TO THE EDITOR:
As a poor-risk group of aggressive B-cell lymphomas (BCLs), high-grade BCLs harboring MYC and BCL2 and/or BCL6 rearrangements high grade B-cell lymphoma-double/triple-hit (HGBL-D/TH) are currently a subject of intense clinical and research interest. New drugs, particularly targeted therapeutic agents, are increasingly being developed and entering the clinic in recent years.1 A panel of well-characterized HGBL-D/TH cell lines will be valuable for preclinical drug screening and development. There are so far few HGBL-D/TH cell lines that have comprehensive genomic data.2-4 Here, we reported 2 new double/triple-hit lymphoma (D/THL) cell lines named “COH-DHL1” and “COH-THL1” with comprehensive characterization.
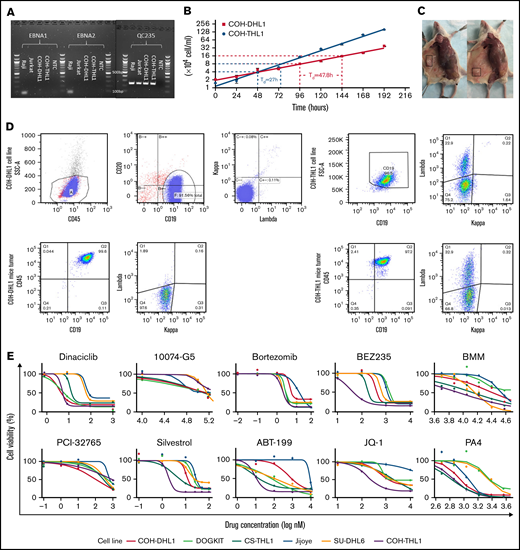
COH-DHL1 and COH-THL1 were derived from 2 male patients at City of Hope, and both were Epstein-Barr virus (EBV)-negative, as shown by the Epstein-Barr nuclear antigen polymerase chain reaction assay (Figure 1A). Their doubling times were 47.8 hours and 27 hours, respectively (Figure 1B). When we injected 10 million cells of each cell line subcutaneously into NSG mice, all 3 replicates developed tumors within 3 weeks (Figure 1C). Fingerprinting showed that the 2 cell lines were unique without any identical cell lines in the Cellosaurus database (supplemental Figure 1 in the data supplement). Flow cytometry showed consistent immunophenotype between the 2 cell lines, their 1- and 2-month cultures, and their mice tumors in terms of CD45, CD19/CD20, and immunoglobulin light chains (supplemental Figure 2 and Figure 1D). For COH-THL1, whose original tumor was available, fingerprinting and flow cytometry also showed consistency with its origin (supplemental Figures 1B and 2B). All animal procedures in the mice experiment were in accordance with the guidelines and approved by the Administrative Panel on Laboratory Animal Care at City of Hope Comprehensive Cancer Center.
Characterization and validation of the D/THL cell lines. (A) EBNA (Epstein–Barr virus nuclear antigen 1) polymerase chain reaction assay of COH cell lines with negative control, an EBV-positive (Raji) and EBV-negative (Jurkat) cell line. QC235 is a positive control separately performed to confirm that DNA samples are amplifiable. (B) Growth plot for COH-DHL1 and COH-THL1 cell lines within 8 days with y-axis log2 transformed and doubling time (Td) labeled. (C) Representative figure of mice with tumor highlighted in the box after subcutaneous injection of COH-DHL1 (left) and COH-THL1 (right). (D) Flow cytometry results of the 2 cell lines with their mice tumors. (E) Sigmoidal dose–response curves showing responses of 6 cell lines to 10 different drugs.
Characterization and validation of the D/THL cell lines. (A) EBNA (Epstein–Barr virus nuclear antigen 1) polymerase chain reaction assay of COH cell lines with negative control, an EBV-positive (Raji) and EBV-negative (Jurkat) cell line. QC235 is a positive control separately performed to confirm that DNA samples are amplifiable. (B) Growth plot for COH-DHL1 and COH-THL1 cell lines within 8 days with y-axis log2 transformed and doubling time (Td) labeled. (C) Representative figure of mice with tumor highlighted in the box after subcutaneous injection of COH-DHL1 (left) and COH-THL1 (right). (D) Flow cytometry results of the 2 cell lines with their mice tumors. (E) Sigmoidal dose–response curves showing responses of 6 cell lines to 10 different drugs.
The 2 cell lines were further characterized with 2 other reported D/THL cell lines (DOGKIT5 and CS-THL16 ) for comparison. We tested the sensitivity of the 4 cell lines along with Jijoye and D/THL cell line (SU-DHL6) to 10 drugs targeting BCL2, MYC, BTK, PI3K, or CDK (Figure 1E). BCL2 is an important antiapoptotic protein that synergizes with other oncogenes, such as MYC, to promote lymphoma development and could be a critical factor for HGBL-D/TH survival. Inhibiting BCL2 by potent BH3 mimetics such as ABT-199 demonstrated a remarkable clinical response in selected patients.7 This is also demonstrated by the higher sensitivity of the 5 HGBL-D/TH cell lines to ABT199, compared with Jijoye, which is a Burkitt lymphoma (BL) lacking BCL2 expression.
MYC, as a key oncogene,8 likely plays a crucial role in HGBL-D/TH, making it an excellent therapeutic target. Although it is yet impossible to directly inhibit MYC function, many strategies have been employed to indirectly inhibit MYC activity. In this study, we attempted to: (1) impair MYC transcription by inhibiting the BRD4 (bromodomain-containing 4) using JQ-19 ; (2) interfere with MYC mRNA translation using the eukaryotic initiation factor (eIF) 4A inhibitor silvestrol10 ; (3) reduce MYC protein stability using Berbamine11 and its derivative PA4 through inhibition of the Ca2+/calmodulin-dependent protein kinase γ12 ; and (4) inhibit the transcriptional activity of MYC by disrupting its binding site using 10074-G5.13 Silvestrol was highly potent at the nM range. PA4 was quite promising (supplemental Table 1), whereas JQ1 and 10074-G5 were disappointing. Silvestrol is a flavagline that inhibits the activity of eIF4A subunit of the eIF4F complex. By inhibiting eIF4F, which is important for efficient translation of RNA containing G-quadruplex structures such as MYC and BCL2, silvestrol has shown powerful activities in human breast and prostate cancer xenograft models,14 as well as MYC-induced lymphomagenesis.15 All D/THL cell lines studied here were highly sensitive to silvestrol, indicating its potential as a powerful treatment option for D/THL.
Interestingly, the proteasome inhibitor bortezomib16 was a potent drug at the nM level, whereas the BTK inhibitor PCI-32765 and PI3K inhibitor BEZ235 required substantially higher concentrations. Dinaciclib, a novel drug that inhibits cyclin-dependent kinases CDK1, 2, 5, and 9,17 was also highly effective, especially for COH-DHL1, COH-THL1, and DOGKIT. This is consistent with the reported essential role of CDK9-mediated transcriptional elongation for tumor maintenance18 in a genetically defined MYC-driven model of hepatocellular carcinoma and downregulation of MCL-119 and MYC20 after CDK9 inhibition. In general, the COH cell lines were more responsive to most of the drugs. DHL6 tended to be more resistant, and Jijoye, which is a BL, had a different pattern of responses and tended to be the most resistant.
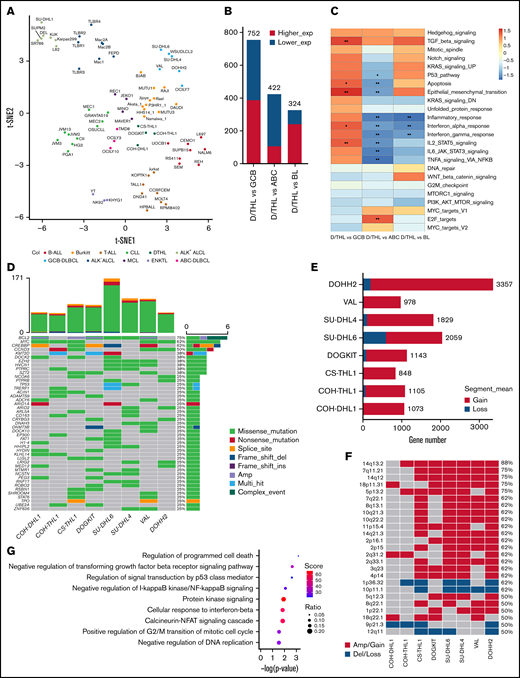
The 4 D/THL cell lines showed an average of 76 mutations with the most frequent alterations of MYC, BCL2, and CREBBP (supplemental Figure 3 and supplemental Table 2). Gene expression profiling of the 4 cell lines along with 74 other lymphoma/leukemia cell lines (supplemental Table 3) showed that their transcriptome profiles were close to BL, diffuse large B-cell lymphoma (DLBCL), and mantle cell lymphoma lines (Figure 2A and supplemental Figure 4A) with the highest similarity to BL cell lines (Figure 2B and supplemental Table 4). Gene set enrichment analysis showed enrichment of reported double-hit signature genes21 in D/THL compared with DLBCL and BL (supplemental Figure 4B-C). Differentially expressed genes in D/THL vs BL were significantly (P < .05) negatively enriched in the inflammatory response and interferon α and γ response, which was also true vs activated B-cell like-DLBCL. However, the opposite was observed when compared with germinal center B-cell like (GCB)-DLBCL, in which enrichment in multiple pathways such as the TP53 pathway, apoptosis, epithelial–mesenchymal transition, TGFB, and IL2/STAT5 signaling was also observed for D/THL (Figure 2C). The more prominent activation of the interferon pathways in BL could be related to the presence of EBV22 in many BL lines or more activation of endogenous retroviral elements in the BL cells.23
Gene characterization of 4 D/THL cell lines compared with the other lymphoma/leukemia cell lines. (A) t-SNE clustering of the COH cell lines along with other 74 cell lines of 11 types. (B) The number of differentially expressed genes (|log2fold change| >1, false discovery rate <0.1) for contrasts between D/THL and activated B-cell like (ABC)-DLBCL, GCB-DLBCL, or BL. (C) Heatmap showing gene set enrichment analysis results with hallmark gene sets for differentially expressed genes between D/THL , ABC-DLBCL, GCB-DLBCL, and BL. Gradient colors represent the net enrichment score. *P < .05; **P < .01. (D) Waterfall plot showing the recurrent mutations in the 4 D/THL cell lines and the other 4 double-hit GCB-DLBCL cell lines. Different colors represent different types of mutations or CNA. The top bar plot shows the total number of mutations in each sample. The right bar plot shows the number of different alterations for each gene labeled by the percentage of samples that have genetic alteration in the gene. “Multi_Hit” means genes with cooccurring mutations of different types. “Complex_Event” means genes with both mutations and CNAs. (E) Bar plot showing expressed gene number covered by regions with copy number gain or loss of D/THL cell lines studied here along with another 4 double-hit cell lines. (F) Heatmap showing the distribution of the MCRs identified by GISTIC among the 8 cell lines. (G) The significant gene ontology (GO) terms (P < .05) enriched by the expressed genes in the MCRs ranked according to −log10 (P values) from top to bottom. The color and size of the dots indicate the enrichment score and ratio between gene number in MCRs and gene number in each GO term, respectively.
Gene characterization of 4 D/THL cell lines compared with the other lymphoma/leukemia cell lines. (A) t-SNE clustering of the COH cell lines along with other 74 cell lines of 11 types. (B) The number of differentially expressed genes (|log2fold change| >1, false discovery rate <0.1) for contrasts between D/THL and activated B-cell like (ABC)-DLBCL, GCB-DLBCL, or BL. (C) Heatmap showing gene set enrichment analysis results with hallmark gene sets for differentially expressed genes between D/THL , ABC-DLBCL, GCB-DLBCL, and BL. Gradient colors represent the net enrichment score. *P < .05; **P < .01. (D) Waterfall plot showing the recurrent mutations in the 4 D/THL cell lines and the other 4 double-hit GCB-DLBCL cell lines. Different colors represent different types of mutations or CNA. The top bar plot shows the total number of mutations in each sample. The right bar plot shows the number of different alterations for each gene labeled by the percentage of samples that have genetic alteration in the gene. “Multi_Hit” means genes with cooccurring mutations of different types. “Complex_Event” means genes with both mutations and CNAs. (E) Bar plot showing expressed gene number covered by regions with copy number gain or loss of D/THL cell lines studied here along with another 4 double-hit cell lines. (F) Heatmap showing the distribution of the MCRs identified by GISTIC among the 8 cell lines. (G) The significant gene ontology (GO) terms (P < .05) enriched by the expressed genes in the MCRs ranked according to −log10 (P values) from top to bottom. The color and size of the dots indicate the enrichment score and ratio between gene number in MCRs and gene number in each GO term, respectively.
We included 4 additional reported HGBL-D/TH cell lines (SU-DHL6, SU-DHL4, VAL, and DOHH2)2 for genetic analysis. These cell lines showed more genetic abnormalities than the COH cell lines (Figure 2D-F), indicating that many acquired changes may have occurred because of in vitro culture, and it is important to use lines with low passage numbers for study to avoid the noise from the artifactual changes. A few frequently mutant genes (eg, CREBBP, EZH2, KMT2D, and EP300) in these cell lines were shared with follicular lymphoma and GCB-DLBCL, consistent with the tumor derivation from GCB cells and the hypothesis that after acquiring the BCL2 translocation, the cells may evolve toward a GCB-like lymphoma with the acquisition of these mutations24 (Figure 2D). The acquisition of the MYC translocation then drives the evolution to HGBL-D/TH in concert with the BCL2 translocation. There are other detected mutations (eg, TP53, PIM1, BAX, and CCND3) that may arise before or after the MYC translocation and further enhance MYC-driven cell cycle progression and cell survival. Importantly, our recent study found that HGBL-D/TH cases with TP53 abnormalities have a much worse prognosis.25 GISTIC (Genomic Identification of Significant Targets in Cancer) identified 25 minimal common regions (MCRs) with genomic copy number abnormalities in ≥4 samples (Figure 2F). Among these, loss of 9p21 harboring CDKN2A and CDKN2B and gain of 2q31 were found in both COH cell lines; gains of 18p11 and 18q22 were in COH-DHL1; gains of 5p13 and 14q13 with NFKBIA; and loss of 1p36 containing TNFRSF14 were in COH-THL1. Genes in these MCRs are enriched in biological processes, including apoptosis, cell cycle and cytokine, TP53, TGFβ (transforming growth factorβ) receptor, NF-κB (nuclear factor kappa B), protein kinase A, and IFN-β (interferon β) signaling (Figure 2G). We can envision that with a sufficient number of cell lines, we can further dissect different genetic/biological subgroups in vitro.
In conclusion, we established 2 new D/THL cell lines, COH-DHL1 and COH-THL1, and described their genetic characteristics as well as those of 2 additional cell lines, CS-THL1 and DOGKIT. We studied the sensitivity to 10 targeted agents using a D/THL cell line panel and demonstrated the potential of cell line models in preclinical studies of novel therapeutic agents that may benefit patients with HGBL-D/TH.
Acknowledgments: The authors thank Zhaohui Gu at the Department of Computational and Quantitative Medicine, City of Hope, for his valuable suggestions on data analysis and the staff of the Flow Cytometry Core, the Integrative Genomics Core, and Animal Tumor Model Core for their experimental assistance.
This work was supported by the National Cancer Institute of Health under grant number P30CA033572. It was also partly supported by Start-up funds from the City of Hope National Medical Center, the Dr. Norman and Melinda Payson Professorship in Hematologic Cancers, the Toni Stephenson Lymphoma Center, and the Nesvig fund.
Contribution: J.Z., T.W., K.S., Y.L., A.F.H., J.Y.S., and W.C.C. drafted the manuscript and prepared the figures; W.C.C. designed and supervised the study; R.K.P., W.H., J.Y.S., H.-G.W., and S.A. provided cell lines and drugs; T.W., K.S., S.X., and Y.L. conducted cell culture and drug response experiments; J.Y.S. and K.S. conducted flow cytometry and PCR experiments; K.S. and X.L. conducted mouse study; J.Z., Q.G., and Z.H. are involved in the bioinformatic analysis; and the manuscript has been read and approved for submission by all authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Wing C. Chan, Department of Pathology, City of Hope National Medical Center, Duarte, CA 91010; e-mail: jochan@coh.org; Joo Y. Song, Department of Pathology, City of Hope National Medical Center, Duarte, CA 91010; e-mail: josong@coh.org; and Raju K. Pillai, Department of Pathology, City of Hope National Medical Center, Duarte, CA 91010; e-mail: rpillai@coh.org.
References
Author notes
J.Z., T.W., and K.S. contributed equally to this study as joint first authors.
The sequencing data have been deposited in NCBI’s Sequence Read Archive database with accession number PRJNA818709 (https://www.ncbi.nlm.nih.gov/sra/PRJNA818709).
The full-text version of this article contains a data supplement.