Key Points
Activating anti-Spacer or anti-CUB antibodies disrupt the Spacer-CUB interaction causing ADAMTS13 to adopt an open conformation.
Disruption of the Spacer-CUB interaction induces conformational changes in the metalloprotease domain that augment proteolytic function.
Abstract
Plasma ADAMTS13 circulates in a folded conformation that is stabilized by an interaction between the central Spacer domain and the C-terminal CUB (complement components C1r and C1s, sea urchin protein Uegf, and bone morphogenetic protein-1) domains. Binding of ADAMTS13 to the VWF D4(-CK) domains or to certain activating murine monoclonal antibodies (mAbs) induces a structural change that extends ADAMTS13 into an open conformation that enhances its function. The objective was to characterize the mechanism by which conformational activation enhances ADAMTS13-mediated proteolysis of VWF. The activating effects of a novel anti-Spacer (3E4) and the anti-CUB1 (17G2) mAbs on the kinetics of proteolysis of VWF A2 domain fragments by ADAMTS13 were analyzed. mAb-induced conformational changes in ADAMTS13 were investigated by enzyme-linked immunosorbent assay. Both mAbs enhanced ADAMTS13 catalytic efficiency (kcat/Km) by ∼twofold (3E4: 2.0-fold; 17G2: 1.8-fold). Contrary to previous hypotheses, ADAMTS13 activation was not mediated through exposure of the Spacer or cysteine-rich domain exosites. Kinetic analyses revealed that mAb-induced conformational extension of ADAMTS13 enhances the proteolytic function of the metalloprotease domain (kcat), rather than augmenting substrate binding (Km). A conformational effect on the metalloprotease domain was further corroborated by the finding that incubation of ADAMTS13 with either mAb exposed a cryptic epitope in the metalloprotease domain that is normally concealed when ADAMTS13 is in a closed conformation. We show for the first time that the primary mechanism of mAb-induced conformational activation of ADAMTS13 is not a consequence of functional exosite exposure. Rather, our data are consistent with an allosteric activation mechanism on the metalloprotease domain that augments active site function.
Introduction
The plasma metalloprotease ADAMTS13 consists of 14 domains: a metalloprotease domain (MP), a disintegrin-like domain (Dis), a first thrombospondin type-1 repeat (TSP1), a cysteine-rich domain (Cys-rich), a Spacer domain, 7 additional thrombospondin type-1 repeats (TSP2-TSP8), and 2 CUB (complement components C1r and C1s, sea urchin protein Uegf, and bone morphogenetic protein-1) domains (CUB1-CUB2).1 ADAMTS13 functions by specifically proteolyzing von Willebrand factor (VWF) multimers in a shear-dependent manner. In this way, it regulates the multimeric size and platelet-tethering function of VWF. Proteolysis involves multiple conformation-dependent exosite interactions between the enzyme and substrate that determine both the timing and efficiency of proteolysis.2
VWF normally circulates in a folded conformation that conceals the GPIbα-binding site in its A1 domain. The adjacent A2 domain that harbors the ADAMTS13 cleavage site is also folded such that the scissile bond and exosite interaction sites are concealed, making it resistant to ADAMTS13 proteolysis. However, when VWF unravels in response to elevated shear forces (at sites of vessel damage, upon secretion, and so forth), it unfolds to facilitate platelet capture. This process can also cause the VWF A2 domain to unravel into a linear conformation that exposes the ADAMTS13 cleavage site and exosite binding regions.2,3
Structural investigation of the native ADAMTS13 conformation by small-angle X-ray scattering4 and by electron microscopy4-6 has revealed a closed or hairpin-like conformation, in which the C-terminal TSP2-CUB2 domains fold back over the N-terminal domains of the molecule. This conformation is stabilized by interactions between the CUB domain and the Spacer domain, involving both Y661 and Y665.7 Three linker regions in the TSP2-CUB2 domains likely provide the necessary flexibility to the C-terminal tail of ADAMTS13 to enable this hairpin-like conformation.5 The MP domain of ADAMTS13 also adopts a latent conformation that prevents off-target proteolysis.8
Closed ADAMTS13 first interacts with VWF via the D4(-CK) region, which opens ADAMTS13 up. This enhances ADAMTS13 enzyme activity by ∼twofold against short VWF A2 domain fragment substrates.4,6,7,9 The Spacer and Cys-rich domains then interact with the unfolded VWF A2 domain, bringing the enzyme and cleavage site into proximity. The Dis domain exosite then engages VWF, which transduces an allosteric change in the MP domain that converts the latent form into its active conformation. This allosteric, substrate-assisted activation of the MP domain enhances ADAMTS13 proteolytic function by allowing accommodation of the scissile bond into the active site cleft.8
Disruption of the intramolecular Spacer-CUB interaction that stabilizes the closed ADAMTS13 conformation causes a structural transition to an open/extended form. This so-called conformational activation of ADAMTS13, which enhances the activity of ADAMTS13 ∼two- to 10-fold,4,6,7 can be induced by low pH (pH 6),4 mutations in the Spacer domain that disrupt the interaction with the CUB domains, or certain murine monoclonal antibodies (mAbs) that induce ADAMTS13 opening.5,6,10 Truncated ADAMTS13 that lacks the C-terminal T2-CUB2 domains (MDTCS) is therefore hyperactive compared with full-length ADAMTS13 because of the loss of the CUB domain interaction.4,5 Binding of ADAMTS13 to the VWF D4(-CK) domains4,6,7,11 was originally considered to colocalize ADAMTS13 and VWF. However, the finding that this interaction also induces conformational activation suggests that this also contributes to the specificity and efficiency of ADAMTS13 function.
The activating effect of opening ADAMTS13 has been proposed to unmask the functional exosite in the Spacer domain thought to be otherwise sterically hindered by the Spacer-CUB interaction and, in turn, increase the binding affinity for the VWF A2 domain.10,12 However, this has not been definitively characterized. A reason for this is that the opening of ADAMTS13 by the isolated D4 domain requires very high concentrations of D4, suggestive of a low-affinity interaction.4,9 To circumvent this, we generated anti-ADAMTS13 mAbs and screened them for their ability to conformationally activate ADAMTS13 for use as tools to characterize the functional effects upon ADAMTS13 opening.
Materials and methods
Anti-ADAMTS13 mAbs 3H9, 17G2, and 6A6
The mAbs 3H9, 17G2, and 6A6 have been described previously.5,13 The 3H9 mAb is directed against an epitope in the MP domain. The 6A6 mAb recognizes a conformationally sensitive epitope in the MP domain that is cryptic in the closed conformation of ADAMTS13 and is specifically exposed on unfolding. The activating anti-CUB1 17G2 mAb induces the open conformation of ADAMTS13.5
Expression and purification of recombinant ADAMTS13 variants VWF96, VWF87(ΔSpacer), and VWF96-Cys
Expression and purification of the different ADAMTS13 variants (supplemental Table 1; supplemental Figure 1) used for immunization and epitope mapping and expression of ADAMTS13 and MDTCS, VWF96, VWF87(ΔSpacer), and VWF96-Cys variants (supplemental Table 1; supplemental Figure 2) used for kinetic analysis are described in the supplemental Data.
Immunization strategy, generation, and epitope mapping of anti-MDTCS mAbs
Anti-MDTCS mAbs were developed by immunizing BALB/c mice (Janvier Labs) with MDTCS(V5-6xHis). mAbs were screened for MDTCS binding, and additional epitope mapping was carried out as described in the supplemental Data.
ADAMTS13 proteolysis of FRETS-VWF73
The effect of the anti-MDTCS mAbs on the activity of plasma ADAMTS13 was measured using the FRETS-VWF73 substrate (Peptides International),14 as described in the supplemental Data.
Kinetics of VWF96, VWF96-Cys, and VWF87(ΔSpacer) proteolysis
Purified ADAMTS13 (0.6-11 nM) or MDTCS(6xHis) in concentrated conditioned media (estimated ∼1.2 nM) were incubated with or without the mAbs (final concentration, 10 µg/mL) in reaction buffer (20 mM Tris at pH 7.55, 50 mM NaCl, 5 mM CaCl2, 1% bovine serum albumin) at 37°C for 15 minutes. Reactions were initiated by adding VWF96, VWF96-Cys, or VWF87(ΔSpacer) (final concentration, 0.5 μM). Reaction subsamples were stopped between 0 and 90 minutes in stopping buffer (20 mM Tris at pH 7.55, 50 mM NaCl, 25 mM EDTA, 1% bovine serum albumin). Stopped reaction subsamples were analyzed by enzyme-linked immunosorbent assay (ELISA),8 as described in the supplemental Data. The catalytic efficiency of proteolysis was calculated, and Michaelis Menten kinetic analysis8 was performed as described in the supplemental Data.
Analysis of conformational changes in the ADAMTS13 MP domain induced by mAb binding
mAb-induced conformational changes in the ADAMTS13 MP domain were studied by ELISA,5 as described in the supplemental Data. This assay uses the 6A6 mAb, which detects a conformationally sensitive epitope in the MP domain of ADAMTS13 that is only available when in the open conformation, and that is cryptic in the closed conformation.5
Statistical analysis
Statistical analysis was performed using 1-way analysis of variance with multiple comparison test. Probabilities of P < .05 were considered statistically significant. All statistical analysis was performed using GraphPad Prism software (GraphPad Software, Inc., San Diego, CA). Data are expressed as mean ± standard deviation (SD).
Results
Development and characterization of a murine activating anti-Spacer mAb
mAb-induced disruption of the Spacer-CUB domain interaction opens ADAMTS13, and in so doing, enhances its proteolytic activity against VWF.4-6 Multiple anti-ADAMTS13 mAbs that activate the enzyme have been identified. These are all directed against either the C-terminal TSP repeats or the CUB domains of ADAMTS13.4,5 However, to date, no activating anti-Spacer mAbs have been identified despite the Spacer domain directly contributing to the interdomain interactions that dictate the conformational activation of ADAMTS13. For this reason, we aimed to generate anti-Spacer mAbs and screen for those that induce conformational activation of ADAMTS13.
After immunization of BALB/c mice with MDTCS(V5-6xHis), 43 murine mAbs were purified that detected immobilized MDTCS(V5-6xHis) in ELISA assays (Figure 1A). Domain specificity of these mAbs was analyzed by ELISA, using the MP(FLAG), MD(FLAG), MDT(FLAG), and MDTCS(FLAG) variants (data not shown), which revealed 8 anti-MP domain mAbs, 4 anti-TSP1 domain mAbs, and 31 anti-Cys/Spacer domain mAbs (Figure 1A).
Generation of anti-MDTCS mAbs. (A) Binding of newly developed mAbs to immobilized MDTCS(V5-6xHis) was investigated by ELISA. Bound anti-MDTCS mAbs were detected using HRP-labeled goat anti-mouse antibodies. Data (mean ± SD; n = 3) were expressed relative to the binding of the anti-MP domain mAb 3H9 to MDTCS(V5-6xHis), which was set as 1 (data not shown). (B) The influence of anti-Cys/Spacer domain mAbs on plasma ADAMTS13 activity was studied using the FRETS-VWF73 assay (mean ± SD; n = 3). Differences between ADAMTS13 activity were statistically analyzed using analysis of variance with multiple comparison test. *P < .01. Activating and inhibitory mAbs are shown as purple and light green bars, respectively. Nonactivating/noninhibitory mAbs are shown as blue bars.
Generation of anti-MDTCS mAbs. (A) Binding of newly developed mAbs to immobilized MDTCS(V5-6xHis) was investigated by ELISA. Bound anti-MDTCS mAbs were detected using HRP-labeled goat anti-mouse antibodies. Data (mean ± SD; n = 3) were expressed relative to the binding of the anti-MP domain mAb 3H9 to MDTCS(V5-6xHis), which was set as 1 (data not shown). (B) The influence of anti-Cys/Spacer domain mAbs on plasma ADAMTS13 activity was studied using the FRETS-VWF73 assay (mean ± SD; n = 3). Differences between ADAMTS13 activity were statistically analyzed using analysis of variance with multiple comparison test. *P < .01. Activating and inhibitory mAbs are shown as purple and light green bars, respectively. Nonactivating/noninhibitory mAbs are shown as blue bars.
We assessed the influence of the 31 anti-Cys/Spacer domain mAbs on plasma ADAMTS13 activity, using the FRETS-VWF73 substrate. Twenty of 31 anti-Cys/Spacer domain mAbs (1C4, 1D11, 3A5, 3B11, 4E2, 5A3, 5D6, 8H6, 9F1, 9F5, 10F1, 11D2, 13A10, 14A10, 15D1, 15D11, 15F3, 17G9, 17H12, and 19A6) exerted no effect on the activity of ADAMTS13 (Figure 1B, blue bars), whereas 2 of the 31 anti-Cys/Spacer domain mAbs (1C9 and 7D5) were inhibitory, significantly reducing ADAMTS13 activity against FRETS-VWF73 (Figure 1B, light green bars). Interestingly, 9 of the 31 anti-Cys/Spacer domain mAbs (3B8, 3E4, 8G1, 9A3, 11C11, 11D5, 13C3, 14B6, and 15A6) significantly enhanced proteolysis of FRETS-VWF73 by 1.4- to twofold (Figure 1B, purple bars), consistent with these mAbs inducing conformational activation.
Further epitope mapping was performed, using the Spacer-CUB2(V5-6xHis) variant in ELISA assays, in search of activating and nonactivating/noninhibitory mAbs that specifically target the Spacer domain of ADAMTS13 (data not shown). From this, the activating anti-Spacer domain mAb 3E4 (Figure 1B; 170.4% ± 16.7% activity; P < .001) was selected to analyze the kinetics of ADAMTS13 conformational activation. The nonactivating, noninhibitory anti-Spacer mAb 15D1 (Figure 1B; 97.6% ± 6.3% activity; P = ns) was selected as a negative control for kinetic studies.
The anti-Spacer mAb 3E4 and anti-CUB1 mAb 17G2 enhance the catalytic efficiency of ADAMTS13
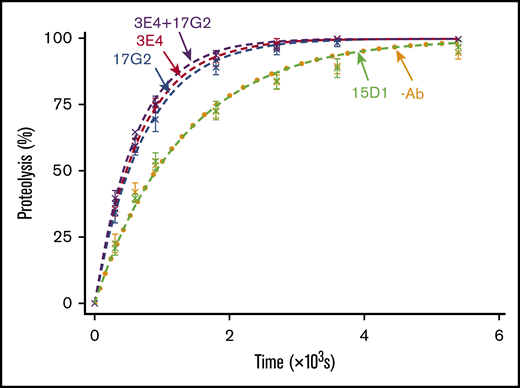
We quantified the mAb-induced enhancement of recombinant ADAMTS13 activity kinetically, using VWF96, a new recently described VWF A2 domain fragment.8 VWF96 spans the VWF A2 domain region Gly1573-Arg1668, which contains the ADAMTS13 scissile bond (Tyr1605-Met1606), and the essential complementary binding sites for ADAMTS13 exosites (supplemental Figure 2). We have previously described an ELISA-based assay that exploits the N-terminal HisG-SUMO and C-terminal HSV tags to detect full-length, uncleaved VWF96. This assay can be used to analyze the time course of VWF96 proteolysis by ADAMTS13 for calculating the catalytic efficiency (kcat/Km).8 The effects of the anti-Spacer domain mAbs 3E4 (activating) and 15D1 (nonactivating, noninhibitory), as well as the previously characterized anti-CUB1 domain mAb 17G2 (activating),5 were studied kinetically using this assay.
In the absence of mAbs, as well as in the presence of the nonactivating, noninhibitory anti-Spacer domain mAb 15D1, VWF96 was proteolyzed by ADAMTS13(V5-6xHis) with a kcat/Km of 1.32 × 106 M−1⋅s−1 and 1.29 × 106 M−1⋅s−1, respectively (n = 7; Figure 2). This is consistent with the previously determined catalytic efficiency for VWF96 proteolysis (kcat/Km = 1.11-1.42 × 106 M−1⋅s−1).8 The anti-Spacer domain mAb 3E4 enhanced the catalytic efficiency of proteolysis by ∼1.9-fold (kcat/Km = 2.52 × 106 M−1⋅s−1; n = 7; Figure 2), consistent with the similar enhancement of FRETS-VWF73 proteolysis (Figure 1B). Addition of the anti-CUB1 domain mAb 17G2 also resulted in a ∼1.8-fold enhancement in kcat/Km (kcat/Km = 2.37 × 106 M−1⋅s−1; n = 7; Figure 2). The presence of both activating mAbs 3E4 and 17G2 in the reaction did not cause further enhancement on the rate of proteolysis (kcat/Km = 2.75 × 106 M−1⋅s−1; n = 3; Figure 2). The absence of synergism suggests that the anti-Spacer domain mAb 3E4 and the anti-CUB1 domain mAb 17G2 activate ADAMTS13 through the same mechanism.
VWF96 proteolysis by ADAMTS13 in the presence and absence of mAbs. After preincubation of recombinant ADAMTS13(V5-6xHis) (0.6 nM) with no mAb (orange; n = 7); the noninhibitory, nonactivating anti-Spacer domain mAb 15D1 (light green; n = 7); the activating anti-Spacer domain mAb 3E4 (red; n = 7); the activating anti-CUB1 domain mAb 17G2 (blue; n = 7); or a cocktail of the 2 activating mAbs 3E4/17G2 (purple; n = 3), VWF96 (0.5 μM) was added to initiate the reaction. Reaction subsamples, stopped between 0 and 90 minutes, were analyzed by ELISA. Fitted time course data show the percentage of VWF96 proteolysis as a function of time (mean ± standard error of the mean).
VWF96 proteolysis by ADAMTS13 in the presence and absence of mAbs. After preincubation of recombinant ADAMTS13(V5-6xHis) (0.6 nM) with no mAb (orange; n = 7); the noninhibitory, nonactivating anti-Spacer domain mAb 15D1 (light green; n = 7); the activating anti-Spacer domain mAb 3E4 (red; n = 7); the activating anti-CUB1 domain mAb 17G2 (blue; n = 7); or a cocktail of the 2 activating mAbs 3E4/17G2 (purple; n = 3), VWF96 (0.5 μM) was added to initiate the reaction. Reaction subsamples, stopped between 0 and 90 minutes, were analyzed by ELISA. Fitted time course data show the percentage of VWF96 proteolysis as a function of time (mean ± standard error of the mean).
mAb-mediated activation is dependent on the opening of ADAMTS13 C-terminal tail
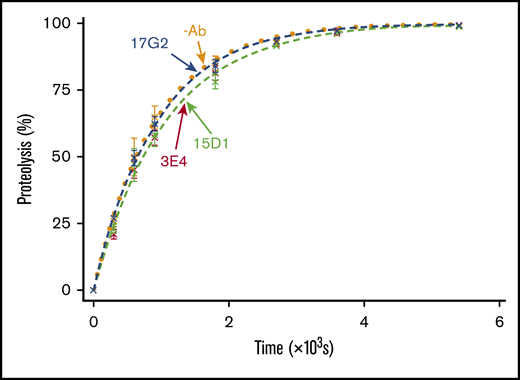
To ascertain whether the mAb-mediated enhancement of ADAMTS13 activity was dependent on the disengagement of the C-terminal tail from the Spacer domain, we assessed the ability of the mAbs to enhance the activity of MDTCS, which lacks the C-terminal tail and therefore cannot be conformationally enhanced. VWF96 proteolysis by MDTCS(6xHis) was monitored in the presence and absence of the mAbs. The anti-CUB1 domain mAb 17G2 served as an additional negative control, as its binding site (CUB1 domain) is not present in MDTCS(6xHis). No significant differences in VWF96 proteolysis MDTCS(6xHis) were associated with the presence of any of the mAbs (Figure 3). These results demonstrate that the activating effect of the mAbs 3E4 and 17G2 is dependent on the presence of the C-terminal tail of ADAMTS13; again, consistent with the effect being mediated by conformational activation.
VWF96 proteolysis by MDTCS in the presence and absence of mAbs. After preincubation of MDTCS(6xHis) with no mAb (orange; n = 3); the noninhibitory, nonactivating anti-Spacer domain mAb 15D1 (light green; n = 3); the activating anti-Spacer domain mAb 3E4 (red; n = 3); or the activating anti-CUB1 domain mAb 17G2 (blue; n = 3), VWF96 (0.5 μM) was added to initiate the reaction. Reaction subsamples, stopped between 0 and 90 minutes, were analyzed by ELISA. Fitted time course data show the percentage of VWF96 proteolysis as a function of time (mean ± standard error of the mean).
VWF96 proteolysis by MDTCS in the presence and absence of mAbs. After preincubation of MDTCS(6xHis) with no mAb (orange; n = 3); the noninhibitory, nonactivating anti-Spacer domain mAb 15D1 (light green; n = 3); the activating anti-Spacer domain mAb 3E4 (red; n = 3); or the activating anti-CUB1 domain mAb 17G2 (blue; n = 3), VWF96 (0.5 μM) was added to initiate the reaction. Reaction subsamples, stopped between 0 and 90 minutes, were analyzed by ELISA. Fitted time course data show the percentage of VWF96 proteolysis as a function of time (mean ± standard error of the mean).
The activating effect of the mAbs is independent of the ADAMTS13 Spacer and Cys-rich domain exosite interactions with VWF
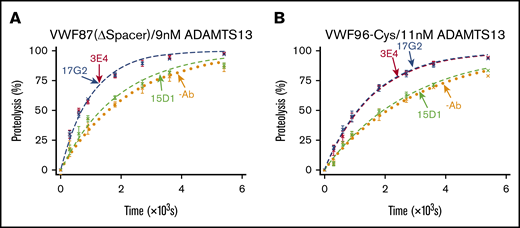
The current model of conformational activation of ADAMTS13 suggests that disruption of the Spacer-CUB interaction unmasks the Spacer domain exosite, enabling its interaction with the VWF A2 domain. To study the dependency of the conformational activation mechanism on the ADAMTS13 Spacer domain exosite interaction, we assessed the ability of the activating mAbs to enhance the rate of VWF87(ΔSpacer) variant proteolysis by ADAMTS13(V5-6xHis). This substrate has the entire Spacer domain exosite interaction ablated, as it lacks the functional binding site (Glu1660-Arg1668) for the Spacer domain exosite of ADAMTS13 (supplemental Figure 2).8 In the absence of mAbs, the VWF87(ΔSpacer) variant was cleaved by ADAMTS13(V5-6xHis) with a kcat/Km of 0.05 × 106 M−1⋅s−1 (n = 3; Figure 4A). The ∼26-fold lower kcat/Km compared with the proteolysis of VWF96 (kcat/Km = 1.32 × 106 M−1⋅s−1; Figure 2) is consistent with the expected reduction in the rate of proteolysis as a result of the ablation of the Spacer domain exosite interaction. The nonactivating, noninhibitory anti-Spacer domain mAb 15D1 did not exert any influence on the rate of VWF87(ΔSpacer) proteolysis (kcat/Km = 0.06 × 106 M−1⋅s−1; n = 3; Figure 4A). Surprisingly, a ∼2.2-fold activating effect was still evident in the presence of the activating anti-Spacer domain mAb 3E4 (kcat/Km = 0.11 × 106 M−1⋅s−1) and the activating anti-CUB1 domain mAb 17G2 (kcat/Km = 0.11 × 106 M−1⋅s−1; n = 3; Figure 4A). These data demonstrate that conformational activation of ADAMTS13 is not manifest through exposure of the Spacer domain exosite.
VWF87(ΔSpacer) and VWF96-Cys proteolysis by ADAMTS13 in the presence and absence of mAbs. After preincubation of ADAMTS13(V5-6xHis) with no mAb (orange; n = 3); the noninhibitory, nonactivating anti-Spacer domain mAb 15D1 (light green; n = 3); the activating anti-Spacer domain mAb 3E4 (red; n = 3); or the activating anti-CUB1 domain mAb 17G2 (blue; n = 3), the substrate (0.5 μM) was added to initiate the reaction. Reaction subsamples, stopped between 0 and 90 minutes, were analyzed by ELISA. Fitted time course data show the percentage of VWF87(ΔSpacer) (A) or VWF96-Cys (B) proteolysis as a function of time (mean ± standard error of the mean).
VWF87(ΔSpacer) and VWF96-Cys proteolysis by ADAMTS13 in the presence and absence of mAbs. After preincubation of ADAMTS13(V5-6xHis) with no mAb (orange; n = 3); the noninhibitory, nonactivating anti-Spacer domain mAb 15D1 (light green; n = 3); the activating anti-Spacer domain mAb 3E4 (red; n = 3); or the activating anti-CUB1 domain mAb 17G2 (blue; n = 3), the substrate (0.5 μM) was added to initiate the reaction. Reaction subsamples, stopped between 0 and 90 minutes, were analyzed by ELISA. Fitted time course data show the percentage of VWF87(ΔSpacer) (A) or VWF96-Cys (B) proteolysis as a function of time (mean ± standard error of the mean).
We therefore examined a potential role for the Cys-rich domain exosite interaction in this mechanism. We used the VWF96-Cys substrate, which has the Cys-rich domain exosite interaction mutated (I1642Q/W1644Y/A1647S/I1649Q/L1650Q/I1651Q).8 ADAMTS13(V5-6xHis) proteolyzed VWF96-Cys with a kcat/Km of 0.03 × 106 M−1⋅s−1 (n = 3; Figure 4B). The ∼42-fold reduced rate of VWF96-Cys proteolysis compared with the proteolysis VWF96 (kcat/Km = 1.32 × 106 M−1⋅s−1) was a result of the ablation of the Cys-rich domain exosite interaction. As with VWF87(ΔSpacer), the presence of the nonactivating, noninhibitory anti-Spacer domain mAb 15D1 did not affect the rate of VWF96-Cys proteolysis (kcat/Km = 0.03 × 106 M−1⋅s−1), whereas both the activating anti-Spacer domain mAb 3E4 (kcat/Km = 0.06 × 106 M−1⋅s−1; n = 3) and the activating anti-CUB1 domain mAb 17G2 (kcat/Km = 0.06 × 106 M−1⋅s−1; n = 3) enhanced the catalytic efficiency of proteolysis by ∼1.9-fold (Figure 4B). Again, these data suggest that unmasking of the Cys-rich domain exosite is not involved in the conformational activation of ADAMTS13.
mAb-induced activation of ADAMTS13 is manifest through increased kcat
Our data demonstrate that mAb-induced conformational activation of ADAMTS13 depends on the unfolding of the C-terminal tail of ADAMTS13, but is not manifest through full exposure of the Spacer or Cys-rich domain exosites. These findings raise the question as to whether the enhanced kcat/Km of open ADAMTS13 is indeed a result of enhanced affinity for the VWF A2 domain, as previously suggested. Increases in kcat/Km can be the result of either enhanced substrate binding (measured as reduction in the Km) or enhanced proteolytic function and substrate turnover (measured as increase in kcat). Therefore, to delineate the effect of conformational activation of ADAMTS13 kinetically, Michaelis Menten kinetic analysis of VWF96 proteolysis in the presence and absence of the activating mAbs was performed.
Initial rates of proteolysis (at <15% proteolysis) were calculated and plotted as a function of VWF96 concentration (0-25μ Μ). Figure 5 shows the data fitted to the Michaelis Menten equation. In the absence of mAbs, VWF96 was proteolyzed with a kcat of 2.5 s−1 and a Km of 2.6 μM (n = 31; Figure 5A). Surprisingly, both activating mAbs influenced the kcat more than the Km. In the presence of the anti-CUB1 domain mAb 17G2 (Figure 5B) or the anti-Spacer domain mAb 3E4 (Figure 5C), the kcat was ∼1.7-fold higher (kcat = 4.3 s−1 and kcat = 4.2s−1, respectively) (Figure 5D). In contrast, the Km of proteolysis was unaffected by the presence of the anti-CUB1 mAb 17G2 (Km = 2.6 μM) and only modestly (∼1.2-fold) increased (Km = 3.0 μM) in the presence of the anti-Spacer domain mAb 3E4. Consistent with our earlier results (Figure 4), the presence of the anti-Spacer mAb 3E4 and the anti-CUB1 mAb 17G2 increased the kcat/Km by ∼1.5-fold (kcat/Km = 1.42 × 106 M−1⋅s−1; n = 26) and ∼1.7-fold (kcat/Km = 1.66 × 106 M−1⋅s−1; n = 31), respectively.
The influence of the activating mAbs on the kinetics of VWF96 proteolysis by ADAMTS13. The initial rates (nMs−1) of VWF96 proteolysis (per nanomole of ADAMTS13(V5-6xHis)) in the absence of mAbs (orange) (A) or in the presence of the activating mAbs 17G2 (blue) (B) or 3E4 (red) (C) were plotted as a function of VWF96 concentration (0-25 μM) and fitted to the Michaelis Menten equation, for independent derivation of the kcat and Km of proteolysis. (D) The fitted Michaelis Menten plots for VWF96 proteolysis in the absence (orange, dashed line) or presence of the activating mAbs 3E4 (red, solid line) and 17G2 (blue, solid line), are plotted on the same axes.
The influence of the activating mAbs on the kinetics of VWF96 proteolysis by ADAMTS13. The initial rates (nMs−1) of VWF96 proteolysis (per nanomole of ADAMTS13(V5-6xHis)) in the absence of mAbs (orange) (A) or in the presence of the activating mAbs 17G2 (blue) (B) or 3E4 (red) (C) were plotted as a function of VWF96 concentration (0-25 μM) and fitted to the Michaelis Menten equation, for independent derivation of the kcat and Km of proteolysis. (D) The fitted Michaelis Menten plots for VWF96 proteolysis in the absence (orange, dashed line) or presence of the activating mAbs 3E4 (red, solid line) and 17G2 (blue, solid line), are plotted on the same axes.
Conformational activation of ADAMTS13 by the anti-Spacer domain mAb 3E4 induces a conformational change in the MP domain
The finding that mAb-mediated conformational activation of ADAMTS13 primarily increases substrate turnover (kcat) suggests a mechanism involving changes in the functionality of the MP domain active site. We previously demonstrated that conformational activation of ADAMTS13 by other mAbs, including the mAb 17G2, leads to exposure of a conformationally sensitive epitope in the MP domain.5 The normally/otherwise cryptic epitope in the MP domain is specifically recognized by our anti-MP domain mAb 6A6.5 We assessed the ability of the anti-Spacer domain mAbs 3E4 and 15D1 and the anti-CUB1 domain mAb 17G2 to enhance capture of plasma ADAMTS13 by the mAb 6A6 in an ELISA assay. The activating mAbs 3E4 and 17G2 significantly enhanced capture by the mAb 6A6, demonstrating exposure of the cryptic epitope in the MP domain, whereas the nonactivating, noninhibitory mAb 15D1 did so to a much lesser extent (Figure 6).
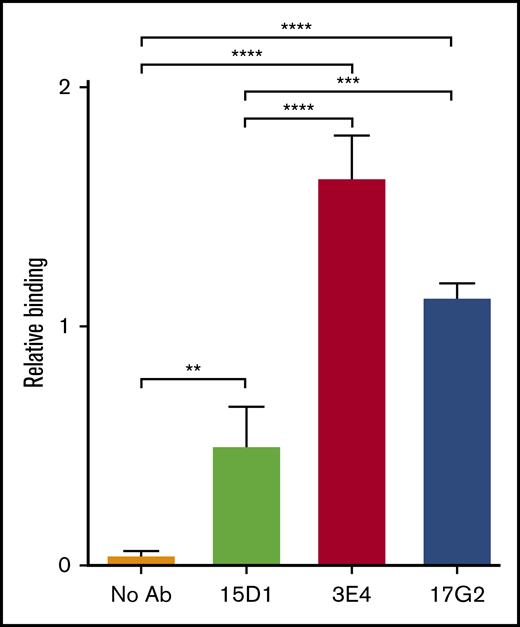
mAb-induced exposure of a cryptic epitope in the MP domain of ADAMTS13. In the open conformation, ADAMTS13 exposes a cryptic epitope in the MP domain, which is specifically detected by the anti-MP domain mAb 6A6. The binding of plasma ADAMTS13, after preincubation in the absence of mAbs (no Ab, orange) or in the presence (1.25 μg/mL) of the mAbs 15D1 (light green), 3E4 (red), and 17G2 (blue), to immobilized anti-MP domain mAb 6A6 was assessed by ELISA. Bound ADAMTS13 was detected using the biotinylated anti-T8 domain mAb 19H4 and HRP-labeled high-sensitivity streptavidin. Binding was calculated relative to binding of plasma ADAMTS13 in the presence of the activating anti-CUB1 mAb 17G25 (2.5 µg/mL), which was set to 1. Differences (mean ± SD; n = 3) were compared using analysis of variance with multiple comparison. **P < .01; ***P < .005; ****P < .001.
mAb-induced exposure of a cryptic epitope in the MP domain of ADAMTS13. In the open conformation, ADAMTS13 exposes a cryptic epitope in the MP domain, which is specifically detected by the anti-MP domain mAb 6A6. The binding of plasma ADAMTS13, after preincubation in the absence of mAbs (no Ab, orange) or in the presence (1.25 μg/mL) of the mAbs 15D1 (light green), 3E4 (red), and 17G2 (blue), to immobilized anti-MP domain mAb 6A6 was assessed by ELISA. Bound ADAMTS13 was detected using the biotinylated anti-T8 domain mAb 19H4 and HRP-labeled high-sensitivity streptavidin. Binding was calculated relative to binding of plasma ADAMTS13 in the presence of the activating anti-CUB1 mAb 17G25 (2.5 µg/mL), which was set to 1. Differences (mean ± SD; n = 3) were compared using analysis of variance with multiple comparison. **P < .01; ***P < .005; ****P < .001.
The conformational change in the MP domain of ADAMTS13 as a result of the mAb binding, coupled to the enhanced substrate turnover (kcat), is consistent with an allosteric activation mechanism, whereby conditions that favor disruption of the Spacer-CUB1 domain interaction enhance the ability of the MP domain active site to cleave VWF.
Discussion
Plasma ADAMTS13 circulates in a folded conformation stabilized by an interaction between the central Spacer domain with the C-terminal CUB domains that limits ADAMTS13 function.4-6 Physiologically, conformational activation of ADAMTS13 through its binding to VWF D4, or VWF D4-CK, appears to disrupt the Spacer-CUB interaction.4-6 This induces a structural change that extends ADAMTS13 into an open conformation and enhances its proteolytic function by ∼twofold.4,6,7 Pathophysiologically, certain anti-ADAMTS13 autoantibodies associated with acquired, antibody-mediated TTP also induce conformational activation of ADAMTS13.4 Although this might seem paradoxical, opening of ADAMTS13 likely also exposes cryptic regions in ADAMTS13, facilitating binding of further autoantibodies against previously hidden epitopes and likely enhancing enzyme inhibition and/or clearance of the enzyme.15 Therefore, understanding the conformational activation of ADAMTS13 provides insights into not only the normal function of the metalloprotease but also into the pathogenicity of acquired TTP.
From previous studies, a model for conformational activation of ADAMTS13 was proposed in which disengagement of the Spacer-CUB interaction induced exposure of either hidden or partially hidden VWF-binding exosites in the ADAMTS13 Spacer domain. In so doing, this would increase the affinity between the enzyme and substrate, and thus the rate of proteolysis.7 This mechanism was suggested on the basis of the observations that the isolated CUB domains could bind to the Spacer domain, and that the binding of the CUB1 to the Spacer domain could be ablated by mutating the previously proposed ADAMTS13 Spacer exosite residues (R568K/F592Y/R660K/Y661F/Y665F).6,7 However, interrogation of conformational activation through mutagenesis of ADAMTS13 can potentially cause independent conformational effects. Conformational activation of ADAMTS13 with the isolated VWF D4 domain has been reported, but this requires very high (μM) concentrations of D4.4,9 Although appreciably lower concentrations of VWF D4-CK have been used to induce the same activating effect, we have been unable to replicate this, likely because of the low-affinity nature of this interaction. Conformational activation of ADAMTS13 can be mimicked in vitro with mAbs that disrupt the Spacer-CUB interaction, and these mAbs bind with much higher affinity; therefore, such mAbs provide appreciably more robust/reproducible tools to kinetically characterize the conformational activation of ADAMTS13.
Generation and screening of novel mAbs recognizing MDTCS revealed that 31 of 43 clones recognized the Cys-rich/Spacer domains of ADAMTS13. Two of these 31 clones were inhibitory, whereas 9 enhanced full-length ADAMTS13 function. It remains unclear at this time whether these mAbs recognize the same/overlapping epitopes in ADAMTS13 or are duplicates of the same clone. It is intriguing to note that more mAbs enhance ADAMTS13 function than inhibit the enzyme. All activating anti-Cys/Spacer mAbs augmented ADAMTS13 function in a similar manner. We therefore selected 1 activating anti-Spacer mAb (3E4) and 1 nonactivating/noninhibitory anti-Spacer mAb (15D1) for kinetic analyses and to compare with the previously described activating anti-CUB1 (17G2) mAb.5
Kinetic analysis of VWF96 proteolysis revealed that both activating mAbs (3E4 and 17G2) augmented ADAMTS13 function by ∼twofold when compared with no mAb or the control nonactivating mAb (15D1). Activation of ADAMTS13 by mAbs 3E4 and 17G2 was not synergistic, suggesting that the augmentation of ADAMTS13 function was occurring through the same mechanism, likely through disruption of the Spacer-CUB interaction. Consistent with this, the activity of MDTCS, which lacks the C-terminal domains, was neither enhanced in the presence of mAb 3E4 nor influenced by the other mAbs.
The VWF87(ΔSpacer) and VWF96-Cys variants, which ablate the Spacer and Cys-rich domain exosite interactions, respectively, were used to investigate whether the activating effects of the anti-Spacer mAb 3E4 and anti-CUB1 mAb 17G2 are dependent on exposure of either of the exosites. Using these VWF96 variants, we found that both activating mAbs still augmented proteolysis of both of these substrates. This approach (of disrupting the substrate interaction site) is somewhat preferable to mutating the exosite in the ADAMTS13 Spacer domain6,16 as it avoids interfering with the structure/conformation of the enzyme, which could potentially influence its activity in an exosite-independent manner. Our results demonstrate that the mAb-induced activation of ADAMTS13 by mAbs 3E4 and 17G2 is not mediated by the exposure of either the Spacer or Cys-rich domain exosites. If exposure of these exosites were a part of the enhancement mechanism, disruption of the exosite binding site in VWF96 would abolish the activating effect. Consistent with this, kinetic analyses revealed that the Km for proteolysis of VWF96 was minimally affected in the presence of the activating mAbs (3E4 and 17G2). Both the ADAMTS13 Spacer domain and Cys-rich domain exosites are known to primarily function in substrate binding/recognition. Disruption of these exosite interactions therefore appreciably increases the Km for proteolysis (ie, diminishes substrate binding).8 Rather, the enhancement of VWF96 proteolysis was primarily manifest through an increase in the kcat, which is a measure of MP domain function, as opposed to substrate binding. For this to occur, we hypothesized that disruption of the Spacer-CUB interaction must therefore conformationally alter the MP domain. We confirmed this using an anti-MP domain mAb (6A6) that recognizes a cryptic epitope in this domain. Whereas in the absence of a mAb, no binding of plasma ADAMTS13 was detected, both activating mAbs (3E4 and 17G2) appreciably enhanced the affinity of mAb 6A6 for ADAMTS13, consistent with a structural shift in the MP domain. Interestingly, the nonactivating mAb 15D1 also induced significant exposure of the cryptic MP domain exosite recognized by mAb 6A6, but to a much lesser extent than with mAbs 3E4 or 17G2. Thus, exposure of the mAb 6A6 epitope appears to be related to, but is not a true marker of, MP domain enhancement, inasmuch as the nonactivating mAb (15D1) must itself induce some conformational change in ADAMTS13, but this has no effect on enzyme function.
Using the mAbs 1C4 and 6A6, which are directed against epitopes that are cryptic in folded ADAMTS13, we previously showed that mAb-induced conformational activation leads to exposure of these cryptic epitopes.5,15 The anti-Spacer mAb 1C4 recognizes a cryptic epitope in the Spacer domain (residues R568/R660/Y661/Y665).15 We showed that binding of the anti-CUB1 mAb (17G2) to ADAMTS13 induced exposure of the 1C4 cryptic epitope in the Spacer domain.15 We now show that the 17G2-induced conformational change in ADAMTS13 also results in the exposure of the mAb 6A6 cryptic epitope in the MP domain. Although the anti-Spacer mAb (3E4) induced a conformational change in the MP domain, as a result of steric hindrance of the 3E4 and 1C4 mAbs15 (data not shown), we could not ascertain whether the cryptic site in the Spacer domain becomes exposed after mAb 3E4-induced conformational changes in ADAMTS13.
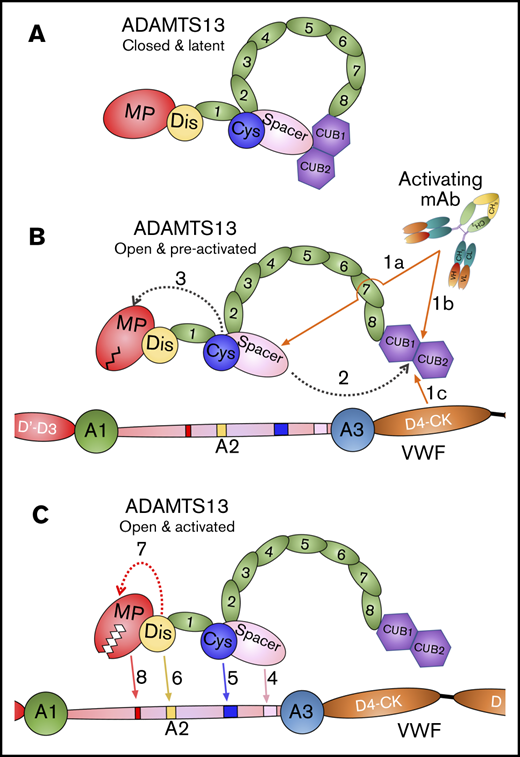
Taken together, our data reveal that activating anti-Spacer or anti-CUB mAbs induce disruption of the Spacer-CUB interaction (Figure 7A-B). This induces a structural change in ADAMTS13 that alters the conformation and functionality of the MP domain (Figure 7B-C). At face value, this may appear surprising, given the spatial separation of the Spacer-CUB interaction site from the MP domain. However, our recent kinetic and structural studies have revealed the importance of ADAMTS13 allostery in enzyme function.8 It appears that in circulation, the ADAMTS13 MP domain adopts a latent fold in which the active site is occluded by the Ca2+-binding loop above the active site (Figure 7B). This is hypothesized to prevent off-target proteolysis by ADAMTS13 while in circulation, and also confer resistance to plasma inhibitors. Only when ADAMTS13 engages with the unfolded VWF A2 domain does the interaction of the Dis domain exosite induce an allosteric change in the MP domain that removes the Ca2+-binding loop from the active site cleft to enable efficient proteolysis of VWF (Figure 7C). On the basis of our data presented here, we propose that the conformational activation of ADAMTS13 involving the disruption of the Spacer-CUB interaction by activating mAbs (and, by inference also, pH and VWF D4 or D4-CK) enhances the flexibility/plasticity of the MP domain in a manner that promotes this Dis exosite-dependent allosteric activation. The domain interfaces in MDTCS are considerable, which provides mechanistic insight into how this long-range structural cross-talk may be manifest.
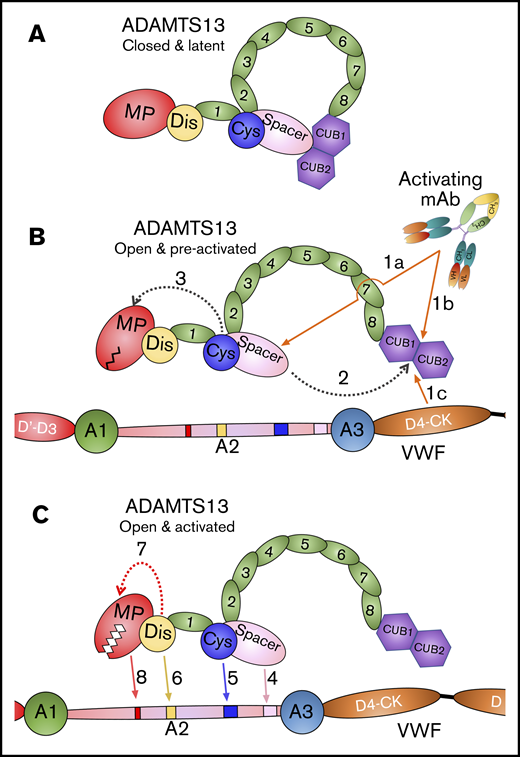
Mode of action of ADAMTS13. (A) Under normal circumstances, ADAMTS13 circulates in a closed conformation stabilized by the interaction of the C-terminal CUB domains with the central Spacer domain. The MP domain of ADAMTS13 naturally favors a latent conformation in which the active site cleft is occluded, preventing off-target proteolysis and conferring resistance to inhibition. (B) When ADAMTS13 is bound by activating mAbs that recognize either the Spacer domain (1a) or CUB domains (1b), or when it binds to VWF via the D4-CK domains of VWF (1c), the CUB-Spacer interaction is disrupted (2) causing ADAMTS13 to adopt an open conformation. This opening of ADAMTS13 induces a structural shift in the MP domain into a preactivated state (3) that, ultimately, enhances the proteolytic function of the enzyme. (C) ADAMTS13 recognizes unfolded VWF A2 domain through multiple interactions. The Spacer (4) and (5) Cys-rich domain exosites recognize the C-terminal region of the unfolded VWF A2 domain bringing enzyme and substrate into close proximity. Thereafter, the Dis domain exosite engages VWF (6), which induces a further allosteric change in the MP domain (7). This conformational change opens the active site cleft to enable accommodation and proteolysis of the cleavage site (8). The preactivation of ADAMTS13 augments this final activation step by ∼twofold.
Mode of action of ADAMTS13. (A) Under normal circumstances, ADAMTS13 circulates in a closed conformation stabilized by the interaction of the C-terminal CUB domains with the central Spacer domain. The MP domain of ADAMTS13 naturally favors a latent conformation in which the active site cleft is occluded, preventing off-target proteolysis and conferring resistance to inhibition. (B) When ADAMTS13 is bound by activating mAbs that recognize either the Spacer domain (1a) or CUB domains (1b), or when it binds to VWF via the D4-CK domains of VWF (1c), the CUB-Spacer interaction is disrupted (2) causing ADAMTS13 to adopt an open conformation. This opening of ADAMTS13 induces a structural shift in the MP domain into a preactivated state (3) that, ultimately, enhances the proteolytic function of the enzyme. (C) ADAMTS13 recognizes unfolded VWF A2 domain through multiple interactions. The Spacer (4) and (5) Cys-rich domain exosites recognize the C-terminal region of the unfolded VWF A2 domain bringing enzyme and substrate into close proximity. Thereafter, the Dis domain exosite engages VWF (6), which induces a further allosteric change in the MP domain (7). This conformational change opens the active site cleft to enable accommodation and proteolysis of the cleavage site (8). The preactivation of ADAMTS13 augments this final activation step by ∼twofold.
For original data, please contact the authors at karen.vanhoorelbeke@kuleuven.be or j.crawley@imperial.ac.uk.
Acknowledgments
The authors thank Nele Vandeputte and Aline Vandenbulcke for technical assistance.
Funding was provided by The European Framework Program for Research and Innovation (Horizon2020 Marie Sklodowska Curie Innovative training network PROFILE grant 675746) awarded to K.V. and by grants awarded to J.T.B.C. by the British Heart Foundation (FS/14/44/30962 and PG/18/17/33572). A.-S.S. is supported by a grant from the Agency Innovation and Entrepreneurship (VLAIO, www.iwt.be), Flanders, Belgium (141136).
Authorship
Contribution: A.-S.S., A.P., J.T.B.C., and K.V. designed the experiments, analyzed and interpreted the data, and wrote the manuscript; A.-S.S., A.P., E.R., and I.P. performed experiments; E.R., H.D., and S.F.D.M. critically reviewed the manuscript; and A.-S.S., K.V., and J.T.B.C. provided funding.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Karen Vanhoorelbeke, Laboratory for Thrombosis Research, IRF Life Sciences, KU Leuven Campus Kulak Kortrijk, Etienne Sabbelaan 53, B-8500 Kortrijk, Belgium; e-mail: karen.vanhoorelbeke@kuleuven.be; and James T. B. Crawley, Department of Immunology and Inflammation, Imperial College London, 5th Floor Commonwealth Building, Du Cane Rd, London W12 0NN, United Kingdom; e-mail j.crawley@imperial.ac.uk.
References
Author notes
A.-S.S. and A.P. contributed equally to this study.
The full-text version of this article contains a data supplement.