1+) pretreated with 30 mg/kg of Busulfan (Busilvex, Pierre Fabre) 24h before transplantation. Chimerism of engrafted and transduced cells was analyzed by flow cytometry (FACS) in BM and thymus 2 months after transplantation. Donor-derived cells (CD45.2+GFP+) from primary BM were transplanted into secondary recipients pretreated with Busulfan (30 mg/kg). Kinetics of engraftment of transduced cells (% CD45.2+GFP+) was assessed by BM aspirations 2, 8, and 21 months after transplantations.
BM and thymus of recipient mice (CD45.1+) 8 weeks after transplantation were analyzed using antibodies obtained from Pharmingen (BD Biosciences, detailed in supplemental Methods). Cytological analysis was performed on at least 200 purified donor-derived cells (% CD45.2+GFP+) from each sgCTL and sgCadm1 #2 transplanted mice.
Statistics
Statistical analyses were performed using Stata 15.1 as detailed in supplemental Methods.
Results
Patients with del(11q) are preferentially female, have a lower monocyte blood count and bone marrow dysmegakaryopoiesis
A series of 113 MDS or MDS/MPN with del(11q) was collected through the Groupe Francophone de Cytogénétique network, MDS being more prevalent than MDS/MPN (103 vs 10 cases, respectively). Clinical and biological data were compared with a cohort of 88 MDS patients without del(11q) (“other MDS”) established by the Groupe Français des Myélodysplasies clinical network (Table 1). Patients with del(11q) patients had a median age of 74 years (range 31-94), similar to other MDS (P = .24), and were preferentially female with a sex ratio of 0.74, in contrast to 1.26 in other MDS (P = .020, Table 1).
Anemia was the more prevalent cytopenia in del(11q) patients with a median hemoglobin concentration of 9.9 g/dL, similar to other MDS (P = .14, Table 1; supplemental Figure 2). The mean corpuscular volume was slightly but significantly higher in del(11q) patients (median of 101 fL vs 99.5 fL, P = .035, Table 1; supplemental Figure 2). A significant lower monocyte count was found in del(11q) patients with a median of 0.2×109/L compared with 0.4×109/L in other MDS (P = .0004; Table 1; supplemental Figure 2). The white blood cell, neutrophil, and lymphocyte counts and the rate of neutropenia, the platelet count, and the rate of thrombocytopenia were similar to other MDS (Table 1; supplemental Figure 2).
The BM smears were reviewed centrally under the coordination of the Groupe Francophone d’Hématologie Cellulaire. The medullar blast count and dyserythropoiesis were similar in del(11q) patients compared with other MDS. Significantly, dysgranulopoiesis was less frequent and dysmegakaryopoiesis increased in del(11q) MDS patients (Table 1) with frequent presence of multinucleated megakaryocytes (Figures 1A-D). It is noteworthy that small and nonlobated megakaryocytes were present when del(11q) was associated to del(5q) (Figure 1D).
Cytological and cytogenetic presentations of del(11q) MDS. (A-D) May-Grünwald Giemsa staining of del(11q) bone marrow. Megakaryocytes were often large and plurinucleated (A, B, and C, original magnification x×50) giving an aspect of ball bags (C) and, when associated to 5q deletion, presence of small and nonlobated megakaryocytes ([D], original magnification ×100). (E) Cytogenetic presentations of del(11q). The 2 main types of del(11q) on R-banded partial karyotypes from 2 representative cases, from left to right: large deletion, del(11)(q14q24), in case #19 and small deletion, del(11)(q22q24), in case #86.
Cytological and cytogenetic presentations of del(11q) MDS. (A-D) May-Grünwald Giemsa staining of del(11q) bone marrow. Megakaryocytes were often large and plurinucleated (A, B, and C, original magnification x×50) giving an aspect of ball bags (C) and, when associated to 5q deletion, presence of small and nonlobated megakaryocytes ([D], original magnification ×100). (E) Cytogenetic presentations of del(11q). The 2 main types of del(11q) on R-banded partial karyotypes from 2 representative cases, from left to right: large deletion, del(11)(q14q24), in case #19 and small deletion, del(11)(q22q24), in case #86.
Chromosome 11q deletion is mainly interstitial and primary event
The del(11q) appeared mainly as an interstitial deletion extending from 11q13 to 11q24 in 102/113 cases (Figure 1E), either as a large deletion leading to a short del(11q) chromosome (11q shorter than 11p, 49 cases) or as a smaller deletion (11q appearing equal in size or longer than 11p, 53 cases). In 11 cases, the deletion of chromosome 11q was more complex, presenting cytogenetically as an unbalanced translocation, a di- or tri-centric chromosome 11, or a ring chromosome 11.
In approximately half of cases (48%, 54/113), del(11q) was present as an isolated clonal abnormality ([CA]; supplemental Table 1). Additional chromosomal abnormalities (ACA) were present in the remaining 59 cases, mainly as a single ACA (38%) thus leading to a double clonal abnormality karyotype as defined by IPSS-R or less frequently as multiple ACA, thus leading to a complex karyotype (14%). Only 3 cases have more than 3 CA (very complex karyotype as defined by IPSS-R5 ). The del(5q) abnormality was the most prevalent ACA (20%) followed by trisomy 8 (10%) and loss of chromosome Y (14% of male patients, supplemental Table 1). No monosomy 7, del(7q), or 3q26 abnormalities were identified in the 113 del(11q) cases. The del(11q) was mainly primary (59%, 67/113 cases); only 8 cases had del(11q) as a secondary cytogenetic abnormality, including 2 cases with a del(5q) as a primary abnormality. Two cases presented with 2 independent clones (supplemental Table 1).
Patients with del(11q) have a prognosis similar to other MDS
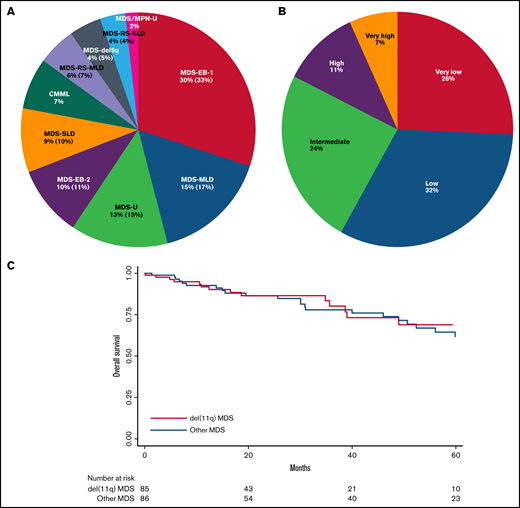
Del(11q) cases were classified according to WHO 2016,2 the most frequent being MDS with excess blasts type 1 (MDS-EB-1, 30%) followed by MDS with multilineage dysplasia (MDS-MLD, 15%), unclassifiable MDS (MDS-U, 13%), and MDS with excess blasts type 2 (MDS-EB-2, 10%; Figure 2A). The IPSS-R risk scoring5,13 combining cytological and cytogenetic data were defined in 74 primary MDS cases with del(11q) as very low in 26%, low in 32%, intermediate in 24%, high in 11%, and very high in 7% (vs 16%, 52%, 16%, 9%, and 6% in other MDS, respectively), with a significant lower frequency of low IPSS-R risk group in del(11q) (32% vs 52%, P = .015; supplemental Table 1; Figure 2B). Recently, the IPSS-R scoring had been shown to apply also to t-MDS cases,17 revealing also in our series a majority of low or very low risk MDS in del(11q) t-MDS (7/18 and 6/18 cases, respectively; supplemental Table 1).
WHO 2016, IPSS-R classifications and overall survival of del(11q) MDS. (A) WHO 2016 classification of del(11q) MDS and MDS/MPN. (B) IPSS-R applied to the primary del(11q) MDS by risk category: very low = risk score ≤1.5; low = risk score >1.5-3; intermediate = risk score >3-4.5; high = risk score >4.5-6; very high = risk score >6. (C) Overall survival of del(11q) MDS compared with other MDS.
WHO 2016, IPSS-R classifications and overall survival of del(11q) MDS. (A) WHO 2016 classification of del(11q) MDS and MDS/MPN. (B) IPSS-R applied to the primary del(11q) MDS by risk category: very low = risk score ≤1.5; low = risk score >1.5-3; intermediate = risk score >3-4.5; high = risk score >4.5-6; very high = risk score >6. (C) Overall survival of del(11q) MDS compared with other MDS.
The clinical follow-up was 22 months for del(11q) patients and 39 months for other MDS, on average. Transformation to AML was seen in 6 del(11q) patients out of 87 (7%) and 11 out of 72 patients for other MDS (15%, P = .089), keeping in mind than the follow-up was longer in the other MDS group. Overall survival was similar between del(11q) MDS and other MDS patients (P = .84, Figure 2C).
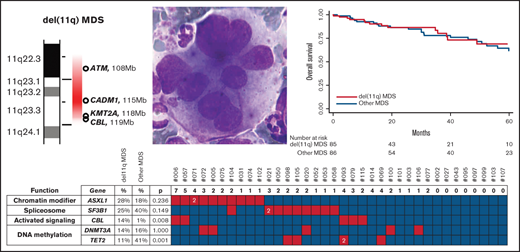
CBL mutation rate is significantly increased in del(11q) MDS
The CBL gene located in the 11q23.3 chromosomal band is recurrently mutated in MDS.6,14,20 CBL is significantly more frequently mutated in del(11q) compared with other MDS (14% vs 1%, respectively, P = .008, Figure 3; supplemental Figure 3; supplemental Table 3). In addition to mutations, the CBL gene was deleted in 84% of del(11q) (49/58; supplemental Figure 3). CBL deletions without CBL mutations were detected in 74% (43 cases), CBL mutations without CBL deletions in 5% (3 cases), and both deletions and mutations of CBL occurred in only 6 cases (10%, supplemental Figure 3). Overall, CBL abnormalities were very frequent but not constant in del(11q) MDS.
Mutations in del(11q) MDS. Mutation screening of 36 del(11q) MDS by next generation sequencing. Genes were classified according to their function (2 first columns). Comparison with other MDS is indicated in the 3 central columns (del[11q] MDS mutation frequency, other MDS mutation frequency, and P value comparing the frequencies between these 2 groups). Mutations are indicated by red squares containing the number of mutations per sample if higher than 1, the absence of mutations by green squares, and the absence of analysis by white squares. Patients are labeled at the top.
Mutations in del(11q) MDS. Mutation screening of 36 del(11q) MDS by next generation sequencing. Genes were classified according to their function (2 first columns). Comparison with other MDS is indicated in the 3 central columns (del[11q] MDS mutation frequency, other MDS mutation frequency, and P value comparing the frequencies between these 2 groups). Mutations are indicated by red squares containing the number of mutations per sample if higher than 1, the absence of mutations by green squares, and the absence of analysis by white squares. Patients are labeled at the top.
In addition to CBL mutations, a targeted resequencing of 36 del(11q) cases revealed frequent mutations in ASXL1 (28%), SF3B1 (25%), DNMT3A (14%), JAK2 (14%), SRSF2 (11%), TET2 (11%), and ZRSR2 (11%). These mutations were present at a similar rate compared with other MDS except for TET2, which was much less frequently mutated in del(11q) compared with other MDS (11% vs 41% respectively, P = .001; Figure 3; supplemental Table 3). SF3B1 mutations were significantly associated within del(11q) MDS with the presence of ring sideroblasts (4/4, P = .002), similar to other MDS (27/30, P < .001, Figure 3).
CADM1 is a candidate gene for the del(11q) CDR
The del(11q) CDR was first investigated by FISH analysis using proximal (CCND1, 11q13.3) and distal probes (D11S1037, subtelomeric, 11q25; supplemental Figure 1B). Probes were retained on both copies of chromosome 11 in 37/37 cases tested for CCND1 and 51/54 cases for D11S1037. The 3 cases with an 11q25 telomeric terminal deletion were the cases with a complex mechanism of del(11q) described above. Thus, FISH analysis confirms that all tested cases with a simple del(11q) are interstitial deletions (supplemental Figure 1B; supplemental Table 1). Inside this large 11q region, ATM and/or KMT2A were deleted in all cases, being both deleted in 86% of the cases (95/110). Fifteen cases lost only 1 of these 2 genes, either ATM but not KMT2A in 11 cases (ATM-/KMT2A+) or KMT2A but not ATM in 4 cases (ATM+/KMT2A-). Consequently, the CDR by FISH analysis is located between these 2 genes, in a 10 Mb region (supplemental Figure 1B; supplemental Table 1).
To delineate the CDR at a much higher resolution, 12 cases that deleted only ATM (8 ATM-/KMT2A+) or KMT2A (4 ATM+/KMT2A-) were analyzed by comparative genomic hybridization analysis allowing to refine the CDR to a 0.4 Mb intergenic region between CADM1 and NXPE2 (Figure 4; supplemental Figure 4). CADM1 (cell adhesion molecule 1), also known as TSLC1 (tumor suppressor in lung cancer 1) or IGSF4 (immunoglobulin superfamily member 4), encodes a protein of 471 amino acids involved in cell-cell adhesion21 and was initially identified as a tumor-suppressor gene in human non–small cell lung22 and nasopharyngeal carcinoma.23 NXPE2 (neurexophilin and PC-esterase domain family member 2) encodes a protein of 559 amino acids without defined function. The reading frames of these 2 genes were deleted in all patients but 2 cases for CADM1 (#43 and #98) and 1 case for NXPE2 (#104; Figure 4). This intergenic region deleted between NXPE2 and CADM1 in those 3 cases contain 4 regulatory elements (e35780, e35781, e35783, and e35784) binding to the Cadm1 promoter according to Encode 3,24 whose deletion could impair its expression (supplemental Figure 5).
CDR of del(11q) MDS defined by high resolution microarray analysis. Detailed map of the chromosome 11 summarizing microarray data. Chromosomal bands are indicated at the top with the genomic location indicated below according to hg19 assembly human genome. Continuous line: presence of the 2 alleles; dashed line: deletion of 1 of the 2 alleles. Genes located in the region are indicated by black dots. The common deleted region was indicated by vertical lines with a magnification of it at the bottom, indicating location of exons of NXPE2 and CADM1, their orientation, and the 3 samples defining the del(11q) MDS CDR.
CDR of del(11q) MDS defined by high resolution microarray analysis. Detailed map of the chromosome 11 summarizing microarray data. Chromosomal bands are indicated at the top with the genomic location indicated below according to hg19 assembly human genome. Continuous line: presence of the 2 alleles; dashed line: deletion of 1 of the 2 alleles. Genes located in the region are indicated by black dots. The common deleted region was indicated by vertical lines with a magnification of it at the bottom, indicating location of exons of NXPE2 and CADM1, their orientation, and the 3 samples defining the del(11q) MDS CDR.
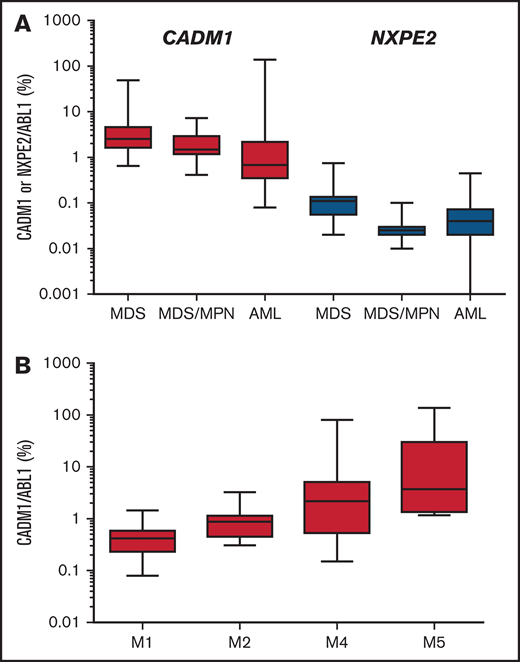
None of these 2 genes was mutated in a cohort of 89 myeloid malignancies, including 20 MDS with del(11q) (Figure 3). Expression of these 2 genes reveals that CADM1 was expressed 71-fold higher (mean expression relative to ABL1: 5.9%, range 0.08% to 138%) than NXPE2 (mean 0.08%, range 0% to 0.75%). NXPE2 expression was not detected in 6 cases (Figure 5A; supplemental Table 2). Expression of CADM1 was higher in acute monoblastic/monocytic (M5a/M5b) and myelomonocytic (M4) leukemias compared with acute myeloblastic leukemias (M1/M2), suggesting a higher expression of CADM1 in monocytic progenitors (Figure 5B; supplemental Table 2). Although we were unable to evaluate the expression of these genes in our del(11q) MDS cases, the high expression of CADM1 in myeloid malignancies and its almost constant deletion in del(11q) strongly suggest that it represents a candidate gene of the del(11q) CDR in MDS.
Expression of CADM1 and NXPE2 in myeloid cells. (A) Expression of CADM1 and NXPE2 in 21 MDS, 10 chronic myelo-monocytic leukemias, and 55 AML samples presented as a box with whiskers. The box extends from the 25th to the 75th percentiles of expression. The line in the middle of the box was the median. The whiskers indicate the 10th and the 90th percentiles. (B) Expression of CADM1 in AML according to their FAB classification.
Expression of CADM1 and NXPE2 in myeloid cells. (A) Expression of CADM1 and NXPE2 in 21 MDS, 10 chronic myelo-monocytic leukemias, and 55 AML samples presented as a box with whiskers. The box extends from the 25th to the 75th percentiles of expression. The line in the middle of the box was the median. The whiskers indicate the 10th and the 90th percentiles. (B) Expression of CADM1 in AML according to their FAB classification.
Genomic editing of Cadm1 perturbs myeloid development
To investigate the effect of Cadm1 loss on hematopoietic development in vivo, we edited Cadm1 gene by clustered regularly interspaced short palindromic repeats-Cas9 (CRISPR-Cas9) strategy. First, we designed 3 sgRNAs targeting the genomic region encoding the exon 2 of Cadm1. To select the sgRNA exhibiting the most editing efficiency of Cadm1 locus, we transduced Lin- cells from the BM of Cas9-expressing mice with lentiviral vectors containing the sgCadm1 #1, #2, #3, or a nontargeted control sgRNA-encoding sequence (sgCTL) (supplemental Figures 6A-B). The sgCadm1 #2 exhibited the highest rate of insertions/deletions (InDels) (supplemental Figures 6C-D). Therefore, sgCadm1 #2 was selected to perform the following in vitro and in vivo functional experiments. Purified Cas9-expressing LSK cells, a population enriched in hematopoietic stem/progenitor cells (HSPCs),25 were transduced with sgCTL or sgCadm1 #2 lentiviral vector, both expressing concomitantly the GFP (Figure 6A). The sgCadm1 #2 allowed for an efficient transduction (87.7%), as revealed by the percentage of GFP+ cells 8 days after the in vitro expansion in methylcellulose (supplemental Figure 7A) and an efficient genomic editing, as revealed by the presence of 89% of InDels in the Cadm1 locus (supplemental Figures 7B-C). The numbers of GFP+ colonies in methylcellulose (total, CFU-GEMM, CFU-GM, and BFU-E) were similar between sgCTL and sgCadm1 #2 (Figure 6B), indicating that Cadm1 deletion did not affect the clonogenic activity of hematopoietic progenitors. In parallel, CD45.2+Cas9-expressing LSK cells transduced with sgCTL or sgCadm1 #2 were transplanted into CD45.1+ syngenic recipient mice (Figure 6A). The analysis of engrafted donor-derived cells (CD45.2+GFP+) revealed that the proportions of the hematopoietic stem/progenitor compartments (LSK, LK, MEP/CMP, and GMP cells) were not altered by Cadm1 deletion (supplemental Figures 8A-B), corroborating our previous observations in clonogenic assay (Figure 6B). In addition, thymic reconstitution of donor-derived cells was not modified by Cadm1 deletion (supplemental Figures 8C-D), indicating that Cadm1 deletion did not alter the multipotent activity of HSPCs. However, we observed that Cadm1 deletion modified the relative proportions of myeloid and B-cells in the BM of recipient mice (Figures 6C-D). Thus, we purified donor-derived cells (CD45.2+GFP+) from sgCTL and sgCadm1 #2 transplanted mice. Cadm1 was edited in almost all transduced CD45.2+ cells in transplanted mice (Figure 6E; supplemental Figure 9), with the presence of only a few distinct InDels in engrafted cells underlying the oligoclonal dynamic of hematopoietic reconstitution from HSPCs.26 In accordance with the immunophenotype of donor-derived cells (Figures 6C-D), Cadm1 deletion perturbed the balance between myeloid and lymphoid cells (Figure 6F). Finally, donor-derived cells (CD45.2+GFP+) from primary BM were transplanted into secondary recipients to assess the impact of Cadm1 deletion on the long-term hematopoietic reconstitution. While secondary recipients did not develop hematopoietic neoplasms even after a period of 21 months (supplemental Figure 10A), the balance between myeloid and lymphoid cells was still perturbed in engrafted sgCadm1 donor-derived cells (CD45.2+GFP+) 2 and 8 months after transplantation (supplemental Figures 10B-C), suggesting an intrinsic differentiation bias in those cells. Together, this functional in vivo approach suggests that Cadm1 loss-of-function perturbs the terminal myeloid cell production.
Functional inactivation of Cadm1 in mice. (A) Experimental procedure to study the effect of Cadm1 edition on murine hematopoietic development (left panel). LSK cells from Cas9-expressing mice were purified by cell sorting (right panel) and transduced with either sgCadm1#2 or control sgRNA (sgCTL) lentiviral vectors. Transduced Cas9-expressing LSK cells (CD45.2+) were either plated in clonogenic assay or transplanted into syngenic recipient mice (CD45.1+, n = 3 for each condition). (B) Number of total CFU-GEMM, CFU-GM, and BFU-E colonies in clonogenic assay. (C) BM chimerism, analyzed by FACS 2 months after transplantation, illustrated by the percentage of transduced donor cells (% CD45.2+GFP+) found in the recipient BM (upper panel). Immunophenotype of engrafted donor-derived cells (CD45.2+GFP+) was analyzed using the Gr1 (Myeloid cells, My) and CD19 (B-cells) markers. FACS profiles and gating strategy were shown for 1 representative mouse of each condition (lower panel). (D) Proportion of myeloid and B cells (%) within total donor-derived CD45.2+GFP+ cells from the BM of recipient mice. (E-F) Donor-derived cells (CD45.2+GFP+) from each sgCTL and sgCadm1 transplanted mouse purified by cell sorting. Genome editing efficiency of targeted Cadm1 region performed on purified donor-derived cells (CD45.2+GFP+) for each condition (E). Cytological analysis was performed on purified CD45.2+GFP+ cells from each condition, and the proportion of myeloid and lymphoid cells was calculated (F).
Functional inactivation of Cadm1 in mice. (A) Experimental procedure to study the effect of Cadm1 edition on murine hematopoietic development (left panel). LSK cells from Cas9-expressing mice were purified by cell sorting (right panel) and transduced with either sgCadm1#2 or control sgRNA (sgCTL) lentiviral vectors. Transduced Cas9-expressing LSK cells (CD45.2+) were either plated in clonogenic assay or transplanted into syngenic recipient mice (CD45.1+, n = 3 for each condition). (B) Number of total CFU-GEMM, CFU-GM, and BFU-E colonies in clonogenic assay. (C) BM chimerism, analyzed by FACS 2 months after transplantation, illustrated by the percentage of transduced donor cells (% CD45.2+GFP+) found in the recipient BM (upper panel). Immunophenotype of engrafted donor-derived cells (CD45.2+GFP+) was analyzed using the Gr1 (Myeloid cells, My) and CD19 (B-cells) markers. FACS profiles and gating strategy were shown for 1 representative mouse of each condition (lower panel). (D) Proportion of myeloid and B cells (%) within total donor-derived CD45.2+GFP+ cells from the BM of recipient mice. (E-F) Donor-derived cells (CD45.2+GFP+) from each sgCTL and sgCadm1 transplanted mouse purified by cell sorting. Genome editing efficiency of targeted Cadm1 region performed on purified donor-derived cells (CD45.2+GFP+) for each condition (E). Cytological analysis was performed on purified CD45.2+GFP+ cells from each condition, and the proportion of myeloid and lymphoid cells was calculated (F).
Discussion
Deletion of chromosome 11 is recurrently found in MDS and other hematological malignancies, such as chronic lymphocytic leukemia (CLL),27,28 mantle cell lymphoma,29 and in solid tumors such as lung cancer,22 nasopharyngeal carcinomas,23 gliomas,30 and neuroblastoma.31 Our analysis of del(11q) MDS represents the largest study of this very rare but recurrent chromosomal deletion. In our MDS cohort, del(11q) was mainly a primary clonal abnormality found predominantly in women presenting with mild cytopenias, a low monocyte blood count, and the frequent presence of multinucleated megakaryocytes. MDS with del(11q) were enriched in CBL gain-of-function mutations in a similar way to acquired uniparental disomy of chromosome 11 in MDS and MDS/MPN.32 ASXL1 and SF3B1 mutations were also frequent in del(11q) MDS, although at a similar rate compared with other MDS.8,33 TET2 mutations, in contrast, were less frequent than in other MDS.20
Our extended analysis of del(11q) MDS identified a unique interstitial CDR between ATM (11q22.3) and CBL (11q23.3), leading to a hemizygous deletions of CADM1 (deleted in 98% of del(11q) MDS), ATM (deleted in 96%), KMT2A (deleted in 90%), and CBL genes (deleted in 84%). The del(11q) MDS CDR is different from the 1 identified in CLL where ATM has been defined as the critical gene.27,28 As shown in other deletions in MDS, the effect of del(11q) might be due to concomitant deletions of ATM, CBL, and KMT2A, in addition to CADM1 leading to MDS and MDS/MPN development. ATM encodes a serine/threonine kinase which is critical to the phosphorylation of numerous proteins during the cell cycle, including p53, Brca1, Chk2, Rad17, and Rad9. Germline mutations in ATM are associated with ataxia telangiectasia, an autosomal recessive disorder.34 CBL encodes an E3 ubiquitin ligase that ubiquitinates signaling proteins, such as the tyrosine kinases Flt3, PDGFRA, Kit, or Jak2. In adult patients, CBL mutations are found in ∼5% of myeloid neoplasms,35 mainly in chronic myelo-monocytic leukemias,36 but are very rare in MDS.37 With a frequency of mutations of 14% in our series and 8% in the Stengel series,8 CBL was more frequently mutated in del(11q) MDS than in other MDS.
The unique minimal CDR seen in del(11q) MDS is centered in an intergenic region between NXPE2 and CADM1. Expression and functional analysis of CADM1 suggest a specific function in hematopoiesis. CADM1 encodes a glycoprotein of 471 amino acids composed of 3 immunoglobulin-like domains involved in cell adhesion: a transmembrane domain and a short cytoplasmic domain with a binding motif homologous to that of glycophorin C.38 It forms homodimers at the cell membrane22 with homophilic39 and heterophilic40 cell-cell adhesion activity. Few publications investigated the mechanism of action of Cadm1 in the development of tumors, rendering it difficult to distinguish a direct effect by haploinsufficiency to an obligatory 2-hit transformation. Non–small cell lung cancer cells present with a frequent loss of heterozygosity of CADM1 associated with a frequent inactivation of the second CADM1 allele by promoter methylation.22 The recurrent deletion of CADM1 is also found in other solid tumors, such as liver, pancreatic, and prostate cancers, and especially in metastatic tumors.38,41 In acute lymphoblastic leukemias, CADM1 promoter is frequently methylated and inactivated.42 Its homozygous inactivation in mice is viable but associated with a male infertility.43-45 The heterozygous mutant mice have no reported abnormalities. Cadm1-null mice die significantly faster than wildtype mice due to spontaneous tumor development.46 Van der Weiden et al challenged Cadm1-null mice by irradiation leading to the development of lymphomas and leukemias.46 Recently, the extracellular domain of CADM1 has been shown to restrict tumor growth and metastases by interacting with HER2 and integrin α6β4 at the cell surface and reducing STAT3 activation.47 The inhibition of this signaling pathway using the JAK1/2 inhibitor ruxolitinib replicates these effects specifically in CADM1-null cells.47 This drug, which is mainly used in myelofibrosis, has been tested in only 1 MDS clinical trial, but none of the 11 patients investigated displayed a del(11q).48 Consequently, ruxolitinib could be an interesting targeted therapy to be specifically evaluated in del(11q) MDS patients.
Here, we used CRISPR strategy to address the effect of Cadm1 deletion on hematopoietic development at the functional level. Although not altering their multilineage hematopoietic reconstitution potential, the genomic editing of Cadm1 of murine LSK cells diminished the myeloid-to-lymphoid ratio in BM after primary and secondary transplantations. However, this phenotype was not associated with the long-term development of a myeloid neoplasm. This observation suggests that Cadm1 deletion is not sufficient by itself to induce clonal transformation in mice and requires collaborative oncogenic events, such as CBL or ASXL1 mutations and additional deletions in the del(11q) region, as observed in del(11q) MDS patients. Nevertheless, the perturbation of myeloid cell production and/or terminal differentiation observed in mice could mimic a preneoplastic state before malignant transformation.
In summary, our report identifies unique features of del(11q) MDS with various combinations of deletions of 4 genes, ATM, CBL, KMT2A, and CADM1, a tumor suppressor gene identified initially in solid tumors.
Acknowledgments
B.G., L.L., V.F., P.-Y.J., L.J., P.B., S.H., C. Broccardo., N.P., S.L., and E.D. were supported by association Laurette FUGAIN (RAK14017BBA), SFCE (Société Française des Cancers de l’Enfant) and the association Capucine (RAK17022BBA), 111 des Arts (RAK18016BBA), la ligue régionale contre le cancer (RAB19007BBA), the région Occitanie (RPH17006BBA), the association CASSANDRA (RAK18008BBA), the association « Constance la petite guerrière astronaute » (R19043BB), and TREC (Toulouse Recherche Enfant Cancer, RAK18006BBA). K.Y. had been funded by Wellcome Trust (WT206194). The Programme Hospitalier de Recherche CliniqueS-04 was supported by the National Institute of Cancer (funding number INCA_9290). M.F., J.H.J., T.T., and D.R. were supported by a European Union's Horizon 2020 research and innovation program S-RIGHT under grant agreement No. 634789.
The authors would like to thank all the members of the Groupe Francophone de Cytogénétique that diagnosed del(11q) cases, provided cytogenetic samples for the study, and participated in the cytogenetic reviewing of cases; present Groupe Francophone de Cytogénétique members (Laurence Baranger, Sophy Laibe, Marie-Joelle Mozziconacci, and Lauren Veronese); and previous (Eric Lippert, Sylvie Taviaux) or retired colleagues (Christian Bastard, Nicole Dastugue, Francine Mugneret, and Pascaline Talmant).
The authors were grateful to Manon Farcé from the cytometry and cell-sorting facility of the Pole Technologique of the CRCT (INSERM U1037) for technical assistance. They thank the Anexplo/Genotoul platforms for technical assistance (UMS006).
The authors thank the physicians taking care of patients, Pierre Fenaux, and all members of the Groupe Français des Myélodysplasies, in particular François Dreyfus (Paris, Cochin), Bruno Quesnel (Lille), Emmanuel Gyan (Tours), Odile Beyne-Rauzy (Toulouse), Norbert Vey (Marseille), Mathilde Hunault (Angers), Borhan Slama (Avignon), Agnès Guerci-Bresler (Nancy-Brabois), Aspasia Stamatoullas (Rouen), Carole Soussain (Saint-Cloud), and Stéphane Chèze (Caen).
The authors also thank Louise Van der Weyden for providing Cadm1 material and Xavier Troussard, Valérie Bardet, and all the members of the Groupe Francophone d’Hématologie Cellulaire for providing cytological data.
Authorship
Contribution: M.L.-P., G.A., C. Barin, A.B., J.B., M.-A.C.-R., C.G., A.I., C.L., I.L., L.M., N.N., H.A.P., I.R.-W., B.R., S.R., S.S., C.T., I.T., D.P., and V.E. provided samples and reviewed cytogenetic data under the supervision of MLP. M.L.-P. performed additional FISH experiments. B.G., L.L., V.F., P.-Y.J., L.J., P.B., S.H., and C. Broccardo performed cellular experiments. V.B. and V.M-D. reviewed cytological smears. O.M.D., K.Y., and G.S.V. provided Cas9 mice. J.H.J., T.T., and D.R. performed MDS RNA sequencing. N.P., L.L., S.L., and E.D. performed molecular analyses. M.F. provide the control cohort of MDS without del(11q). E.D., M.L.-P., B.G., V.B., V.M-D., J.H.J., D.P., V.E., M.F., and C. Broccardo reviewed data and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A full list of study group members appears in “ Appendix.”
Correspondence: Eric Delabesse, Institut Universitaire de Cancérologie de Toulouse, Laboratory of Hematology, 1 avenue Irène Joliot-Curie, 31059 Toulouse cedex 9, France; e-mail: delabesse.eric@iuct-oncopole.fr.
Appendix: study group members
The members of the Groupe Francophone de Cytogénétique Hématologique were Nassera Abermil (Paris), Lucille Altounian (Metz), Nathalie Auger (Villejuif), Estelle Balducci (Villejuif), Zsofia Balogh (Villejuif), Laurence Baranger (Angers), Audrey Basinko (Brest), Beatrice Grange (Lyon), Martine Becker (Rouen), Chrystèle Bilhou-Nabera (Paris), Rossana Bonomi (Montevideo), Claire Borie (Villejuif), Elise Boudry (Lille), Evelyne Callet-Bauchu (Lyon), Helene Cannoni (Marseille), Élise Chapiro (Paris), Lucie Coster (Toulouse), Sophie Cotteret (Villejuif), Wendy Cuccuini (Paris), Laurent Dano (Strasbourg), Agnès Daudignon (Lille), Mathieu Decamp (Caen), Sabine Defasque (Cergy-Pontoise), Barbara Dewaele (Leuven), Nathalie Douet-Guilbert (Brest), Olivier Dupuy (Toulon), Yann Ferret (Amiens), Sandra Fert-Ferrer (Chambéry), Nathalie Gachard (Limoges), Baptiste Gaillard (Reims), Catherine Godon (Nantes), Ellen Hammouche (Tours), Nazha Hda (Casablanca), Catherine Helias (Strasbourg), Catherine Henry (Rennes), Jean-Loup Huret (Poitiers), Éric Jeandidier (Mulhouse), Mélanie Jimenez (Tours), Sophie Kaltenbach (Paris), Stephan Kemeny (Clermont-Ferrand), Emilie Klein (Bordeaux), Elodie Laharanne (Bordeaux), Sophy Laibe (Marseille), Erika Launay (Rennes), Pascale Cornillet-Lefebvre (Reims), Geneviève Lefort (Brest), Odile Maarek (Paris), Mélanie Martin (Nîmes), Catherine Menten (Liège), Marie-Joëlle Mozziconacci (Marseille), Marc Muller (Nancy), Florence Nguyen-Khac (Paris), Marie-Pierre Pages (Lyon), Agathe Paubel (Chambray), Benoit Quilichini (Lyon), Sophie Raynaud (Nice), Lauren Rigollet (Saint-Etienne), Catherine Roche-Lestienne (Lille), Serge Romana (Paris), Véronique Saada (Villejuif), Gwendoline Soler (Clermont-Ferrand), Anne Staal-Viliare (Metz), Andrei Tchirkov (Clermont-Ferrand), Sylvie Tondeur (Grenoble), Saloua Toujani (Rennes), and Lauren Veronese (Clermont-Ferrand).
References
Author notes
M.L.-P., B.G., V.B., C. Broccardo, and E.D. contributed equally to this study.
Requests for data sharing may be submitted to Eric Delabesse (delabesse.eric@iuct-oncopole.fr).
The full-text version of this article contains a data supplement.

![Cytological and cytogenetic presentations of del(11q) MDS. (A-D) May-Grünwald Giemsa staining of del(11q) bone marrow. Megakaryocytes were often large and plurinucleated (A, B, and C, original magnification x×50) giving an aspect of ball bags (C) and, when associated to 5q deletion, presence of small and nonlobated megakaryocytes ([D], original magnification ×100). (E) Cytogenetic presentations of del(11q). The 2 main types of del(11q) on R-banded partial karyotypes from 2 representative cases, from left to right: large deletion, del(11)(q14q24), in case #19 and small deletion, del(11)(q22q24), in case #86.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/6/2/10.1182_bloodadvances.2021005311/5/m_advancesadv2021005311f1.png?Expires=1769244952&Signature=1yMFyvt9m2spSLvshDyiS4QgMol7eSTUgHh~~ilWZ9YZVd091v0bgLv~D8~3qr~XW2ohXuqWveBnTLBOld67Vw~cbfuygNns7dXjFLKX2PI8WZ7oAJ2WeuPo9AGi-S3Q9UOpML6CPJaukl1hi71a5ZqoUTHNO5Go7wQ3lNU68gcK0AWxpS7QbW5S-U6NX21aL5At20zxZ8IfwCppWS4d~-wqlbaXm8foGLQUDsL7fHLdzlRbGj3nxpTkivdDeBBZhIT-Pk6MCoBQfLCaRn~k4vwTLHBXGYhUjCNQp2raAVaky2~8ptq~VeR3uP~FnUuuRh1MkDeEr-lxuR6QloIMFA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Mutations in del(11q) MDS. Mutation screening of 36 del(11q) MDS by next generation sequencing. Genes were classified according to their function (2 first columns). Comparison with other MDS is indicated in the 3 central columns (del[11q] MDS mutation frequency, other MDS mutation frequency, and P value comparing the frequencies between these 2 groups). Mutations are indicated by red squares containing the number of mutations per sample if higher than 1, the absence of mutations by green squares, and the absence of analysis by white squares. Patients are labeled at the top.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/6/2/10.1182_bloodadvances.2021005311/5/m_advancesadv2021005311f3.png?Expires=1769244952&Signature=DZCRNkbrYr0i-td-HnKv9Lrl1aUNjAQRw98270dqLOFvhV-1yW6HKr49V6z3I-p1nOymTFoMQH2K~~uYQ~rMjjTNyTetNfgmzTDKswcHtEru-~42YhQ2cBD87XAQYY8vhOCWk4X7wh9not2cUOVV7TM4jpeFwM-m7cS71ayd0YgBx34Cjuht1bHnnUlL3ufuJZazszETQ1QdVdeCi4xVSxNARGR3Dy73KjxtxXzRZufuzYONV0R4k0GdzNIlbeEq7PQb8ssXqemQCLf-j272ZF4SJd1z70-XjxSIeBSER1Osov4sh5ODGDd3QazbiPBJzmShPx5Fe0yevm4c~gr16g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)