Key Points
Pre-HCT mosaicism is related to increased relapse risk and lower survival after unrelated HCT, independent of cytogenetics at diagnosis.
Pre-HCT mosaicism could be a useful clinical tool to guide risk stratification in acute lymphoblastic leukemia patients.
Introduction
Mosaicism, detectable in peripheral blood, can be an important predictor for the development of a subsequent hematological malignancy.1,2 The potential utility of mosaicism in predicting relapse and survival after allogeneic hematopoietic cell transplant (HCT) in patients with hematologic malignancies is not clear. We sought to determine whether mosaicism contributed independent information on acute lymphoblastic leukemia (ALL) patient survival and risk of relapse 1 year post–allogeneic unrelated HCT.
Methods
HumanOmniExpress-12v1_A BeadChip SNP Array data were used to detect mosaicisms ≥ 2 Mb based on the log2 relative probe intensity ratio (LRR) and B-allele frequency for each single nucleotide polymorphism (SNP) measured in DNA from 773 ALL patients who received HCT between 2000 and 2011. Patients were from Determining the Influence of Susceptibility COnveying Variants Related to one-Year mortality after BMT (DISCOVeRY-BMT) cohorts, described previously.3-6 DNA was extracted from recipient peripheral blood samples taken immediately prior to the start of conditioning before allogeneic HCT for ALL. Mosaicisms were defined as copy-number gain (LRR > 0), copy-number loss (LRR < 0), or copy-neutral loss of heterozygosity when a deviation occurred from B-allele frequency heterozygosity without LRR changes.2,7-9
We compared differences in clinical and transplant-related factors between patients with and without detectable mosaicisms. Overall survival (OS) probability and cumulative incidence of relapse at 1 year post-HCT were calculated using the Kaplan-Meier estimator and cumulative incidence function, respectively. We selected clinical and demographic covariates for inclusion using bidirectional stepwise Cox proportional-hazard models of OS and competing risk models of relapse. The association between mosaicism and OS was evaluated using a multivariable Cox proportional-hazard models. For disease relapse, competing risk models were used, with transplant-related mortality treated as a competing event. Stratified survival analyses were performed by disease status, early and intermediate (in complete remission [CR]) vs advanced (not in CR), and cytogenetic status (normal vs abnormal) at diagnosis, adjusting for other patient and transplant characteristics. Statistical analyses were conducted using R 3.6.1.
The biospecimen protocol was approved by the National Marrow Donor Program’s Institutional Review Board (IRB), as well as the individual transplant and donor centers’ IRBs. Recipients and donors were not compensated for their participation. This study was reviewed and approved by the Roswell Park Comprehensive Cancer Center IRB.
Results and discussion
In our study, the median age at HCT was 27.8 years (interquartile range [IQR], 16.0-43.0); 622 patients (80.5%) were in CR pre-HCT. The 34 patients with mosaicism were older at transplant (median, 35.5 years; IQR, 20.0-50.0) and had advanced disease (47%) compared with those without mosaicisms (18%) (supplemental Table 1). Cytogenetics at ALL diagnosis were available for 669 patients (86.5%); 45% of those had abnormalities. The 1-year post-HCT OS in our study was 58.6%, and the relapse rate was 27.0%. The above clinical data show DISCOVeRY-BMT represents a more aggressive subtype of ALL.6
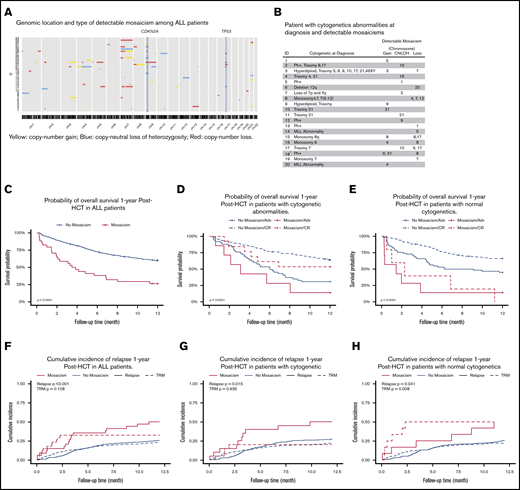
We observed 82 mosaicisms ≥ 2 Mb in 34 ALL patients (median, 1; range, 1-9; Figure 1A). The median proportion of affected cells was 61% (IQR, 39-93), and copy-number loss was the most frequently observed mosaic event (NEvents = 47; 57.3%; supplemental Figure 1). Copy-number losses on chromosomes 7, 9, and 17 were the most frequently detected events, which were largely independent, with the exception of losses on chromosomes 9 and 17, which co-occurred in 3 patients. Although almost all cytogenetic abnormalities are independent of mosaicism, there were some expected correlations. For example, Figure 1B shows a chromosome 21 copy-number gain by mosaicism in 1 patient with trisomy 21 by conventional cytogenetics at diagnosis.
Characteristics of detectable pre-HCT mosaicism in ALL patients. (A) Genomic location and type of detectable mosaicism among ALL patients. (B) Patient with cytogenetic abnormalities at diagnosis and detectable mosaicisms. 1No detailed cytogenetics report at diagnosis. 2Complex chromosome aberration in the major histocompatibility complex region. (C) Probability of OS 1 year post-HCT in ALL patients. (D) Probability of OS 1 year post-HCT in patients with cytogenetic abnormalities. (E) Probability of OS 1 year post-HCT in patients with normal cytogenetics. (F) Cumulative incidence of relapse 1 year post-HCT in ALL patients. (G) Cumulative incidence of relapse 1 year post-HCT in patients with cytogenetic abnormalities. (H) Cumulative incidence of relapse 1 year post-HCT in patients with normal cytogenetics. TRM, transplant-related mortality.
Characteristics of detectable pre-HCT mosaicism in ALL patients. (A) Genomic location and type of detectable mosaicism among ALL patients. (B) Patient with cytogenetic abnormalities at diagnosis and detectable mosaicisms. 1No detailed cytogenetics report at diagnosis. 2Complex chromosome aberration in the major histocompatibility complex region. (C) Probability of OS 1 year post-HCT in ALL patients. (D) Probability of OS 1 year post-HCT in patients with cytogenetic abnormalities. (E) Probability of OS 1 year post-HCT in patients with normal cytogenetics. (F) Cumulative incidence of relapse 1 year post-HCT in ALL patients. (G) Cumulative incidence of relapse 1 year post-HCT in patients with cytogenetic abnormalities. (H) Cumulative incidence of relapse 1 year post-HCT in patients with normal cytogenetics. TRM, transplant-related mortality.
OS at 1 year post-HCT in patients with or without a mosaic event was 26.5% or 60.1%, respectively (P = 8.2 × 10−8; Figure 1C). Patients with mosaicism, stratified by cytogenetics, had significantly lower survival (Figure 1D-E), with the lowest survival among patients with normal cytogenetics compared with all other patients (OS, 8.3% vs 60.6%; P = 3.0 × 10−10). Patients with a pre-HCT mosaicism also had an increased risk for disease relapse compared with those without (50% vs 26%; P = .00067) (Figure 1F). Mosaicisms were associated with relapse risk in patients with cytogenetic abnormalities (P = .015) and normal cytogenetics (P = .041) at diagnosis (Figure 1G-H); this translated into a significant association with a risk for transplant-related mortality in the latter group (P = .008).
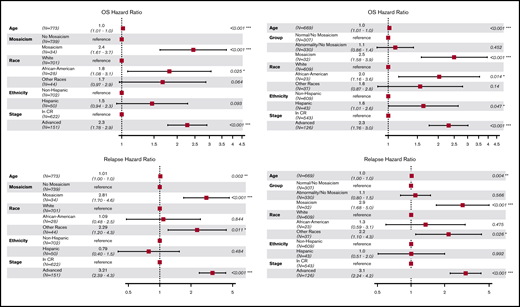
Multivariable models of OS and relapse included age, race, ethnicity, and disease stage (CR or advanced) as covariates. Figure 2 shows that patients with a mosaic event had a 2.5-fold increased risk for death (hazard ratio [HR], 2.50; 95% confidence interval [CI], 1.58-3.90; P = 8.6 × 10−5), and mosaicism remained significant even among patients in CR (HR, 2.24; 95% CI, 1.21-4.14; P = .01). These OS associations are likely driven by the risk of relapse (HR, 2.81; 95% CI, 1.70-4.60; P = 5.9 × 10−5), which remains significant when stratified by cytogenetics (HR, 2.90; 95% CI, 1.68-5.00; P = .0001; Figure 2).
Hazard ratios of OS and disease relapse post-HCT. Forest plot for OS HR (upper left panel) and relapse HR (lower left panel) adjusted for age, race, ethnicity, and disease stages. Forest plot for OS HR (upper right panel) and relapse HR (lower right panel) adjusted for age, race, ethnicity, disease stages, and cytogenetics; only patients with known cytogenetics at diagnosis were included (N = 669). *P < .05, **P < .01, ***P < .001.
Hazard ratios of OS and disease relapse post-HCT. Forest plot for OS HR (upper left panel) and relapse HR (lower left panel) adjusted for age, race, ethnicity, and disease stages. Forest plot for OS HR (upper right panel) and relapse HR (lower right panel) adjusted for age, race, ethnicity, disease stages, and cytogenetics; only patients with known cytogenetics at diagnosis were included (N = 669). *P < .05, **P < .01, ***P < .001.
We identified copy-number loss of regions on chromosomes 7, 9, and 17 (n = 15 patients). Copy-number loss on chromosome 7 was the most frequently observed event in patients who died as a result of disease relapse within 1 year post-HCT (supplemental Figure 2), with most losses on the short arm (p) of chromosome 7. Deletions of 7p have been correlated with worse event-free survival in pediatric patients with ALL.10 Mosaic loss of 7p might also be an important indicator of poor OS across the lifespan; 5 of the 6 patients with loss of 7p were older than 18 years. Higher mortality was also attributable to loss of the 9p21.3 locus, which harbors CDKN2A, a known tumor suppressor and key candidate ALL-susceptibility gene. The 9p21.3 loss was seen in all patients (n = 5) with chromosome 9p losses; all had normal cytogenetics at diagnosis. Studies showed that inactivation of CDKN2A was associated with worse survival in children and adults.11-13 Lastly, we found that all patients with copy-number loss of 17p13.1 (n = 4), which harbors the tumor suppressor TP53, died within 1 year post-HCT (supplemental Figure 2A); TP53 mutations are known to correlate with poor survival in ALL patients.14,15 These 3 mosaicisms, not captured by conventional ALL-specific cytogenetics at diagnosis, may identify patients with more aggressive disease and, subsequently, higher mortality due to genetic diversification.16 However, when excluding these losses, mosaicism remained significantly associated with poor OS and higher relapse risk.
SNP arrays have been used to inform prognosis for a number of pediatric and adult ALL subtypes17,18 Herein, we propose that mosaicism, estimated using SNP array data from pretransplant peripheral blood, is associated with poorer survival and higher disease relapse at 1 year post-HCT, regardless of disease stage; this association is even more pronounced in patients with normal cytogenetics. Although detectable mosaicism is not a surrogate for CR or advanced disease, it may be a plausible marker of measurable residual disease because an elevated risk for relapse was also observed.19,20 Thus, it would be of interest to assess the relationship between measurable residual disease, measured using flow cytometry, and mosaicism in future studies.
This study adds to prior evidence about the possible prognostic value of pre-HCT mosaicisms detected by SNP arrays in HCT outcomes, and it is the first study in ALL to show a relationship with poor survival. However, despite our relatively large sample size and high frequency of detectable mosaicism (4.4% compared with <1% in the general population2,21 ), the complexity of cytogenetic risk in the disease limits our analysis. Although Center for International Blood and Marrow Transplant Research data and biospecimens allow us to capture ∼99% of 8/8 unrelated donor allogeneic bone marrow transplants performed for ALL in the United States, future studies in patients undergoing related or haploidentical transplants across different races/ethnicities are needed to better classify patient risk post-HCT that is attributable to mosaicisms.
All deidentified individual participant data that underlie the reported results are readily available at the Center for International Blood and Marrow Transplant (www.cibmtr.org) by e-mailing info-request@mcw.edu or sucheston-campbell.1@osu.edu. Informed consent is not compliant with the Genomic Data Sharing Policy for 140 samples. Samples that adhere to the Genomic Data Sharing policy are in the process of being uploaded to the database of Genotypes and Phenotypes.
Acknowledgments
The authors acknowledge the research contributions of the Cancer Genomics Research Laboratory, which is funded by federal funds from the National Institutes of Health National Cancer Institute under NCI Contract 75N910D00024.
This work was supported by the intramural program of the National Cancer Institute National Institutes of Health and by grants from the National Institutes of Health National Heart Lung and Blood Institute (1R01HL102278) and National Institutes of Health National Cancer Institute (1R03CA188733).
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Authorship
Contribution: Yiwen Wang analyzed and interpreted data and wrote the manuscript; W.Z. performed mosaicism analyses of data; J.W., E.K., H.T., Youjin Wang, L.J.M., H.A.K., M.J.M., and M.Y. analyzed and interpreted data; C.A.H., X.S., D.V.D.B., and L.P. performed genotyping and quality control; S.J.C., S.J.L, A.I.C.-G., and S.M.G. provided feedback on the approach and analyses; T.E.H. acquired data and provided feedback on the manuscript; L.E.S.-C. designed the study, acquired, analyzed, and interpreted data, and wrote the manuscript; and all authors revised the manuscript, contributed critically important intellectual content, and approved the final version of the manuscript.
Conflict-of-interest disclosure: L.J.M. is an Associate Editor for Molecular Genetics & Genomic Medicine and receives honoraria for that work and is a US government employee. The remaining authors declare no competing financial interests.
Correspondence: Lara E. Sucheston-Campbell, The Ohio State University, College of Pharmacy, 604 Riffe Building, Columbus, OH 43210; e-mail: lara.sucheston-campbell@osumc.edu.
References
Author notes
The full-text version of this article contains a data supplement.