Key Points
Midostaurin significantly improves event-free survival in patients with FLT3-TKD mutated AML.
NPM1 mutations and CBF rearrangements were favorable independent predictors of OS and EFS in the multivariable analysis.
Abstract
The results from the RATIFY trial (ClinicalTrials.gov: NCT00651261; CALGB 10603) showed that midostaurin combined with standard chemotherapy significantly improved outcomes in patients with FMS-like tyrosine kinase 3 (FLT3)–mutated acute myeloid leukemia (AML), compared with placebo. In this post hoc subgroup analysis from the trial, we evaluated the impact of midostaurin in 163 patients with FLT3-tyrosine kinase domain (TKD) mutations. At a median follow-up of 60.7 months (95% CI, 55.0-70.8), the 5-year event-free survival (EFS) rate was significantly higher in patients treated with midostaurin than in those treated with placebo (45.2% vs 30.1%; P = .044). A trend toward improved disease-free survival was also observed with midostaurin (67.3% vs 53.4%; P = .089), whereas overall survival (OS) was similar in the 2 groups. Patients with AML and NPM1mut/FLT3-TKDmut or core binding factor (CBF)–rearranged/FLT3-TKDmut genotypes had significantly prolonged OS with or without censoring at hematopoietic cell transplantation (HCT), compared with NPM1WT/CBF-negative AMLs. The multivariable model for OS and EFS adjusted for allogeneic HCT in first complete remission as a time-dependent covariable, revealed NPM1 mutations and CBF rearrangements as significant favorable factors. These data show that NPM1 mutations or CBF rearrangements identify favorable prognostic groups in patients with FLT3-TKD AMLs, independent of other factors, also in the context of midostaurin treatment.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease with unsatisfactory long-term outcomes, despite significant progress in its diagnosis, monitoring, and treatment.1 Major advances in genomic characterization have recently improved patient stratification and led to the identification of molecular targets to help individualized treatment approaches. A paradigm for this are activating mutations in FMS-like tyrosine kinase 3 (FLT3), which characterize ∼35% of AML cases and have a decreasing prevalence in elderly patients.2-4 FLT3 mutations include internal tandem duplications (ITDs), present in about 25% to 30% of younger patients with AML, and tyrosine kinase domain (TKD) mutations (eg, point mutations in D835 and I836 or deletion of codon I836), present in ∼8% of patients.2-4
FLT3-ITD mutations are a known unfavorable prognostic marker associated with poor overall survival (OS) and relapse-free survival5-7 ; as such, testing for these mutations is part of the routine workup for patients with AML and is recommended by current guidelines at the time of diagnosis and at relapse.8,9 The leukemogenic relevance and prognostic impact of FLT3-TKD mutations are more uncertain, with conflicting evidence observed across studies.10-15 Other common mutations often co-occurring with FLT3 affect the nucleophosmin 1 (NPM1) gene; these are present in ∼30% of patients with AML and are associated with a favorable to intermediate prognosis, depending on the presence of concomitant FLT3-ITD mutations.7,8,16 Core binding factor (CBF) rearrangements are also frequent in FLT3-TKDmut AML, and occur in ∼15% of patients.3,10
Midostaurin is a multikinase inhibitor, originally developed for the treatment of solid tumors. In AML, it inhibits FLT3-kinase in patients with both ITD and TKD mutations, as well as other targets involved in the disease pathogenesis.17 In the Cancer and Leukemia Group B (CALGB) 10603/RATIFY trial, which enrolled patients with a FLT3-mutated (FLT3mut) AML, midostaurin in combination with standard induction and consolidation chemotherapy, followed by single-agent maintenance therapy, significantly improved outcomes vs placebo.17 In this study, patients were stratified according to the presence of TKD mutations and ITD allelic ratios (AR). Benefits of midostaurin were seen across all FLT3 stratification subgroups, with a significantly longer survival for patients treated with midostaurin as compared with those receiving placebo (median, 74.7 months vs 25.6 months, P = .009).17 The data from this trial led to the approval of midostaurin in 2017 for the treatment of adults with newly diagnosed, FLT3mut AML.
In this post hoc analysis from the RATIFY trial, we further investigated the role of midostaurin in the treatment of FLT3-TKDmut AML, also taking into account karyotype and NPM1 mutation status.
Patients and methods
Study design
Results from the RATIFY trial were published previously.17 In brief, patients aged 18 to 59 years with newly diagnosed AML and a FLT3 mutation were randomly assigned 1:1 to receive midostaurin or placebo together with standard induction and consolidation therapy. Randomization was stratified by FLT3 mutation subtype: TKD, ITDlow with AR between 0.05 and 0.7, and ITDhigh with AR >0.7. During induction and the 4 28-day cycles of consolidation therapy, 50 mg midostaurin or placebo were administered orally twice per day on days 8 to 21. Patients who remained in remission after consolidation therapy received 12 28-day cycles of maintenance therapy, with 50 mg oral midostaurin or placebo twice per day daily. The RATIFY study (ClinicalTrials.gov number: NCT00651261) was approved by the ethics committees of all participating centers. Patients gave written informed consent for participating in the study.
FLT3-TKD mutations were detected by polymerase chain reaction (PCR) followed by EcoRV digestion and capillary electrophoresis.18 A TKD mutation-to-wild-type signal ratio of 0.05 was required for positivity.
A total of 162 patients with a FLT3-TKDmut AML were included in the original RATIFY report,17 on the basis of randomization stratum, whereas 163 patients are included in this analysis, on the basis of the actual FLT3-TKD mutation status. None of the cases had a concomitant FLT3-ITD as tested by capillary electrophoresis. A total of 134 patients gave informed consent for additional NPM1 mutation analysis and are considered in this report. NPM1 mutation status was analyzed at the participating reference laboratories, according to published methods, and after interlaboratory crossvalidation procedures. Analysis of PCR products allowed us to distinguish between wild-type (WT) (236 bp) and mutated NPM1 (NPM1mut), which is characterized by an insertion of 4 nucleotides.7,19,20 Conventional karyotyping for recurrent translocations was performed in patients at the time of initial diagnosis.
Statistical analysis
Demographic and baseline data including disease characteristics were summarized descriptively by treatment group. Categorical data are presented as frequencies and percentages. For continuous data, median and interquartile range are shown.
Complete remission (CR) was defined by standard criteria,8 and assessed after 1 to 2 induction cycles. Definitions of OS and event-free survival (EFS) were as recommended,8 and were calculated from the date of randomization. Disease-free survival (DFS) and cumulative incidence of relapse (CIR) were calculated from the date of CR. Additional sensitivity analyses were performed censoring OS, EFS, and DFS at the time of transplantation. The median follow-up for survival and the distribution of time-to-event endpoints (OS, DFS, and EFS) were estimated using the Kaplan-Meier method.21 Differences between the groups were analyzed using 2-sided log-rank tests. Kaplan-Meier rates at 5 years with 95% confidence interval (CI) are presented. Hazard ratios (HR) with 95% CI were estimated using the Cox proportional hazards model. CIR was evaluated using the cumulative incidence method, considering death in CR as a competing risk. The aforementioned analyses were repeatedly performed to evaluate treatment effect (midostaurin vs placebo) and the prognostic effect of NPM1 mutation status (mutated vs WT). To examine the effect of allogeneic hematopoietic cell transplantation (HCT), univariable and multivariable Cox models with allogeneic HCT as a time-dependent variable were applied.21
Multivariable analyses for OS and EFS were performed using the Cox model, and included the following covariates: NPM1 vs CBF rearrangements vs other genetic alterations, treatment, sex, age (numeric variable), white blood cell counts (numeric variable), cytogenetics (normal karyotype vs CBF or vs other abnormalities), and allogeneic HCT as time-dependent covariate.
Results
Patient demographics and baseline characteristics
In total, 717 patients with AML and a FLT3 mutation were enrolled in the RATIFY trial.17 Of these, 163 were FLT3-TKDmut. Table 1 shows baseline characteristics of the 163 patients. Median FLT3-TKD AR was 0.44. Of 135 patients with available cytogenetic data, 74 (55%) had a normal karyotype and 20 (15%) had a CBF-AML, including 11 patients with inv(16) and 9 with t(8;21).
Characteristics of patients with a FLT3-TKDmut AML included in the RATIFY trial (n = 163)
| . | Total patient population (N = 163) . | Midostaurin (n = 83) . | Placebo (n = 80) . | P, mido vs placebo . |
|---|---|---|---|---|
| Sex, males/females, n | 79/84 | 45/38 | 34/46 | .180 |
| Age, median (range), y | 48.8 (19.3-59.9) | 48.0 (19.8-59.8) | 49.4 (19.3-59.9) | .827 |
| ECOG PS 2, % | 13 (8) | 6 (7.2) | 7 (8.8) | .945 |
| WBC count, median (range), × 109/L | 28.80 (1.20-300.00) | 29.70 (1.20-298.40) | 28.15 (1.30-300.00) | .480 |
| Platelet count, median (range), × 109/L | 49.00 (10.00-328.00) | 53.00 (11.00-328.00) | 46.50 (10.00-237.00) | .430 |
| Bone marrow blasts, median (range), % | 75.00 (0.00-100.00) | 75.00 (0.00-100.00) | 77.00 (0.00-100.00 | .817 |
| Peripheral blood blasts, median (range), % | 55.00 (0.00-99.00) | 56.50 (0.00-99.00) | 55.00 (0.00-98.00) | .594 |
| Prior myelodysplastic syndrome, n (%) | 4 (2.5) | 2 (2.4) | 2 (2.5) | >.99 |
| Extramedullary disease, n (%) | 31 (19.0) | 13 (15.7) | 18 (22.5) | .362 |
| NPM1, n (%) (n = 134) | ||||
| Mutated | 79 (59.0) | 45 (64.3) | 34 (53.1) | .256 |
| WT | 55 (41) | 25 (35.7) | 30 (46.9) | |
| Karyotype, n (%) (n = 135) | ||||
| CBF* | 20 (14.8) | 10 (15.6) | 10 (14.1) | .140 |
| Normal | 74 (54.8) | 39 (60.9) | 35 (49.3) | |
| Others† | 41 (30.4) | 15 (23.4) | 26 (36.6) | |
| CR after induction, n (%) | 105 (64.4) | 57 (68.6) | 48 (60) | .321 |
| Allogeneic HCT in first CR, n (%) | 62 (38.0) | 32 (38.6) | 30 (37.5) | >.99 |
| Midostaurin maintenance, n (%) | 58 (35.6) | 32 (38.6) | 26 (32.5) | .52 |
| . | Total patient population (N = 163) . | Midostaurin (n = 83) . | Placebo (n = 80) . | P, mido vs placebo . |
|---|---|---|---|---|
| Sex, males/females, n | 79/84 | 45/38 | 34/46 | .180 |
| Age, median (range), y | 48.8 (19.3-59.9) | 48.0 (19.8-59.8) | 49.4 (19.3-59.9) | .827 |
| ECOG PS 2, % | 13 (8) | 6 (7.2) | 7 (8.8) | .945 |
| WBC count, median (range), × 109/L | 28.80 (1.20-300.00) | 29.70 (1.20-298.40) | 28.15 (1.30-300.00) | .480 |
| Platelet count, median (range), × 109/L | 49.00 (10.00-328.00) | 53.00 (11.00-328.00) | 46.50 (10.00-237.00) | .430 |
| Bone marrow blasts, median (range), % | 75.00 (0.00-100.00) | 75.00 (0.00-100.00) | 77.00 (0.00-100.00 | .817 |
| Peripheral blood blasts, median (range), % | 55.00 (0.00-99.00) | 56.50 (0.00-99.00) | 55.00 (0.00-98.00) | .594 |
| Prior myelodysplastic syndrome, n (%) | 4 (2.5) | 2 (2.4) | 2 (2.5) | >.99 |
| Extramedullary disease, n (%) | 31 (19.0) | 13 (15.7) | 18 (22.5) | .362 |
| NPM1, n (%) (n = 134) | ||||
| Mutated | 79 (59.0) | 45 (64.3) | 34 (53.1) | .256 |
| WT | 55 (41) | 25 (35.7) | 30 (46.9) | |
| Karyotype, n (%) (n = 135) | ||||
| CBF* | 20 (14.8) | 10 (15.6) | 10 (14.1) | .140 |
| Normal | 74 (54.8) | 39 (60.9) | 35 (49.3) | |
| Others† | 41 (30.4) | 15 (23.4) | 26 (36.6) | |
| CR after induction, n (%) | 105 (64.4) | 57 (68.6) | 48 (60) | .321 |
| Allogeneic HCT in first CR, n (%) | 62 (38.0) | 32 (38.6) | 30 (37.5) | >.99 |
| Midostaurin maintenance, n (%) | 58 (35.6) | 32 (38.6) | 26 (32.5) | .52 |
Mido, midostaurin; ECOG-PS, Eastern Cooperative Oncology Group performance status; WBC, white blood cell.
CBF rearrangements: inv(16), n = 11, and t(8;21), n = 9
Includes 8 patients with adverse and 7 with intermediate karyotype in the midostaurin arm, vs 7 and 19 patients, respectively, in the placebo arm.
NPM1 mutation status was available for 134 patients: 79 (59%) had NPM1mut, and 55 (41%) had NPM1WT. Patient characteristics according to NPM1 mutation status are shown in supplemental Table 1. The median white blood cell count was higher in patients with NPM1mut (median, 34.1 × 109/L; range, 16.5 × 109/L to 76.0 × 109/L) than in those with NPM1WT (median, 15.5×109/L; range, 8.5 × 109/L to 34.9 × 109/L; P = .003). As expected, normal karyotype was significantly more frequent in patients with NPM1mut (90.5% vs 20.0%; P < .001), whereas 28.0% percent of patients with NPM1WT had a CBF-AML (supplemental Table 1).
Outcome analysis
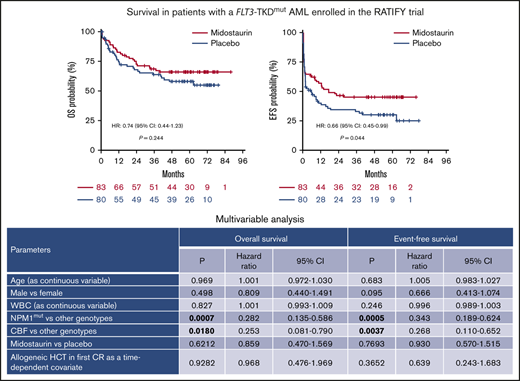
The median follow-up for the FLT3-TKDmut patient cohort was 60.7 months (95% CI, 55.0-70.8 months; n = 163 patients). After induction chemotherapy, CR rates were similar in patients treated with midostaurin and in those treated with placebo (68.6% vs 60.0%; P = .321) (Table 1). Similarly, 5-year OS rates were comparable between the 2 treatment arms (65.9% vs 58.0%; HR, 0.74; 95% CI, 0.44-1.23; P = .244; Figure 1A). However, the 5-year EFS rate was significantly higher in patients treated with midostaurin than in those treated with placebo (45.2% vs 30.1%; HR, 0.66; 95% CI, 0.45-0.99; P = .044), and there was a trend toward improved DFS at 5 years (67.3% vs 53.4%; HR, 0.60; 95% CI, 0.34-1.08; P = .089) (Figure 1B-C), as well as a trend for reduced CIR (26.5% with midostaurin vs 38.5% with placebo; HR, 0.63; 95% CI, 0.33-1.20; P = .156; Figure 1D). FLT3-TKD AR, grouping patients according to the median value (<0.4 vs ≥0.4), did not play a significant prognostic role for survival or CIR (data not shown).
Survival in patients with FLT3-TKDmutAML, according to treatment arm (n = 163 patients). HR and 95% CI are shown. OS (A), EFS (B), DFS (C), CIR (D).
Survival in patients with FLT3-TKDmutAML, according to treatment arm (n = 163 patients). HR and 95% CI are shown. OS (A), EFS (B), DFS (C), CIR (D).
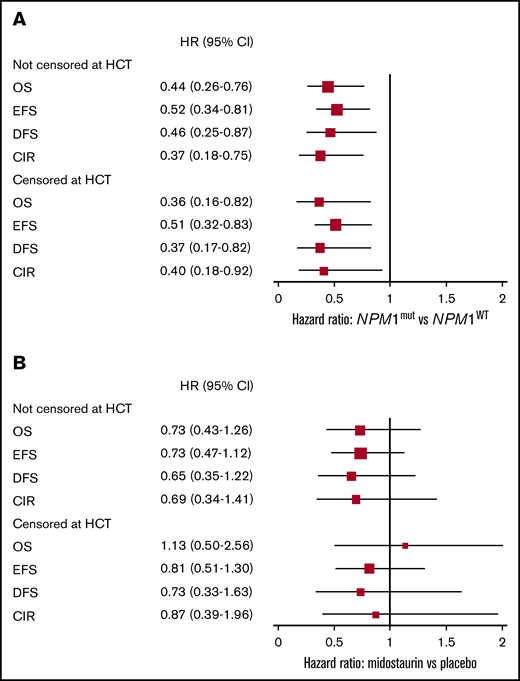
We then analyzed the activity of midostaurin in 134 patients with a TKD mutation and available NPM1 mutation status (supplemental Table 1). Patients with FLT3-TKD/NPM1mut represented a prognostically favorable subgroup, with a significant survival advantage in all endpoints (Table 2; Figure 2A) compared with NPM1wt patients. In patients with NPM1mut, the OS rate was 69.8%, the EFS rate was 48.4%, the DFS rate was 69.5%, and the CIR rate was 21.0% at 5 years. The corresponding values in patients with NPM1WT were 45.7%, 24.6%, 50.3%, and 44.5% (OS, P = .003; EFS, P = .003; DFS, P = .001; CIR, P = .006). Relative to treatment, there were no differences in CR rates between patients with NPM1mut (77.8% with midostaurin, vs 64.7% with placebo; P = .303), and those with NPM1WT (52.0% vs 63.3%, respectively; P = .566). Figure 3 shows survival outcomes in 134 patients according to NPM1 mutation status and treatment arm. The impact of midostaurin on all survival endpoints was not significant (Figures 2B and 3; Table 3).
Five-year survival outcomes by NPM1 mutation status (n = 134 patients)
| . | Overall . | Censored at HCT in 1st CR . | ||||
|---|---|---|---|---|---|---|
| . | NPM1mut (n = 79) . | NPM1WT (n = 55) . | P . | NPM1mut (n = 79) . | NPM1WT (n = 55) . | P . |
| OS, estimates at 5 y (95% CI), % | 69.8 (60.2-80.9) | 45.7 (34.1-61.3) | .003 | 82.6 (72.9-93.5) | 56.8 (38.0-84.8) | .015 |
| EFS, estimates at 5 y (95% CI), % | 48.4 (38.4-61) | 24.6 (15.3-39.4) | .003 | 50.7 (39.7-64.6) | 26.4 (15.3-45.5) | .007 |
| DFS, estimates at 5 y (95% CI), % | 69.5 (58.9-81.9) | 50.3 (36.6-69.1) | .001 | 70.5 (57.8-86.1) | 44.9 (27.2-74.1) | .015 |
| CIR, estimates at 5 y (95% CI), % | 21.0 (20.1-21.9) | 44.5 (43.1-45.9) | .006 | 22.0 (21.3-22.6) | 34.8 (33.2-36.3) | .030 |
| . | Overall . | Censored at HCT in 1st CR . | ||||
|---|---|---|---|---|---|---|
| . | NPM1mut (n = 79) . | NPM1WT (n = 55) . | P . | NPM1mut (n = 79) . | NPM1WT (n = 55) . | P . |
| OS, estimates at 5 y (95% CI), % | 69.8 (60.2-80.9) | 45.7 (34.1-61.3) | .003 | 82.6 (72.9-93.5) | 56.8 (38.0-84.8) | .015 |
| EFS, estimates at 5 y (95% CI), % | 48.4 (38.4-61) | 24.6 (15.3-39.4) | .003 | 50.7 (39.7-64.6) | 26.4 (15.3-45.5) | .007 |
| DFS, estimates at 5 y (95% CI), % | 69.5 (58.9-81.9) | 50.3 (36.6-69.1) | .001 | 70.5 (57.8-86.1) | 44.9 (27.2-74.1) | .015 |
| CIR, estimates at 5 y (95% CI), % | 21.0 (20.1-21.9) | 44.5 (43.1-45.9) | .006 | 22.0 (21.3-22.6) | 34.8 (33.2-36.3) | .030 |
Significant differences are indicated in bold.
Estimated prognostic effect of NPM1 status and treatment effect. The figures show HR and 95% CI, censored at allogeneic HCT or not, for 134 FLT3-TKD mutated patients, grouped for NPM1 mutation status (mutated vs WT) (A) and according to treatment (midostaurin vs placebo) (B). The size of the boxes indicates the precision of the estimates.
Estimated prognostic effect of NPM1 status and treatment effect. The figures show HR and 95% CI, censored at allogeneic HCT or not, for 134 FLT3-TKD mutated patients, grouped for NPM1 mutation status (mutated vs WT) (A) and according to treatment (midostaurin vs placebo) (B). The size of the boxes indicates the precision of the estimates.
Survival outcomes (not censored at allogeneic HCT) by NPM1 mutation status and treatment arm (n = 134 patients). OS (A), EFS (B), DFS (C), CIR (D).
Survival outcomes (not censored at allogeneic HCT) by NPM1 mutation status and treatment arm (n = 134 patients). OS (A), EFS (B), DFS (C), CIR (D).
Five-year survival outcomes in patients with a FLT3-TKD mutation, by NPM1 mutation status and treatment
| . | NPM1mut (n = 79) . | NPM1WT (n = 55) . | ||||
|---|---|---|---|---|---|---|
| . | Placebo . | Midostaurin . | HR (95% CI) . | Placebo . | Midostaurin . | HR (95% CI) . |
| OS | ||||||
| n | 34 | 45 | 30 | 25 | ||
| Estimates at 5 y (95% CI), % | 68.9 (54.5-87.0) | 70.5 (58.2-85.3) | 0.94 (0.41-2.15) | 41.4 (26.8-63.8) | 51.3 (34.9-75.6) | 0.70 (0.34-1.46) |
| EFS | ||||||
| n | 34 | 45 | 30 | 25 | ||
| Estimates at 5 y (95% CI), % | 39.7 (26.0-60.7) | 55.0 (42.1-71.8) | 0.63 (0.34-1.18) | 25.7 (13.8-47.8) | 22.4 (10.4-48.2) | 1.04 (0.57-1.92) |
| DFS | ||||||
| n | 18 | 34 | 18 | 11 | ||
| Estimates at 5 y (95% CI), % | 65.3 (48.5-88.0) | 71.9 (59.2-87.5) | 0.81 (0.32-2.01) | 46.4 (29.1-74.2) | 54.3 (35.2-83.9) | 0.64 (0.26-1.57) |
| CIR | ||||||
| n | 18 | 34 | 18 | 11 | ||
| Estimates at 5 y (95% CI), % | 26.1 (24.4-27.9) | 17.9 (17.2-18.7) | 0.67 (0.23-1.94) | 43.7 (41.1-46.2) | 45.7 (42.6-48.8) | 0.93 (0.37-2.34) |
| . | NPM1mut (n = 79) . | NPM1WT (n = 55) . | ||||
|---|---|---|---|---|---|---|
| . | Placebo . | Midostaurin . | HR (95% CI) . | Placebo . | Midostaurin . | HR (95% CI) . |
| OS | ||||||
| n | 34 | 45 | 30 | 25 | ||
| Estimates at 5 y (95% CI), % | 68.9 (54.5-87.0) | 70.5 (58.2-85.3) | 0.94 (0.41-2.15) | 41.4 (26.8-63.8) | 51.3 (34.9-75.6) | 0.70 (0.34-1.46) |
| EFS | ||||||
| n | 34 | 45 | 30 | 25 | ||
| Estimates at 5 y (95% CI), % | 39.7 (26.0-60.7) | 55.0 (42.1-71.8) | 0.63 (0.34-1.18) | 25.7 (13.8-47.8) | 22.4 (10.4-48.2) | 1.04 (0.57-1.92) |
| DFS | ||||||
| n | 18 | 34 | 18 | 11 | ||
| Estimates at 5 y (95% CI), % | 65.3 (48.5-88.0) | 71.9 (59.2-87.5) | 0.81 (0.32-2.01) | 46.4 (29.1-74.2) | 54.3 (35.2-83.9) | 0.64 (0.26-1.57) |
| CIR | ||||||
| n | 18 | 34 | 18 | 11 | ||
| Estimates at 5 y (95% CI), % | 26.1 (24.4-27.9) | 17.9 (17.2-18.7) | 0.67 (0.23-1.94) | 43.7 (41.1-46.2) | 45.7 (42.6-48.8) | 0.93 (0.37-2.34) |
Estimated Kaplan-Meier rates at 5 years (with 95% CI) and corresponding HR. This analysis was not censored at allogeneic hematopoietic cell transplant. HR <1 indicates a benefit with midostaurin.
Allogeneic HCT in first CR was performed in 51 patients, including 28 of 79 with NPM1mut (35.4%) and 23 of 55 with NPM1WT (41.8%, P = .571). In the sensitivity analyses, patients were censored at HCT, confirming significantly improved outcomes for all survival endpoints in the NPM1mut group across the 2 treatment arms, with survival rates exceeding 80% at 5 years (Figure 2; Table 2).
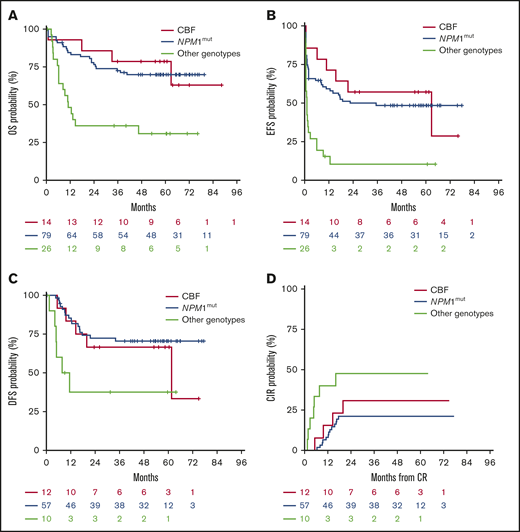
Multivariable analyses validated the independent prognostic role of the NPM1mut genotype in OS (P = .0007) and EFS (P = .0005) and of CBF rearrangements (OS, P = .0180; EFS, P = .0037), whereas midostaurin treatment had no prognostic impact on OS and EFS (Table 4). Other factors analyzed (age, sex, white blood cell count at baseline, HCT as time-dependent covariate) did not reach significance in the Cox model. A prior history of myelodysplastic syndrome, extramedullary disease, and Eastern Cooperative Oncology Group performance status of 2 were not included in the analysis, because of the low numbers of patients with these characteristics. Figure 4 shows the Kaplan-Meier estimates for OS, EFS, DFS, and CIR, grouping patients according to NPM1mut, CBF rearrangements, and other genotypes.
Multivariable analysis for survival
| . | OS . | EFS* . | ||||
|---|---|---|---|---|---|---|
| . | P . | HR . | 95% CI . | P . | HR . | 95% CI . |
| Age† | .969 | 1.001 | 0.972-1.030 | .683 | 1.005 | 0.983-1.027 |
| Male vs female | .498 | 0.809 | 0.440-1.491 | .095 | 0.666 | 0.413-1.074 |
| WBC count, ×109/L† | .827 | 1.001 | 0.993-1.009 | .246 | 0.996 | 0.989-1.003 |
| NPM1mut vs other genotypes | .0007 | 0.282 | 0.135-0.586 | .0005 | 0.343 | 0.189-0.624 |
| CBF vs other genotypes | .0180 | 0.253 | 0.081-0.790 | .0037 | 0.268 | 0.110-0.652 |
| Midostaurin vs placebo | .6212 | 0.859 | 0.470-1.569 | .7693 | 0.930 | 0.570-1.515 |
| Allogeneic HCT in first CR as a time-dependent covariate | .9282 | 0.968 | 0.476-1.969 | .3652 | 0.639 | 0.243-1.683 |
| . | OS . | EFS* . | ||||
|---|---|---|---|---|---|---|
| . | P . | HR . | 95% CI . | P . | HR . | 95% CI . |
| Age† | .969 | 1.001 | 0.972-1.030 | .683 | 1.005 | 0.983-1.027 |
| Male vs female | .498 | 0.809 | 0.440-1.491 | .095 | 0.666 | 0.413-1.074 |
| WBC count, ×109/L† | .827 | 1.001 | 0.993-1.009 | .246 | 0.996 | 0.989-1.003 |
| NPM1mut vs other genotypes | .0007 | 0.282 | 0.135-0.586 | .0005 | 0.343 | 0.189-0.624 |
| CBF vs other genotypes | .0180 | 0.253 | 0.081-0.790 | .0037 | 0.268 | 0.110-0.652 |
| Midostaurin vs placebo | .6212 | 0.859 | 0.470-1.569 | .7693 | 0.930 | 0.570-1.515 |
| Allogeneic HCT in first CR as a time-dependent covariate | .9282 | 0.968 | 0.476-1.969 | .3652 | 0.639 | 0.243-1.683 |
FLT3-ITD mutation status was available for 107 patients; 10 were NPM1mut/FLT3-ITDmut and 11 were NPM1wt/FLT3-ITDmut. Significant differences are indicated in bold.
EFS, considering complete remission within 1 or 2 induction cycles (n = 134).
Age and WBC were considered as continuous variables.
Survival outcomes, according to CBF rearrangements (n = 20 patients), NPM1mut(n = 79 patients), and other genotypes (n = 26 patients). Other genotypes includes patients with intermediate (n = 11) and adverse karyotypes (n = 15 patients). OS (A), EFS (B), DFS (C), CIR (D).
Survival outcomes, according to CBF rearrangements (n = 20 patients), NPM1mut(n = 79 patients), and other genotypes (n = 26 patients). Other genotypes includes patients with intermediate (n = 11) and adverse karyotypes (n = 15 patients). OS (A), EFS (B), DFS (C), CIR (D).
Discussion
This post hoc analysis from the RATIFY trial confirms the advantage in EFS and DFS of the addition of midostaurin to standard chemotherapy in patients with a FLT3-TKDmut AML. At a median follow-up of 5 years, the EFS rate was 45% with midostaurin vs 30% with placebo, whereas the DFS rate was 67% vs 53%, respectively. However, this did not translate into improved OS. The ability to salvage patients after relapse in this chemoresponsive group could explain the discrepancy between the benefit in EFS and the lack of survival benefit. Also, allogeneic HCT performed in first CR in about 38% of patients and midostaurin maintenance may be confounding factors in this setting and will be the subject of a separate analysis. Results of the AMLSG 16-10 trial showed that the addition of midostaurin as single-agent maintenance after allogeneic HCT or after high-dose cytarabine improved EFS, compared with a historical control patient cohort that received similar treatment combinations without tyrosine-kinase inhibitor maintenance.22
NPM1 mutations are a known favorable prognostic factor in AML and are associated with FLT3-ITD mutations in ∼50% of patients. NPM1 mutations mitigate the negative prognostic effect of FLT3-ITD mutations, and, according to the 2017 European LeukemiaNet (ELN) recommendations, these 2 mutations when combined define favorable- or intermediate-risk AML, depending on low (<0.5) or high (≥0.5) ITD AR.8 The prognostic role of FLT3-TKD mutations is established less clearly and appears to be diminished relative to karyotype or concomitant mutations of other genes.4,10-15 Actually, the favorable impact of the combined NPM1mut/FLT3-TKD or CEBPAmut/FLT3-TKD genotypes has been previously reported in a study of 3082 patients with newly diagnosed AML treated with conventional chemotherapy, with the strongest effects seen in de novo and normal karyotype AML, after excluding FLT3-ITD–positive patients.23 These data were confirmed by a retrospective study conducted in France in 126 patients with newly diagnosed AML, in which patients with the NPM1mut/FLT3-TKDmut profile had a high survival probability overall, and after censoring at the time of HCT.24
The results from our study confirm the favorable prognostic impact of the NPM1mut/FLT3-TKD genotype, with a 5-year OS rate of ∼70%. In this study, we identified another favorable prognostic subgroup, represented by patients with a CBF-AML exhibiting a FLT3-TKD mutation. Following AML with an NPM1 mutation, CBF-AML was the second most frequent disease entity, representing 28% of NPM1WT patients. In the multivariable analysis, both NPM1mut/FLT3-TKD and CBF/FLT3-TKD genotypes were independent prognostic factors for prolonged survival when including allogeneic HCT performed in first CR as a time-dependent covariate, whereas treatment arm did not play a significant role.
Although the trial was not designed to compare the role of different postconsolidation strategies and did not include assessment of measurable residual disease, in patients with a molecular marker, allogeneic HCT could reasonably be considered in cases with persistent or increasing measurable residual disease levels, as previously proposed.15 Treatment decisions in these situations can rely on the quantification of NPM1mut transcripts by quantitative PCR, which has been shown to consistently predict outcome, with persistent negativity being associated with prolonged remission status and cure.25,26 Similarly, in the context of CBF-AML, levels of RUNX1-RUNX1T1 and CBFB-MYH11 transcripts, overall or as log reduction in transcript number after consolidation therapy, were shown to be predictive of relapse.27,28
Evaluation of other disease features may further guide treatment stratification, as the characterization mutations of the CEBPA gene, which have been reported in 7% to 8% of patients with TKD mutations.4,15,23,29 Data deriving from the ongoing genomic profiling of FLT3-mutated AML included in the RATIFY trial may further improve the stratification model.
Relative to treatment, midostaurin had no significant impact on achievement of CR after in the overall group, or in NPM1mut patients. Similarly, in the recently published subanalysis of the RATIFY trial, in 318 patients with NPM1mut/FLT3-ITD mutation grouped according to the 2017 ELN classification, we found that rates of CR were similar and independent of treatment arm in the favorable- and intermediate-risk categories, which include most patients with NPM1 mutations.6,7 Neither FLT3-ITD nor FLT3-TKD mutations had an impact on achievement of CR in a large patient series treated with conventional chemotherapy followed by allogeneic HCT in about 20% of the patients.30
The major limitation of our analysis was the relatively low number of patients with a FLT3-TKD mutation derived from the RATIFY trial, which was not powered for subgroup analyses. In this patient subgroup, midostaurin showed a beneficial effect in terms of EFS. Patients with combined NPM1mut/FLT3-TKD and CBF/FLT3-TKD genotypes were identified as prognostically favorable patient subgroups, with an overall significant survival advantage, when compared with patients with NPM1WT AML or to those with other karyotype abnormalities.
Send data sharing requests via e-mail to the corresponding author, Maria Teresa Voso (voso@med.uniroma2.it).
Acknowledgments
The authors acknowledge the support of the National Institutes of Health, National Cancer Institute grants U24 CA196171 and U10 CA180821 (Alliance for Clinical Trials in Oncology), UG1 CA233338 (C.D.B.), and U10 CA180867 (Dana-Farber Cancer Institute); AIRC 5x1000 call “Metastatic disease: the key unmet need in oncology to MYNERVA” project, #21267 (MYeloid NEoplasms Research Venture AIRC) (M.T.V.); the Deutsche Forschungsgemeinschaft (SFB 1074, project B3) (K.D.); and Catalan and Spanish government grants (2017 SGR 1395 and PERIS SLT002/16/00433 and ISCIII FIS PI17/01246) (J.S.). Study CALGB (Alliance) 10603 was funded in part by Novartis (CALGB is now part of the Alliance for Clinical Trials in Oncology).
The authors regretfully acknowledge the passing of 2 of the senior authors (C.D.B. and F.L.-C.), who made immense contributions to the design and analysis of the study, and to the writing of the manuscript.
Authorship
Contribution: M.T.V., H.D., C.D.B., and F.L.-C. analyzed data and wrote the paper; R.A.L., D.J., G.M., T.P., J.K., M.H., S.L., J.N., S.M.G., A.W., A.W., J.S., M.A.S., J.M.B., T.M.d.W., J.H.J., D.N., F.R.A., B.C.M., M.S.T., R.F.S., A.G., S.A., G.E., J.B., C.T., K.D., and R.M.S. performed research and reviewed the paper; and Y.C., Y.M.C., C.P., L.D., and A.P. analyzed data and reviewed the paper.
Conflict-of-interest disclosure: M.T.V. has received research funding and is a member of the Speakers Bureau for Celgene. R.A.L. has served on independent data safety monitoring committees and acted as a consultant or advisor and has received clinical research support from Novartis. D.J. has received clinical research support from Novartis. G.M. has acted as a consultant or advisor for trials supported by Novartis. J.K. has acted as a consultant or advisor for Amgen, Astellas, Bristol-Myers Squibb, Daiichi Sankyo, and Pfizer. M.H. has acted as a consultant or advisor for AbbVie, Daiichi Sankyo, Novartis, Pfizer, and Bayer Pharma and has received clinical research support from Pfizer, Daiichi Sankyo, Karyopharm, BerGenBio, Bayer Pharma, Novartis, and Astellas. J.N. has acted as consultant or advisor for Novartis. A.W. has received clinical research support from Novartis. A.H.W. has acted as a consultant or advisor for Novartis, Astellas, Pfizer, MacroGenics, AbbVie, Genentech, Servier, Celgene, Takeda, and Argenix; has consulted for Amgen, AstraZeneca, and Janssen; is a member of the speakers bureau for AbbVie/Genentech and Novartis; has received research funding from Novartis, Celgene, AbbVie, Servier, AstraZeneca, and Amgen; and is a former employee of the Walter and Eliza Hall Institute and receives a fraction of its royalty stream related to venetoclax. J.S. has acted as a consultant or advisor for Pfizer, Daiichi Sankyo, AbbVie, Novartis, Astellas, and Roche and is a member of the speakers’ bureau for Novartis, Pfizer, Daiichi Sankyo, and AbbVie. M.A.S. has acted as a consultant or advisor for Teva Pharmaceutical Industries, Daiichi Sankyo, Orsenix, AbbVie, Novartis, and Pfizer. J.M.B. has acted as a consultant or advisor for Novartis, Celgene, Pfizer, Astellas, Teva, and Roche and has received research funding from Celgene. T.M.d.W. has acted as a consultant or advisor for Novartis, Celgene, Johnson & Johnson, and Incyte and has received clinical research support from Novartis, Celgene, and Johnson & Johnson. D.N. reports support from Cellectis and Daiichi and has participated in a Novartis speakers’ bureau outside the scope of the submitted work. B.C.M. has acted as a consultant or advisor for Celgene, Novartis, and Astellas and is currently employed by Roche/Genentech. M.S.T. has acted as a consultant or advisor for AbbVie, BioLineRx, Daiichi Sankyo, Orsenix, KAHR Medical, Rigel Pharmaceuticals, Nohla, Delta Fly Pharma, Tetraphase, Oncolyze, and Jazz Pharmaceuticals; has received clinical research funding from AbbVie, Cellerant Therapeutics, Orsenix, ADC Therapeutics, and BioSight; and has received royalties from UpToDate. R.F.S. has acted as a consultant or advisor for Pfizer and Daiichi Sankyo; is a member of the speakers’ bureau for Novartis, Pfizer, and Daiichi Sankyo; and has received clinical research funding from Pfizer, Daiichi Sankyo, PharmaMar, AstraZeneca, and Roche. A.G. has received clinical research support from Novartis. S.A. has acted as a consultant or advisor for Novartis and Daiichi Sankyo. Y.C., Y.M.C., and L.D. disclose employment with Novartis Pharmaceuticals Corporation. C.D. discloses employment with Novartis Pharma AG. C.T. is the Chief Executive Officer and a co-owner of Agendix, a company performing molecular diagnostics; has acted as a consultant or advisor for Novartis and Astellas; and has received clinical research support from Bayer. K.D. has acted as a consultant or advisor for Astellas, Celgene, Daiichi Sankyo, Janssen, Novartis, and Roche and has received clinical research support from Astex, Celgene, and Novartis. R.M.S. has acted as a consultant or advisor for AbbVie, Actinium, and Agios; has received personal fees from Amgen, Arogargenx, AROG, Astellas, AstraZeneca, BiolinerxBioLineRx, Celgene, Cornerstone, Daiichi Sankyo, Fujifilm, Janssen, Juno, MacrogenicsJazz Pharmaceuticals, MacroGenics, Novartis, Ono/Theradex Oncology, Orsenix, Otsuka/Astex, Pfizer, Roche, Stemline, Sumitomo Therapeutics, Takeda, and Trovagene; and has received institutional research support (to the institution) for clinical trials sponsored by AbbVie, Agios, AROG, and Novartis. H.D. has acted as a consultant or advisor for AbbVie, Agios, Amgen, Astellas, Astex Pharmaceuticals, Celgene, Janssen, Jazz Pharmaceuticals, Novartis, Roche, and Seattle Genetics and has received institutional research support from Amgen, AROG Pharmaceuticals, Bristol-Myers Squibb, Celgene, Jazz Pharmaceuticals, Novartis, Pfizer, and Sunesis. F.L.-C. discloses honoraria and a consulting or advisory role from Teva Pharmaceuticals Industries and Lundbeck. The remaining authors declare no competing financial interests.
Clara D. Bloomfield died on 1 March 2020.
Francesco Lo-Coco died on 3 March 2019.
Correspondence: Maria Teresa Voso, Department of Biomedicine and Prevention, University of Rome Tor Vergata, Via Montpellier 1, 00133 Rome, Italy; e-mail: voso@med.uniroma2.it.
References
Author notes
H.D., C.D.B., and F.L.-C. share senior authorship
The full-text version of this article contains a data supplement.