TO THE EDITOR:
Endemic Burkitt Lymphoma (eBL) is one of the most common pediatric cancers in sub-Saharan Africa, and the 1-year survival remains close to 50%.1 The low survival rate is partly attributable to patients lacking access to early diagnosis and presenting with advanced disease.2,3 eBL is often diagnosed via fine-needle aspiration (FNA) cytology, which like regular histology, requires timely staining, a microscope, and trained personnel.4,5 A defining feature of eBL is the presence of Epstein-Barr virus (EBV), and histological detection of this virus in cytological specimens obtained by FNA from tumor biopsies can distinguish EBV-driven tumors from other conditions such as nonmalignant lymphadenopathies.4 We hypothesize that simplified nucleic acid testing for EBV from FNA offers a feasible diagnostic solution in regions where access to pathologists, specialized reagents, and equipment is limited.
We report the development of a loop-mediated isothermal amplification (LAMP) assay6 for detection of EBV and a cells-to-LAMP process that overcomes the need for DNA extraction and purification (described in the supplemental Methods). This system seamlessly integrates into a new point-of-care detection device, called TINY, which was developed specifically for isothermal amplification and can be powered by electricity, flame, or sunlight, and thus far tested for the presence of the Kaposi sarcoma (KS) herpesvirus (KSHV; or human herpesvirus 8 [HHV-8]) in KS in skin biopsies.7 The cells-to-LAMP system shows high specificity in identifying EBV-driven tumors and can be easily deployed at sites with limited infrastructure, opening opportunities for earlier diagnosis and better patient outcomes.
Primer sets toward different regions of the EBV genome were tested for specificity and sensitivity. As LAMP can produce late, nontemplate amplification,8,9 primer sets were screened for early positive amplification and significantly late or no nontemplate control amplification. LAMP was done using DNA extracted from 5 different EBV+ BL cell lines (Kem I, Kem III, Namalwa, Mutu I, and Rael) and one lymphoblastoid cell line (LCL9001) and loaded in 100 ng and 10 ng quantities with DNA from the EBV− cell lines and salmon sperm DNA, used as negative controls. The EBV-encoded RNAs (EBER) primer set showed the highest specificity for EBV and was selected for subsequent analysis.
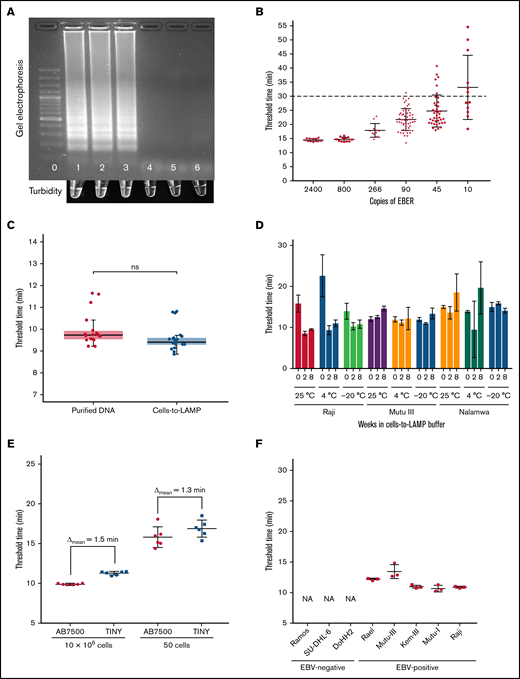
A representative gel image from a standard LAMP reaction run at 67°C for 30 minutes with DNA from a control plasmid and 2 EBV+ cell lines shows positivity inferred by band smearing or turbidity, typical of LAMP6 (Figure 1A). The limit of detection of the assay was determined in real-time with serial diluted plasmid DNA containing the EBER target sequence. An amplification threshold cutoff was set at 30 minutes and confirmed to be more than 2 standard deviations from the limit of the blank. The limit of detection of the assay was 90 copies of EBER per reaction with a sensitivity of 98% across multiple independent experiments (Figure 1B).
Development of LAMP for EBV and cells-to-LAMP Assay. (A) Top: gel electrophoresis results of EBER LAMP reaction run for 30 minutes in a standard thermal cycler. Bottom: in-tube turbidity immediately after reaction. Lane/tube 0: 100bp DNA ladder; lane/tube 1: 480 copies EBER plasmid DNA control; lane/tube 2: Namalwa (EBV+) DNA; lane/tube 3: Raji (EBV+) DNA; lane/tube 4: BC-3 (EBV-) DNA; lane/tube 5: Ramos (EBV-) DNA; lane/tube 6: water. (B) Amplification of serially diluted plasmid DNA amplified with EBER LAMP in real-time. Fifty of 51 samples containing 90 copies per reaction were amplified before the 30-minute threshold cutoff. (C) Amplification threshold results of purified DNA from cells with Qiagen DNeasy and the cells-to-LAMP procedure, no significant difference detected, P = .2222 by Mann-Whitney U test. Each sample contained the same number of cells (1 million cells) in the same final volume across assays. Data show combined results of 4 independent experiments containing ≥3 replicates in each group shown together. (D) Stability assessment of amplification from cell lines stored in cells-to-LAMP solution at room temperature (∼25°C), 4°C, or −20°C at 0, 4, or 8 weeks after extraction. The EBV– control Ramos was also evaluated but is not shown because no amplification was seen before our 30-minute threshold. (E) Amplification threshold times at high (10 million cells) and low (50 cells) loads in a conventional real-time thermal cycler were compared with the TINY system. (F) Cells-to-LAMP amplification times from 1 million cells from EBV+ and EBV- cell lines in TINY show specificity. The EBV– cell lines did not amplify (NA).
Development of LAMP for EBV and cells-to-LAMP Assay. (A) Top: gel electrophoresis results of EBER LAMP reaction run for 30 minutes in a standard thermal cycler. Bottom: in-tube turbidity immediately after reaction. Lane/tube 0: 100bp DNA ladder; lane/tube 1: 480 copies EBER plasmid DNA control; lane/tube 2: Namalwa (EBV+) DNA; lane/tube 3: Raji (EBV+) DNA; lane/tube 4: BC-3 (EBV-) DNA; lane/tube 5: Ramos (EBV-) DNA; lane/tube 6: water. (B) Amplification of serially diluted plasmid DNA amplified with EBER LAMP in real-time. Fifty of 51 samples containing 90 copies per reaction were amplified before the 30-minute threshold cutoff. (C) Amplification threshold results of purified DNA from cells with Qiagen DNeasy and the cells-to-LAMP procedure, no significant difference detected, P = .2222 by Mann-Whitney U test. Each sample contained the same number of cells (1 million cells) in the same final volume across assays. Data show combined results of 4 independent experiments containing ≥3 replicates in each group shown together. (D) Stability assessment of amplification from cell lines stored in cells-to-LAMP solution at room temperature (∼25°C), 4°C, or −20°C at 0, 4, or 8 weeks after extraction. The EBV– control Ramos was also evaluated but is not shown because no amplification was seen before our 30-minute threshold. (E) Amplification threshold times at high (10 million cells) and low (50 cells) loads in a conventional real-time thermal cycler were compared with the TINY system. (F) Cells-to-LAMP amplification times from 1 million cells from EBV+ and EBV- cell lines in TINY show specificity. The EBV– cell lines did not amplify (NA).
Given sample preparation time limits diagnostic tools aimed at the point-of-care, we examined if we could omit the DNA extraction step by experimenting with different buffers. We found that amplification of EBV+ Raji eBL cells in a simple solution of 0.05% TWEEN 20, 0.5 M Betaine, and 1 mg/mL BSA (bovine serum albumin), which we termed TBB buffer, produced amplification threshold times statistically indistinguishable from purified DNA obtained from equivalent starting material using a commercial DNA extraction kit (Figure 1C), showing that this simple buffer obviates the need for DNA extraction before LAMP when starting with cells in suspension.
As it may not be possible to run the LAMP assay immediately at the bedside, we investigated the temporal and thermal stability of samples in the TBB buffer. Our data show that target DNA was amplifiable with LAMP from cells stored in TBB buffer for ≥8 weeks and that amplification threshold time of samples stored at ambient temperature was comparable to refrigerated and frozen samples (Figure 1D).
To bring this assay to the point of care, we assessed if the cells-to-LAMP system could be integrated with TINY7 and detect EBV across high and low sample cellularity. EBV detection with cells-to-LAMP was not significantly different in TINY when compared with detection using a standard real-time thermocycler, from a minimal load of 50 Raji cells (P = .8182, Mann-Whitney U test) to a high load of 10 million cells per reaction (P = .7835, Mann-Whitney U test), with the difference in assay threshold time being <2 minutes, which is likely due to the samples reaching the steady-state temperature more slowly in TINY or minor differences in software thresholding calculations (Figure 1E). Cells-to-LAMP effectively distinguished multiple EBV+ cell lines from EBV− cell lines using TINY, confirming the specificity of the reaction and detection systems (Figure 1F).
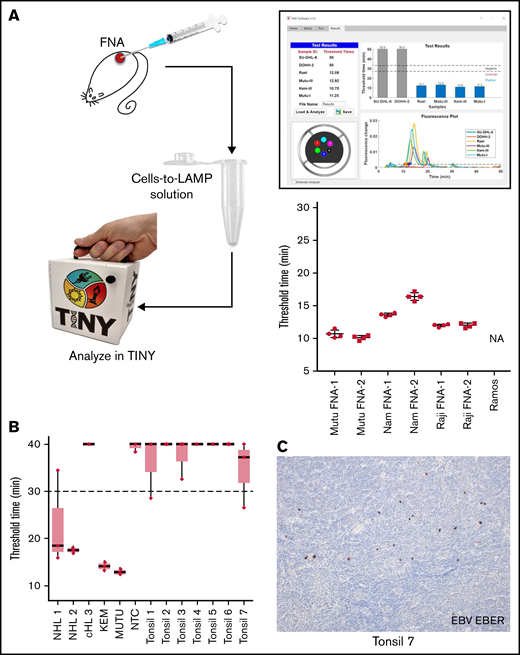
To test the cells-to-LAMP system with diagnostically relevant inputs, clinical samples were mimicked with FNAs from mouse xenografts of EBV+ BL lines. Mouse FNAs were resuspended in cells-to-LAMP solution and run simultaneously in TINY and a realtime thermocycler. Results for each FNA in TINY are shown in Figure 2A. Each FNA produced internally consistent amplification threshold times across independent runs in the TINY and conventional thermocycler alike.
EBV cells-to-LAMP assay is compatible with a point-of-care approach. (A) FNA samples obtained from EBV+ cell line xenografts in mice were tested using cells-to-LAMP followed by analysis with TINY (left), with representative cells-to-LAMP results output by TINY software of EBV+ and EBV- cell lines shown (top right). The blue bars in this image indicate the 4 EBV+ cell lines that amplified before the threshold time. Four independent aspirates were taken from each of 2 tumors per cell line (bottom right). NA denotes that the negative control cell line, Ramos, was not amplifiable. (B) Fresh tonsil specimens were used to perform an ex vivo FNA with cells-to-LAMP followed by TINY. Tissue specimens from 3 EBV+ lymphoma specimens were used for DNA extraction and tested in parallel by LAMP and analyzed with TINY. These included 2 non-Hodgkin lymphomas (NHL1 and NHL 2) and a classical Hodgkin lymphoma (cHL 3). Results are from a representative experiment with triplicate reactions for each sample, out of 6 to 8 independent experiments for each specimen (data shown in aggregate in supplemental Figure 1 and supplemental Table 2). All “undetermined” values, meaning no amplification, were falsely set to 40 minutes for the purpose of visualization and averaging. (C) EBER in situ hybridization was performed to evaluate positivity for EBV in the tonsil specimens. A representative example from tonsil 7 is shown. Original magnification is ×10.
EBV cells-to-LAMP assay is compatible with a point-of-care approach. (A) FNA samples obtained from EBV+ cell line xenografts in mice were tested using cells-to-LAMP followed by analysis with TINY (left), with representative cells-to-LAMP results output by TINY software of EBV+ and EBV- cell lines shown (top right). The blue bars in this image indicate the 4 EBV+ cell lines that amplified before the threshold time. Four independent aspirates were taken from each of 2 tumors per cell line (bottom right). NA denotes that the negative control cell line, Ramos, was not amplifiable. (B) Fresh tonsil specimens were used to perform an ex vivo FNA with cells-to-LAMP followed by TINY. Tissue specimens from 3 EBV+ lymphoma specimens were used for DNA extraction and tested in parallel by LAMP and analyzed with TINY. These included 2 non-Hodgkin lymphomas (NHL1 and NHL 2) and a classical Hodgkin lymphoma (cHL 3). Results are from a representative experiment with triplicate reactions for each sample, out of 6 to 8 independent experiments for each specimen (data shown in aggregate in supplemental Figure 1 and supplemental Table 2). All “undetermined” values, meaning no amplification, were falsely set to 40 minutes for the purpose of visualization and averaging. (C) EBER in situ hybridization was performed to evaluate positivity for EBV in the tonsil specimens. A representative example from tonsil 7 is shown. Original magnification is ×10.
We also assessed whether this method could distinguish tumor-associated EBV, which occurs in a large proportion of cells in a biopsy, from EBV that may be present as spurious infection in reactive lymphadenopathies. We performed cells-to-LAMP using primary patient samples where EBV status was determined in tissue sections by in situ hybridization for EBER (supplemental Table 2). The samples included: 7 human hyperplastic tonsils, 2 non-Hodgkin lymphomas (NHLs 1 and 2) with many EBV+ cells, and 1 classical Hodgkin lymphoma (cHL 3) with few EBV+ Hodgkin/Reed Sternberg cells. While NHLs 1 and 2 showed consistent amplification at <30 minutes, cHL 3 showed variable to no amplification (Figure 2B; supplemental Figure 1; supplemental Table 2). The tonsil specimens showed either no amplification in multiple independent experiments or variable amplification, but with a mean amplification exceeding the 30-minute threshold. Some tonsils had scattered EBV+ cells (Figure 2C), explaining sporadic amplification. We conclude EBV+ NHL can be distinguished from reactive lymphadenopathy, even if containing EBV+ cells. However, given spurious amplification in the EBV+ tonsils, at least triplicate reactions would better predict whether a given case is eBL or a reactive lymphadenopathy.
In summary, we developed a simple method for detecting EBV in FNA samples without complex equipment, where EBV can be detected from as few as 50 cells without DNA extraction using a simple, thermostable buffer. We evaluated the cells-to-LAMP approach using an innovative diagnostic device recently developed and clinically tested by our groups, showing high sensitivity and specificity for the detection of KSHV/HHV-8 in KS tissue biopsies.7 Since KS most frequently affects the skin, biopsy tissue digestion and DNA extraction were required when using TINY for KSHV detection, making time to diagnosis ∼3 hours.7 Because eBL cells can be obtained by FNA instead of a tissue biopsy, the cells-to-LAMP assay avoids DNA extraction and allows diagnosis of an EBV+ lymphoma in <30 minutes. This makes integration of the cells-to-LAMP assay with detection by TINY a highly attractive approach for a rapid, decentralized preliminary diagnosis of BL in endemic regions. Assessment of sensitivity and specificity for eBL in diagnostic settings in Africa will be necessary before clinical implementation, but this technology should allow prompt referral and earlier treatment of patients. Our data provide a practical approach for rapid point-of-care diagnosis of eBL based on EBV detection, and this method may be extendable to additional pathogens present in cytological specimens.
Acknowledgments
This study was supported in part by the National Institutes of Health/National Cancer Institute grant UH2/UH3CA202723 to E.C. and D.E.
Contribution: E.C. and A.G. conceived the project; A.G. designed experiments; A.G., J.A., and C.G. performed experiments; R.S. and D.E. designed and built TINY; R.S. and V.K. developed TINY software; and D.E. and E.C. advised all personnel.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ethel Cesarman, Department of Pathology & Laboratory Medicine, Weill Cornell Medicine, 1300 York Ave, Rm C-410, New York, NY 10065; e-mail: ecesarm@med.cornell.edu.
References
Author notes
Requests for data sharing may be submitted to Ethel Cesarman (ecesarm@med.cornell.edu).
The full-text version of this article contains a data supplement.