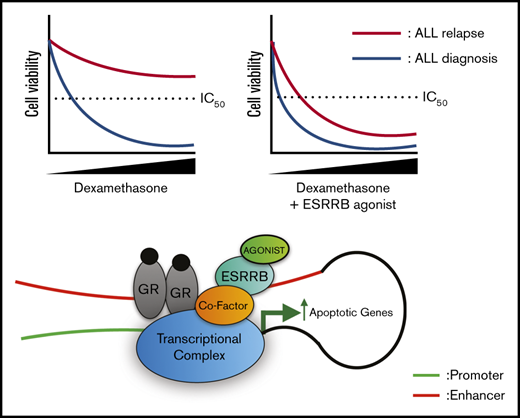
Key Points
Repression of ESRRB results in glucocorticoid resistance in ALL, and glucocorticoid-regulated transcription depends on ESRRB expression.
ESRRB agonists synergize with dexamethasone to induce human leukemic cell death.
Abstract
Synthetic glucocorticoids (GCs), such as dexamethasone and prednisone, remain key components of therapy for patients with lymphoid malignancies. For pediatric patients with acute lymphoblastic leukemia (ALL), response to GCs remains the most reliable prognostic indicator; failure to respond to GC correlates with poor event-free survival. To uncover GC resistance mechanisms, we performed a genome-wide, survival-based short hairpin RNA screen and identified the orphan nuclear receptor estrogen-related receptor-β (ESRRB) as a critical transcription factor that cooperates with the GC receptor (GR) to mediate the GC gene expression signature in mouse and human ALL cells. Esrrb knockdown interfered with the expression of genes that were induced and repressed by GR and resulted in GC resistance in vitro and in vivo. Dexamethasone treatment stimulated ESRRB binding to estrogen-related receptor elements (ERREs) in canonical GC-regulated genes, and H3K27Ac Hi-chromatin immunoprecipitation revealed increased interactions between GR- and ERRE-containing regulatory regions in dexamethasone-treated human T-ALL cells. Furthermore, ESRRB agonists enhanced GC target gene expression and synergized with dexamethasone to induce leukemic cell death, indicating that ESRRB agonists may overcome GC resistance in ALL, and potentially, in other lymphoid malignancies.
Introduction
Glucocorticoids (GCs) are critical components of multiagent chemotherapy for lymphoid malignancies. Of the lymphoid malignancies, acute lymphoblastic leukemia (ALL) is the most common one that occurs in childhood and involves transformation of B- or T-lymphoid progenitors.1,2 A patient’s response to GCs is the most reliable prognostic indicator in pediatric ALL, and GC resistance remains an obstacle to improving the outcomes of these patients.3,4 In lymphoid cells, synthetic GCs such as dexamethasone induce apoptosis by stimulating GC-receptor (GR) translocation to regulate transcription.5 In lymphoid cells, GC treatment induces proapoptotic genes, including BCL2L11 (BIM). There is also evidence that the GR represses expression of the prosurvival genes BCL2 and BCL-XL.6
GC resistance can result from mutation in NR3C1, which encodes the GR; however, these mutations are rare in patients with relapsed ALL.7 GR activity can also be impaired by AKT-mediated phosphorylation of the GR or by NLRP3 inflammasome activation and caspase 1-mediated cleavage of the GR.8,9 Impaired GC responses can also result from missense mutations in NR3C1 coactivators such as CREBBP.10 Several mechanisms of GC resistance involve activation of prosurvival pathways, including LCK, FLT3, WNT, MAPK, IL-7-JAK/STAT, and mTOR signaling.11-17 An impaired GC response in ALL can also reflect increased DNA methylation of the BCL2L11 locus observed in a subset of GC-resistant patients.18 Last, mutations in NOTCH1 increase HES1 levels, which interfere with GR autoregulation, contributing to GC resistance.19
To further elucidate GC resistance mechanisms in ALL, we performed a short hairpin RNA (shRNA) screen in primary T-ALL cells isolated from a Tal1/Lmo2 mouse T-ALL model.20 We found that shRNAs targeting the GR (Nr3c1) or known GC resistance genes (Rcan1, Mllt10, Smarca1, Smard2, and Btg1) were enriched in the screening. Importantly, shRNAs for Ikzf1, Utx, and the CREBBP paralogue Ep300, known leukemia-suppressor genes in human ALL, conferred dexamethasone resistance in our mouse screening. An shRNA targeting the estrogen related receptor-β (ESRRB) was identified in the screening. ESRRB is a member of the estrogen-related receptor (ERR) family of orphan nuclear receptors and is known to act as a constitutively active transcription factor that binds an estrogen-related response element (ERRE) to regulate gene expression.21 ESRRB has been studied in mouse embryonic stem (ES) cells, where ESRRB maintains self-renewal through activation of Oct-4 and as a member of the NANOG complex.22,23 We revealed novel functions of ESRRB in the control of GR-mediated transcription and showed that an ESRRB agonist potentiates dexamethasone-induced gene expression and apoptosis. The data suggest that ESRRB agonists may provide therapeutic benefit to GC-resistant patients with ALL.
Materials and methods
Mice and cells
Tal1/Lmo2 transgenic mice, generated when M.A.K. was a postdoc at Harvard Medical School, were monitored for leukemia, and mouse and human T-ALL cells were cultured as described.24 Primary human T-ALL samples were expanded in NSG mice and cultured as previously described.25 All animal procedures used in this study were approved by the University of Massachusetts Medical School Institutional Animal Care and Use Committee.
Cell proliferation and death assays
Mouse or human ALL cell lines or samples were cultured in increasing concentrations of dexamethasone (0-10 μM) for 24 to 72 hours, and cell viability was monitored by trypan blue staining and cell proliferation was observed by carboxyfluorescein succinimidyl ester (CFSE) staining followed by flow cytometry. Metabolic activity was also assayed by using CellTiter-Glo chemiluminescence reagent (Promega). Mouse and human leukemic cells were treated with dexamethasone, 2 × 105 cells were stained with annexin V-fluorescein isothiocyanate and 7-aminoactinomycin D (7-AAD), and apoptotic cells were quantified by flow cytometry. The synergistic relationship between dexamethasone and ESRRB agonists was determined by the Chou-Talalay method.26
Quantitative real-time polymerase chain reaction
Total RNA was extracted by using Trizol, and cDNA was synthesized from RNA (2 μg) by using the Superscript First-Strand Synthesis System (Invitrogen). Quantitative real-time- polymerase chain reaction (qRT-PCR) was performed on the AB7300 Detection System (Applied Biosystems), using Power SYBR Green Master Mix (Applied Biosystems) and gene-specific primers. Gene expression was determined using the ΔΔ cycle threshold (ΔΔCT) method and normalized to β-actin. Specific primer sequences are provided in supplemental Table 5.
RNA sequencing and chromatin immunoprecipitation-qPCR
RNA was isolated from mouse T-ALL cells infected with nonsilencing (NS) Nr3c1 or Esrrb shRNAs treated with vehicle or dexamethasone for 6 hours, using the Invitrogen RNA mini kit. RNA was sent to BGI (Shenzhen, China) for library preparation and paired-end sequencing. RSEM was used to quantify RNA-sequencing results.27 For chromatin immunoprecipitation (ChIP), 107 human ALL cells (KOPTK1) were treated with dexamethasone or dimethyl sulfoxide (DMSO) for 12 hours, and ChIP-qPCR was performed as previously described.28 Specific primer sequences are in supplemental Table 5.
Hi-ChIP
T-ALL cells (1 × 107) were cross-linked for 10 minutes in 1% formaldehyde solution. Nuclei were harvested and sonicated on a Covaris ME220 for 6 minutes. Chromatin was incubated with 7.5 µg of H3K27ac antibody overnight at 4°C. The next day, 60 µL of protein G magnetic beads was used to isolate antibody-bound chromatin. DNA was then purified with DNA Clean and Concentrate 5 columns (D4013; Zymo), and PCR amplification was performed. DNA was purified and sequenced by BGISeq500, using a 50-bp paired-end library. Full Hi-ChIP method is available in the supplemental Methods and has been adapted from the original publication.29
Results
The orphan nuclear receptor ESRRB regulates GC-induced cell death in vitro and in vivo
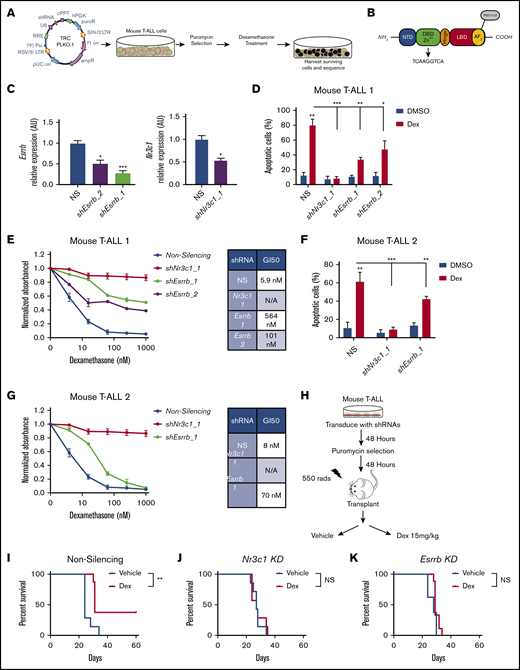
To identify GC resistance genes, we transduced the dexamethasone-sensitive mouse T-ALL cell line 1390 with a whole-genome lentiviral TRC (The RNAi Consortium) library containing ∼75 000 shRNA constructs directed against 16 000 genes. Leukemic cells were infected with a multiplicity of infection of 0.2 to achieve a single shRNA per cell. Infected cells were selected with puromycin for 48 hours and treated with dexamethasone for 4 days to kill >99% of the leukemic cells. Surviving cells were expanded and sequenced to identify shRNAs that confer dexamethasone resistance (Figure 1A). This screen identified an shRNA specific for ESRRB, an orphan nuclear receptor containing a zinc-finger DNA binding domain and a ligand binding domain that contains an activation function (AF)-2 domain for the recruitment of coactivators or corepressors (Figure 1B). We achieved >50% knockdown of Esrrb in mouse T-ALL cells, using 2 independent shRNAs (Figure 1C; supplemental Figure 1A). To determine whether ESRRB alters dexamethasone response, we treated leukemic cells deficient for Esrrb or Nr3c1 encoding the GR with dexamethasone and assessed cell viability by MTS assay and cell death by annexin V/7-AAD staining. Knockdown of Esrrb prevents dexamethasone-induced cell death and shifts the GI50 for dexamethasone ∼96-fold from 6 to 564 nM, or 17-fold when a second, less effective, Esrrb shRNA is used (Figure 1D-E). We examined GC responses in an additional mouse T-ALL cell line (5059) and found that Esrrb knockdown shifted the GI50 from 8 to 70 nM and prevented dexamethasone-induced apoptosis (Figure 1F-G). Although we showed that an ESRRB-deficiency results in dexamethasone resistance in vitro, we recognized that dexamethasone resistance in vivo may be influenced by prosurvival factors. For example, the lymphoid survival factor IL-7 can contribute to dexamethasone resistance.11 Therefore, we examined the effects of Esrrb knockdown on the dexamethasone response in vivo. Mouse T-ALL cells deficient for Esrrb or Nr3c1 were transplanted into sublethally irradiated syngeneic recipients, and the mice were treated with vehicle or dexamethasone (15 mg/kg) for 3 weeks (Figure 1H). In mice receiving transplants of leukemic cells expressing an NS control, dexamethasone significantly prolonged survival and eliminated disease in 4 of 10 mice (Figure 1I). In contrast, dexamethasone treatment had no significant effect on the survival of mice that received Esrrb- or Nr3c1-deficient leukemic cells; all animals succumbed to the disease (Figure 1J-K). These data demonstrate that an ESRRB-deficiency results in GC resistance in vivo.
The orphan nuclear receptor ESRRB regulates GC-induced cell death in vitro and in vivo. (A) ESRRB was identified as a mediator of GC resistance in a whole-genome mouse shRNA screen, using the whole-genome TRC shRNA library. (B) ESRRB contains a ligand and DNA binding domain that binds an ERRE site and mediates transcriptional changes via the recruitment of coactivator or corepressor proteins to its AF2 domain. (C) Mouse T-ALL cells (1390) were transduced with lentiviruses expressing NS control or 2 independent shRNAs targeting mouse Esrrb (1, 2) or Nr3c1 and Esrrb and Nr3c1 mRNA expression was analyzed by qRT-PCR. The copy number was normalized to β-actin using the ΔΔCT method. Knockdown of Esrrb in mouse T-ALL cells 1 (1390) and 2 (5059) resulted in resistance to dexamethasone-induced apoptosis. (D,F) Mouse T-ALL cells expressing NS or Esrrb-shRNAs were cultured with dexamethasone (50 nM) for 2 days, and apoptosis was assayed by annexin V-FITC/7-AAD staining followed by flow cytometry. (E,G) ESRRB repression significantly shifted dexamethasone GI50. NS cells or cells deficient in ESRRB or NR3C1 were cultured in increasing concentrations of dexamethasone for 48 hours, and viability was assayed by MTS. Absorbance values were normalized to vehicle control. (H) Experimental approach to examine dexamethasone response in vivo. Esrrb or Nr3c1 knockdown mediated dexamethasone resistance in vivo. Kaplan-Meier survival curves demonstrate that dexamethasone significantly delayed disease in mice receiving transplants of mouse T-ALL cells transduced with NS shRNA (I) but had no effect on mice receiving transplants of Nr3c1- (J) or Esrrb- (K) gene–specific shRNAs (n = 5-7 mice per group; statistics by log-rank test). The results are averages of 3 independent experiments; error bars represent standard error of the mean (SEM). *P < .05; **P < .01; ***P < .001. NS, not significant.
The orphan nuclear receptor ESRRB regulates GC-induced cell death in vitro and in vivo. (A) ESRRB was identified as a mediator of GC resistance in a whole-genome mouse shRNA screen, using the whole-genome TRC shRNA library. (B) ESRRB contains a ligand and DNA binding domain that binds an ERRE site and mediates transcriptional changes via the recruitment of coactivator or corepressor proteins to its AF2 domain. (C) Mouse T-ALL cells (1390) were transduced with lentiviruses expressing NS control or 2 independent shRNAs targeting mouse Esrrb (1, 2) or Nr3c1 and Esrrb and Nr3c1 mRNA expression was analyzed by qRT-PCR. The copy number was normalized to β-actin using the ΔΔCT method. Knockdown of Esrrb in mouse T-ALL cells 1 (1390) and 2 (5059) resulted in resistance to dexamethasone-induced apoptosis. (D,F) Mouse T-ALL cells expressing NS or Esrrb-shRNAs were cultured with dexamethasone (50 nM) for 2 days, and apoptosis was assayed by annexin V-FITC/7-AAD staining followed by flow cytometry. (E,G) ESRRB repression significantly shifted dexamethasone GI50. NS cells or cells deficient in ESRRB or NR3C1 were cultured in increasing concentrations of dexamethasone for 48 hours, and viability was assayed by MTS. Absorbance values were normalized to vehicle control. (H) Experimental approach to examine dexamethasone response in vivo. Esrrb or Nr3c1 knockdown mediated dexamethasone resistance in vivo. Kaplan-Meier survival curves demonstrate that dexamethasone significantly delayed disease in mice receiving transplants of mouse T-ALL cells transduced with NS shRNA (I) but had no effect on mice receiving transplants of Nr3c1- (J) or Esrrb- (K) gene–specific shRNAs (n = 5-7 mice per group; statistics by log-rank test). The results are averages of 3 independent experiments; error bars represent standard error of the mean (SEM). *P < .05; **P < .01; ***P < .001. NS, not significant.
ESRRB has no effect on the leukemic gene expression signature or on leukemic growth
ESRRB is required for embryonic stem (ES) cell pluripotency and self-renewal of trophoblast stem cells.30 Moreover, ESRRB functions as a mitotic bookmarking factor in ES cells.31 These findings suggest that ESRRB may alter leukemic cell proliferation. We examined ESRRB effects on leukemic growth by labeling mouse leukemic cells expressing an NS control or Esrrb-specific shRNAs with CFSE and monitored proliferation. An ESRRB-deficiency had no significant effect on CFSE loss or dilution, because of proliferation. Furthermore, a similar number of leukemic cells was observed in cultures expressing NS- or Esrrb-specific shRNAs (supplemental Figure 1B-C). To examine the effects of an ESRRB-deficiency on leukemic gene expression, we performed RNA sequencing on NS- and Esrrb-knockdown cells and found that Esrrb knockdown had no effect on basal leukemic gene expression (supplemental Figure 1D). To assess the effects of ESRRB on leukemic growth in vivo, we transplanted mouse leukemic cells expressing NS or Esrrb-specific shRNAs into sublethally irradiated, syngeneic mice and monitored disease progression. Consistent with the gene expression studies, an ESRRB deficiency had no significant effects on leukemic growth in vivo (supplemental Figure 1E; P = .4).
ESRRB cooperates with GR to potentiate dexamethasone-induced gene expression
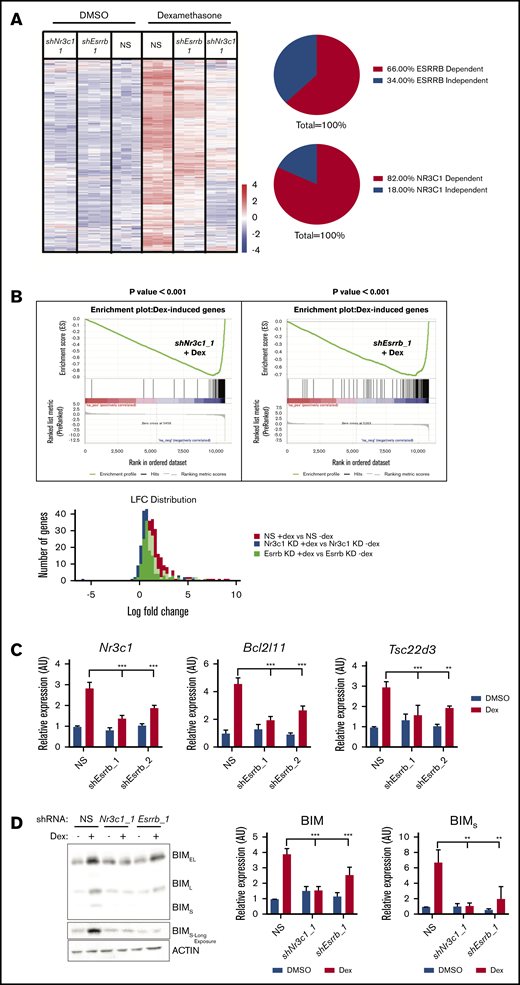
GCs, such as dexamethasone, induce cell death in lymphoid cells by activating GR-mediated transcription.5 We hypothesized that ESRRB deficiency would mediate GC resistance by interfering with expression of GR-regulated genes. We performed RNA sequencing on vehicle or dexamethasone-treated mouse leukemic cells expressing NS control or Nr3c1- or Esrrb-specific shRNAs. Our RNA-sequencing studies revealed 339 genes of diverse functions that were altered by dexamethasone treatment of T-ALL cells and, as expected, gene expression was significantly abrogated by Nr3c1 knockdown (Figure 2A-B; supplemental Figure 2). Surprisingly, we found that dexamethasone-regulated gene expression was also abrogated by Esrrb knockdown, with 66% of dexamethasone-induced genes dependent on ESRRB (Figure 2A). Gene set enrichment analysis revealed that dexamethasone-regulated gene expression correlated negatively with Esrrb or Nr3c1 knockdown when compared to leukemic cells expressing the NS control (Figure 2B, top). Analysis of the log-fold change in NS or Esrrb- or Nr3c1-silenced leukemic cells showed that the greatest induction of dexamethasone-regulated genes occurred in the NS control leukemic cells, and Esrrb knockdown reduced the expression of 143 of the 214 dexamethasone-induced genes (66%; Figure 2B, bottom). These data reveal that, similar to NR3C1-deficiency, Esrrb knockdown interfered with dexamethasone-induced gene expression in mouse T-ALL cells.
ESRRB cooperates with GR to potentiate dexamethasone-induced gene expression. RNA was isolated from mouse T-ALL cells infected with NS, Nr3c1, or Esrrb shRNAs treated with vehicle (DMSO) or dexamethasone (100 nM) for 6 hours and sequenced using BGI-seq 500. RSEM was used to quantify RNA-sequencing results. (A) The heat map represents differentially expressed genes between vehicle- and dexamethasone-treated NS cells using log 1.5-fold cutoff (n = 3 replicates). (B) Gene set enrichment analysis (top) on dexamethasone-induced gene set shows significant negative correlation in Esrrb-knockdown leukemic cells treated with dexamethasone (P < .001). Distribution of log fold change across detected genes (bottom). The NS cells exhibit the highest average log fold change when treated with dexamethasone, whereas Nr3c1 or Esrrb knockdown interferes with optimal changes in gene expression. Nr3c1, Bcl2l11, and Tsc22d3 are GR-regulated genes dependent on ESRRB for their expression. (C) Control or Esrrb-knockdown cells (2 independent shRNAs) were treated with dexamethasone for 6 hours, and Nr3c1, Bcl2l11, and Tsc22d3 mRNA expression was analyzed by qRT-PCR. The copy number was normalized to β-actin using the ΔΔCT method. (D) BIM protein expression was analyzed by immunoblot after 12 hours of dexamethasone (100 nM) treatment in NS or Nr3c1- or Esrrb-knockdown cells. Protein was quantified using densitometry and normalized to actin. The results are averages of 3 independent experiments; error bars represent SEM. **P < .01; ***P < .001.
ESRRB cooperates with GR to potentiate dexamethasone-induced gene expression. RNA was isolated from mouse T-ALL cells infected with NS, Nr3c1, or Esrrb shRNAs treated with vehicle (DMSO) or dexamethasone (100 nM) for 6 hours and sequenced using BGI-seq 500. RSEM was used to quantify RNA-sequencing results. (A) The heat map represents differentially expressed genes between vehicle- and dexamethasone-treated NS cells using log 1.5-fold cutoff (n = 3 replicates). (B) Gene set enrichment analysis (top) on dexamethasone-induced gene set shows significant negative correlation in Esrrb-knockdown leukemic cells treated with dexamethasone (P < .001). Distribution of log fold change across detected genes (bottom). The NS cells exhibit the highest average log fold change when treated with dexamethasone, whereas Nr3c1 or Esrrb knockdown interferes with optimal changes in gene expression. Nr3c1, Bcl2l11, and Tsc22d3 are GR-regulated genes dependent on ESRRB for their expression. (C) Control or Esrrb-knockdown cells (2 independent shRNAs) were treated with dexamethasone for 6 hours, and Nr3c1, Bcl2l11, and Tsc22d3 mRNA expression was analyzed by qRT-PCR. The copy number was normalized to β-actin using the ΔΔCT method. (D) BIM protein expression was analyzed by immunoblot after 12 hours of dexamethasone (100 nM) treatment in NS or Nr3c1- or Esrrb-knockdown cells. Protein was quantified using densitometry and normalized to actin. The results are averages of 3 independent experiments; error bars represent SEM. **P < .01; ***P < .001.
To identify the biological processes regulated by the GR and ESRRB, we performed gene ontology analysis using DAVID32,33 and found genes enriched for cell death. In fact, dexamethasone induced expression of 42 genes related to cell death, 26 of which were dependent on ESRRB (supplemental Table 1). The ESRRB-dependent proapoptotic genes include Tsc22d3, which encodes GC-induced leucine zipper, known to trigger thymocyte apoptosis when overexpressed,34 and the proapoptotic Bcl2l11 gene, which encodes BIM (supplemental Table 1). Although Bcl2l11 scored just below the 1.5-fold cutoff, qRT-PCR with 2 independent shRNAs confirmed that Bcl2l11, Tsc22d3, and Nr3c1 expression significantly decreased in dexamethasone-treated Esrrb-knockdown leukemic cell lines (Figure 2C). Additional ESRRB-dependent genes include the death-associated protein kinase Stk17b, the Litaf gene implicated in B-cell apoptosis, and the p53 targets Epha2 and Acer2.35-38 Induction of proapoptotic BIM is critical for dexamethasone-induced cell death, and we confirmed that Esrrb knockdown reduces BIM protein levels by approximately twofold.18,39 Importantly, we saw induction of the short BIM isoform, the most potent inducer of apoptosis, reduced by more than threefold (Figure 2D).40 These data suggest that the dexamethasone resistance observed in ESRRB-deficient leukemic cells can be explained in part by a failure to induce proapoptotic genes and sufficient BIM protein levels.
Our RNA-sequencing data also revealed a set of GC-repressed genes that were influenced by ESRRB. Approximately 58% of the dexamethasone-repressed genes were affected by Nr3c1 knockdown and 54% of these repressed genes appeared to be ESRRB dependent (supplemental Figure 3A-B). Consistent with anti-inflammatory functions of the GR, we found that the repressed genes were related to immune regulation (supplemental Table 2). Interestingly, we found that genes important in thymocyte differentiation, including Myb, Rag1, Rasgrp1, and Runx1, are repressed by dexamethasone in an ESRRB-dependent manner. These data are consistent with published B-ALL studies, which implicate GC repression of B-cell development genes in the therapeutic response.41 We validated the repressive effects of ESRRB on 3 randomly selected candidate genes, Bcl2l1, Rag1, and Klf3, in mouse leukemic cells by qRT-PCR and found expression of these genes to be unaffected by dexamethasone in ESRRB-knockdown cells (supplemental Figure 3C). This finding is consistent with studies in human ALL cells, where dexamethasone repressed BCL2 and BCL2L1.6 Because Bcl2l1 (encoding BCL-XL) expression was not significantly repressed in ESRRB-knockdown cells, we used the ABT-263 inhibitor of BCL-XL and found that ABT-263 treatment resensitized ESRRB-deficient cells to dexamethasone-induced apoptosis (supplemental Figure 3D). Collectively, these data reveal that ESRRB cooperates with the GR to regulate gene expression and is consistent with known functions, where ESRRB recruits coactivators and corepressors to regulate gene expression.21
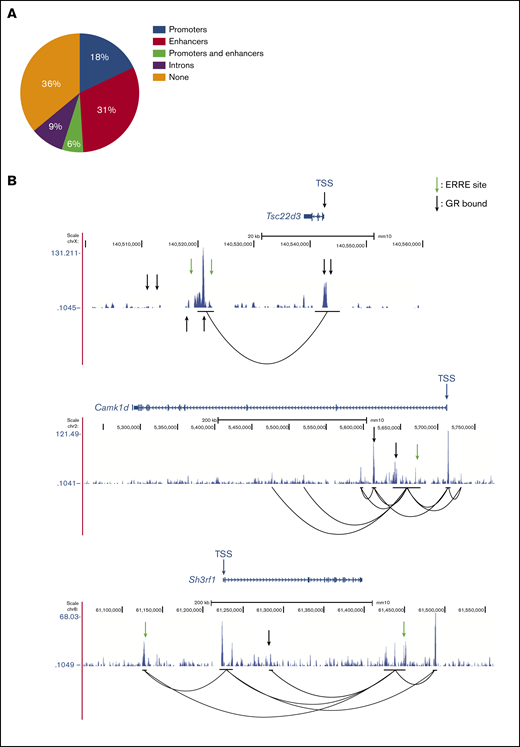
GBS- and ERRE-containing cis-regulatory elements interact with promoter regions of GR- and ESRRB-dependent genes in mouse T-ALL cells
Our RNA-sequencing data revealed that most dexamethasone-regulated genes in mouse T-ALL cells depend on ESRRB. We examined the GR- and ESRRB-dependent genes for the presence of ERREs. Motif analysis revealed ERREs in 49% of these genes, with 18% of the genes harboring ERREs in promoters, 31% in putative enhancer regions, and 6% in both promoters and enhancers (Figure 3A). We then performed H3K27Ac Hi-ChIP in the mouse T-ALL cell line (2553) and detected preestablished interactions between putative enhancer regions and looping between promoter-enhancer elements in the GR/ESRRB-dependent genes examined (Figure 3B). Using published GR ChIP-sequencing data42 we confirmed GR binding to the promoter and/or enhancer regions of Tsc22d3, Camk1d, and Sh3rf1 (Figure 3B; arrows). We then validated these findings in 2 additional mouse T-ALL cell lines (720 and 1390; supplemental Figure 4).
GBS- and ERRE-containing cis-regulatory elements interact with promoter regions of ESRRB- and GR-regulated genes in mouse T-ALL cells. (A) Distribution of ERREs in GR-regulated, ESRRB-dependent genes. (B) H3K27Ac Hi-ChIP tracks of 3 ESRRB/GR-dependent mouse genes show preexisting promoter interactions with active ERRE-containing enhancers known to bind the GR. Black lines indicate the anchors of interaction.
GBS- and ERRE-containing cis-regulatory elements interact with promoter regions of ESRRB- and GR-regulated genes in mouse T-ALL cells. (A) Distribution of ERREs in GR-regulated, ESRRB-dependent genes. (B) H3K27Ac Hi-ChIP tracks of 3 ESRRB/GR-dependent mouse genes show preexisting promoter interactions with active ERRE-containing enhancers known to bind the GR. Black lines indicate the anchors of interaction.
ESRRB regulates gene expression in ESCs through complex formation with NANOG,23 suggesting that an ESRRB/GR complex may regulate GC gene expression. Co-IP experiments, however, failed to detect an ESRRB/GR complex in ALL cells or in transfected HEK293T cells (supplemental Figure 5A-B). Our analysis of the GR/ESRRB-dependent genes in mouse T-ALL cells found GR and ERRE motifs to be nonoverlapping (Figure 3A), leading us to hypothesize that ESRRB and GR bind DNA independently; however, 36% of genes harbored sites that were separated by >10 kb, suggesting that ESRRB and GR may interact to mediate chromatin looping.
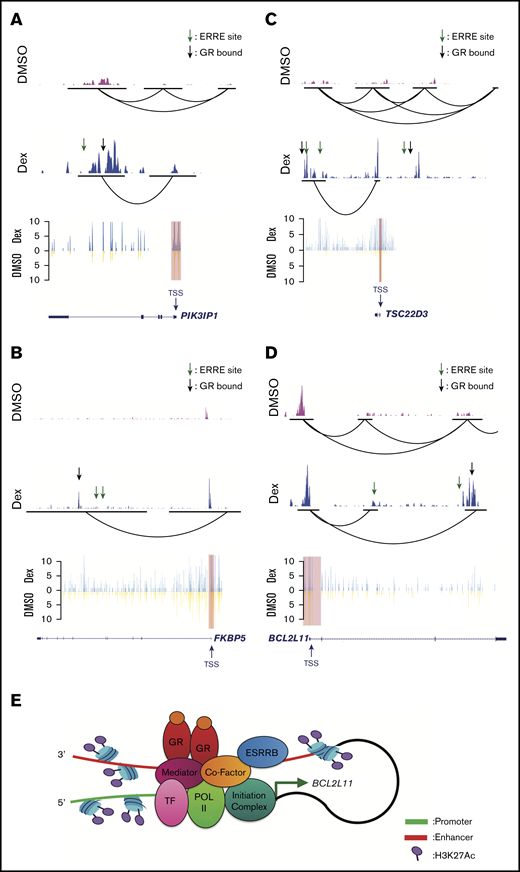
We next examined the effects of dexamethasone on H3K27Ac levels and chromatin looping in human T-ALL cells, by using the canonical GR target genes PIK3IP1, FKBP5, and TSC22D3 identified by Piovan et al.8 Similar to our Hi-ChIP studies in mouse T-ALL cells (Figure 3), we detected preexisting promoter/enhancer interactions in PIK3IP1, TSC22D3, and BCL2L11 (Figure 4A,C-D). We used virtual 4C analysis of our Hi-ChIP data to quantify interaction frequency. Using the transcription start site of these GR-regulated genes as the viewpoint (red highlight), we find that dexamethasone significantly increased the frequency of promoter interactions with GR-bound and ERRE-containing enhancer elements; furthermore, dexamethasone increased H3K27Ac levels at these putative enhancers (Figure 4A-C). Similarly, we detected increases in active histone marks and promoter/enhancer interactions at the BCL2L11 locus, where our Hi-ChIP data validated the presence of a dexamethasone-responsive intronic enhancer (Figure 4D), previously observed in GC-sensitive ALL cells.39 Together, these data suggest that GR and ESRRB binding support these promoter/enhancer interactions to potentiate gene expression (Figure 4E).
Dexamethasone increases H3K27Ac levels and promoter/enhancer interactions in canonical GC-regulated genes in human T-ALL cells. H3K27Ac Hi-ChIP tracks (top) showing interaction between H3K27Ac regions in canonical GC target genes PIK3IP1 (A), FKBP5 (B), TSC22D3 (C), and BCL2L11 (D), treated with DMSO (pink track) or dexamethasone (blue track). Black lines indicate the anchors of interaction. Virtual 4C extract from Hi-ChIP (bottom) shows interaction frequency from the viewpoint (red highlight) of GC target genes PIK3IP1 (A), FKBP5 (B), TSC22D3 (C), and BCL2L11 (D), treated with DMSO (yellow) or dexamethasone (blue). ESRRB binding sites were determined using the ERRE consensus site AAGGTCA. GR binding sites were determined using published ChIP sequencing data in T-ALL patient samples.39 (E) Hi-ChIP data for BCL2L11 was used to model ESRRB regulation of GC-induced gene expression. Dexamethasone triggered GR and ESRRB and coactivator binding to the BCL2L11 enhancer (red) which looped back to the promoter (green) to recruit RNA polymerase II and the transcription initiation complex, thereby inducing BCL2L11 expression.
Dexamethasone increases H3K27Ac levels and promoter/enhancer interactions in canonical GC-regulated genes in human T-ALL cells. H3K27Ac Hi-ChIP tracks (top) showing interaction between H3K27Ac regions in canonical GC target genes PIK3IP1 (A), FKBP5 (B), TSC22D3 (C), and BCL2L11 (D), treated with DMSO (pink track) or dexamethasone (blue track). Black lines indicate the anchors of interaction. Virtual 4C extract from Hi-ChIP (bottom) shows interaction frequency from the viewpoint (red highlight) of GC target genes PIK3IP1 (A), FKBP5 (B), TSC22D3 (C), and BCL2L11 (D), treated with DMSO (yellow) or dexamethasone (blue). ESRRB binding sites were determined using the ERRE consensus site AAGGTCA. GR binding sites were determined using published ChIP sequencing data in T-ALL patient samples.39 (E) Hi-ChIP data for BCL2L11 was used to model ESRRB regulation of GC-induced gene expression. Dexamethasone triggered GR and ESRRB and coactivator binding to the BCL2L11 enhancer (red) which looped back to the promoter (green) to recruit RNA polymerase II and the transcription initiation complex, thereby inducing BCL2L11 expression.
Knockdown of ESRRB prevents dexamethasone-induced cell death and gene expression in human ALL cells
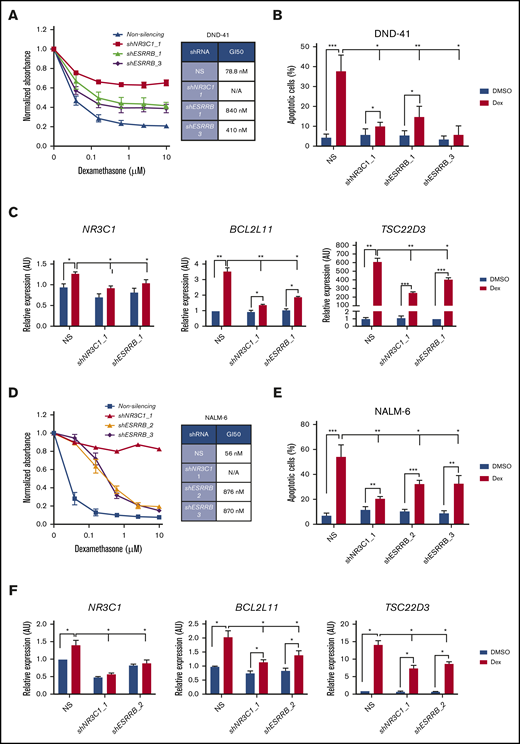
We then generated ESRRB-deficient human T- and B-ALL cells and assayed the dexamethasone response. We validated ESRRB knockdown by qRT-PCR and by immunoblot analysis (supplemental Figure 5C-E). Similar to our findings in mouse T-ALL cells, ESRRB knockdown in the human T-ALL cell line DND41 shifted the dexamethasone GI50 from 79 to 840 nM or 410 nM when a second shRNA was used (Figure 5A). As with NR3C1 knockdown, both ESRRB shRNAs prevented dexamethasone-induced apoptosis of human T-ALL cells and interfered with dexamethasone-induction of the GR target genes NR3C1, BCL2L11, and TSC22D3 (Figure 5B-C). ESRRB knockdown in the human T-ALL cell line KOPTK1 also prevented dexamethasone-induced gene expression and apoptosis (supplemental Figure 5F-G). Similarly, a deficiency in ESRRB in NALM-6 cells shifted the dexamethasone GI50 from 56 to 870 nM or 876 nM when a second independent ESRRB shRNA was used (Figure 5D; supplemental Figure 5D). ESRRB knockdown significantly reduced the percentage of apoptotic leukemic cells and interfered with dexamethasone-induced gene expression in NALM-6 cells (Figure 5E-F). These data suggest that ESRRB contributes to GC resistance in human T- and B-ALL and indicate that, although performed in mouse T-ALL cells, the genes identified in our screening may be relevant to steroid resistance in other lymphoid malignancies.
ESRRB regulates dexamethasone-induced gene expression and cell death in human ALL cell lines. (A,D) The human T-ALL cell line DND-41 and the human pre-B-ALL cell line NALM-6, transduced with lentiviruses expressing NS control or shRNAs targeting ESRRB or NR3C1, were cultured in increasing concentrations of dexamethasone for 72 hours, and viability was assayed by MTS. Absorbance values were normalized to vehicle control. (B,E) ESRRB knockdown in human T-ALL cells conferred resistance to dexamethasone-induced apoptosis. Silenced cells were cultured with dexamethasone for 2 days, and apoptosis was assayed by annexin V-FITC/7-AAD staining followed by flow cytometry. (C,F) Optimal dexamethasone-induced NR3C1, BCL2L11, and TSC22D3 gene expression required NR3C1 and ESRRB. Control or knockdown cells were treated with dexamethasone for 24 hours, and NR3C1, BCL2L11, and TSC22D3 mRNA expression was analyzed by qRT-PCR. The copy number was normalized to β-actin by the ΔΔCT method. The results are averages of 3 independent experiments; error bars represent SEM. *P < .05; **P < .01; ***P < .001.
ESRRB regulates dexamethasone-induced gene expression and cell death in human ALL cell lines. (A,D) The human T-ALL cell line DND-41 and the human pre-B-ALL cell line NALM-6, transduced with lentiviruses expressing NS control or shRNAs targeting ESRRB or NR3C1, were cultured in increasing concentrations of dexamethasone for 72 hours, and viability was assayed by MTS. Absorbance values were normalized to vehicle control. (B,E) ESRRB knockdown in human T-ALL cells conferred resistance to dexamethasone-induced apoptosis. Silenced cells were cultured with dexamethasone for 2 days, and apoptosis was assayed by annexin V-FITC/7-AAD staining followed by flow cytometry. (C,F) Optimal dexamethasone-induced NR3C1, BCL2L11, and TSC22D3 gene expression required NR3C1 and ESRRB. Control or knockdown cells were treated with dexamethasone for 24 hours, and NR3C1, BCL2L11, and TSC22D3 mRNA expression was analyzed by qRT-PCR. The copy number was normalized to β-actin by the ΔΔCT method. The results are averages of 3 independent experiments; error bars represent SEM. *P < .05; **P < .01; ***P < .001.
Dexamethasone increases ESRRB expression and binding to GR-regulated genes
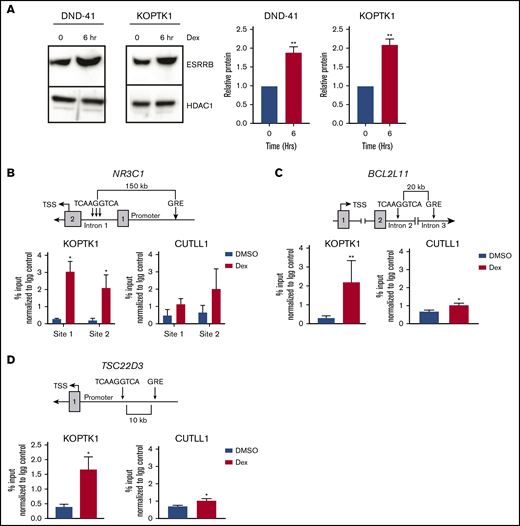
The effects of ESRRB repression on dexamethasone-induced gene expression suggests that ESRRB cooperates with the GR to induce optimal transcriptional responses to dexamethasone. Therefore, we hypothesized that dexamethasone may induce ESRRB expression to amplify the GC response. We treated human T-ALL cells with dexamethasone for 6 hours and detected significant increases in ESRRB protein levels (Figure 6A). We analyzed the genomic sequence of canonical GC-regulated genes including NR3C1, BCL2L11, and TSC22D3 and found an ERRE (TCAAGGTCA) in intronic regions of all 3 genes. We performed ChIP-qPCR in the human T-ALL cell line KOPTK1 and assayed ESRRB binding to these regions. We detect increased ESRRB binding to 3 ERRE sites in intron 1 of the NR3C1 gene after dexamethasone treatment (Figure 6B). These data are consistent with our gene expression data and qRT-PCR analyses showing that ESRRB is required for GR autoinduction (Figures 2C and 5C,F). ESRRB recruitment to the ERRE in intron 2 of BCL2L11 and a TSC22D3 enhancer was also observed in human T-ALL cells treated with dexamethasone (Figure 6C-D). The direct binding of ESRRB to dexamethasone-regulated genes in human T-ALL cells further implicated ESRRB in GR-mediated transcription. Interestingly, we detected significant ESRRB recruitment to BCL2L11 and TSC22D3 in the GC-sensitive human T-ALL cell line KOPTK1, whereas significantly less ESRRB recruitment to these genes was observed in the GC-resistant human T-ALL cell line CUTLL1, which also does not upregulate ESRRB expression in response to dexamethasone (Figure 6B-D; supplemental Figure 6A). Importantly, we did not detect increased ESRRB binding to regions of BCL2L11 or TSC22D3 that did not contain an ERRE or exhibit GR binding (supplemental Figure 6B-C). 39 These findings are consistent with recent ATAC-seq studies that show increased chromatin accessibility in GC-regulated genes in GC-sensitive vs GC-resistant ALL cells.18 Taken together, these data demonstrate that in addition to changes in H3K27 acetylation and promoter-enhancer interactions (Figure 4), dexamethasone increased ESRRB binding to the ERRE in GC-sensitive ALL cells and suggest direct regulation of GR target genes by ESRRB.
Dexamethasone increases ESRRB expression and binding to GR-regulated genes. (A) The human T-ALL cell lines DND-41 and KOPTK1 were treated with dexamethasone for 6 hours, and ESRRB expression was quantified in nuclear lysates by immunoblot analysis with an ESRRB antibody. ESRRB protein levels were quantified by densitometry. ESRRB bound GC target genes in dexamethasone-sensitive (KOPTK1) but not dexamethasone-resistant (CUTLL1) human T-ALL cells. (B-D) The schematics (top) show putative ERRE sites in introns of GR target genes. ChIP (bottom) was performed with an ESRRB antibody or an IgG control, and DNA was quantified by qPCR and the ΔΔCT method, with primers to the putative ESRRB binding sites. The percentage input was normalized to IgG control. The results are averages of 3 independent experiments; error bars represent SEM. *P < .05; **P < .01.
Dexamethasone increases ESRRB expression and binding to GR-regulated genes. (A) The human T-ALL cell lines DND-41 and KOPTK1 were treated with dexamethasone for 6 hours, and ESRRB expression was quantified in nuclear lysates by immunoblot analysis with an ESRRB antibody. ESRRB protein levels were quantified by densitometry. ESRRB bound GC target genes in dexamethasone-sensitive (KOPTK1) but not dexamethasone-resistant (CUTLL1) human T-ALL cells. (B-D) The schematics (top) show putative ERRE sites in introns of GR target genes. ChIP (bottom) was performed with an ESRRB antibody or an IgG control, and DNA was quantified by qPCR and the ΔΔCT method, with primers to the putative ESRRB binding sites. The percentage input was normalized to IgG control. The results are averages of 3 independent experiments; error bars represent SEM. *P < .05; **P < .01.
The ESRRB agonist GSK4716 synergizes with dexamethasone to induce human leukemic cell death
The role of ESRRB in the regulation of GR target genes suggests that ESRRB expression is reduced in samples from relapsed ALL. We compared ESRRB expression in paired ALL samples obtained at diagnosis and upon relapse.43 We found ESRRB expression significantly reduced in 24 of 49 ALL samples at the time of relapse (supplemental Figure 7A; P < .05) further implicating ESRRB in dexamethasone resistance. We did not, however, detect evidence of ESRRB promoter methylation in samples from relapsed patients (supplemental Figure 7B). We also analyzed ESRRB expression in GC-sensitive and -resistant samples from patients with ALL and found ESRRB significantly reduced in GC-resistant samples in 1 of 2 data sets examined (supplemental Figure 7C-D).9,39 We examined additional ALL data sets to determine whether ESRRB levels correlate with genetic subtypes of ALL. Although no significant differences were detected among B-ALL subtypes (supplemental Figure 7E), ESRRB expression was significantly reduced in TLX1-positive T-ALL samples when compared to TAL1-positive samples. Interestingly, TLX1 is a genetic subtype associated with steroid resistance (supplemental Figure 7F).44,45 Although reduced ESRRB expression was observed in paired ALL samples at the time of relapse and in GC-resistant ALL samples (supplemental Figure 7A,D,F), we confirmed that the ESRRB protein was detected in GC-resistant (50% inhibitory concentration >1 μM) primary T-ALL patient samples (supplemental Figure 7G), thereby validating ESRRB as a potential therapeutic target for GC resensitization in relapsed ALL. Finally, we demonstrated that dexamethasone significantly increased ESRRB mRNA levels in the human T-ALL cell line KOPTK1 and in 2 relapsed T-ALL patient samples examined (supplemental Figure 7A,H). Consistent with these data, dexamethasone increased GR binding to the human ESRRB gene in a T-ALL patient sample (supplemental Figure 7I).39 Collectively, these data suggest that ESRRB expression is induced by dexamethasone to feedback and potentiate GR activity and that ESRRB suppression results in GC resistance.
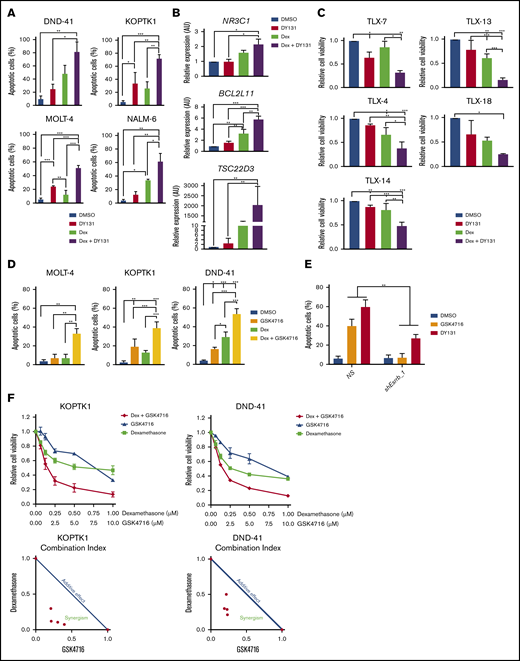
Although ERRs have no known endogenous ligands, synthetic phenolic acyl hydrazones increase the activity of these receptors by stabilizing the AF-2 domain and increasing coactivator recuitment.46 ESRRB and ESRRG agonists suppress gastric cancer and breast cancer growth in vitro.47,48 Initially, we tested the effects of the ESRRB/G agonist DY131 on GC responses in human T-ALL cell lines. We detected significant increases in dexamethasone-induced apoptosis and gene expression when dexamethasone and the ESRRB/G agonist DY131 were used in combination (Figure 7A-B). The combination therapy resulted in significant decreases in the viability of 5 T-ALL patient samples examined, including 2 samples from patients with relapsed leukemia (TLX-14 and -13) who were resistant to dexamethasone (Figure 7C). We also examined the effects of a second ESRRB agonist GSK4716 and found similar increases in apoptotic leukemic cells after combination treatment (Figure 7D). We evaluated agonist selectivity in mouse T-ALL cells, where treatment with agonist alone induced cell death (Figure 7E). GSK4716, and to a lesser extent DY131, induced cell death that was dependent on the expression of ESRRB (Figure 7E). The combined effect of GSK4716 and dexamethasone was evaluated by the combination index (CI) method in 2 human T-ALL cell lines: KOPTK1 and DND-41. This analysis resulted in a CI < 1, indicating that GSK4716 and dexamethasone synergized to induce leukemic cell death (Figure 7F), providing proof of principle that ESRRB activation potentiates dexamethasone-induced cell death in human T-ALL cells.
The ESRRB agonist GSK4716 synergizes with dexamethasone to induce human leukemic cell death. (A) Dexamethasone and DY131 dual treatment increase GC-induced apoptosis in human ALL cells in vitro. Human ALL cell lines were treated with vehicle (DMSO), dexamethasone (1 μM), DY131 (10 μM), or dexamethasone+DY131 for 72 hours, and apoptosis was assayed by annexin V-FITC/7-AAD staining followed by flow cytometry. Dexamethasone and DY131 dual treatment increase dexamethasone-induced gene expression in the human T-ALL cell line KOPTK1 in vitro. (B) Cells were treated with vehicle, dexamethasone (1 μM), DY131 (10 μM), or dexamethasone+DY131 for 24 hours, and NR3C1, BCL2L11, and TSC23D3 expression was analyzed by qRT-PCR. The copy number was normalized to β-actin by the ΔΔCT method. T-ALL patient samples are sensitive to dexamethasone and DY131 dual treatment. (C) Cells were treated with vehicle or dexamethasone (50 nM) and/or DY131 (50 μM), and cell viability was assayed by CellTiterGlo. Treatment with dexamethasone and GSK4716 increased GC-induced apoptosis in human ALL cells in vitro. (D) Human ALL cell lines were treated with vehicle (DMSO), dexamethasone (1 μM), GSK4716 (10 μM), or dexamethasone+GSK4716 for 72 hours, and apoptosis was assayed by annexin V-FITC/7-AAD staining followed by flow cytometry. The ESRRB agonists GSK4716 and DY131 require ESRRB to induce leukemic cell death. (E) NS or ESRRB-deficient mouse T-ALL cells were treated with DY131 or GSK4716 (10 μM) for 48 hours, and apoptosis was assayed by annexin V-FITC/7-AAD staining followed by flow cytometry. GSK4716 synergized with dexamethasone to induce human leukemic cell death. The human T-ALL cell lines KOPTK1 and DND-41 were treated with increasing doses of GSK4716 and dexamethasone, and cell viability was determined by CellTiterGlo. (F) The CI was calculated by using the formula a/A + b/B. Synergisms, additive effect, and antagonism of combined treatment assays were defined as CI < 1, CI = 1, and CI > 1, respectively. The results are averages of 3 independent experiments; error bars represent SEM. *P < .05; **P < .01; ***P < .001.
The ESRRB agonist GSK4716 synergizes with dexamethasone to induce human leukemic cell death. (A) Dexamethasone and DY131 dual treatment increase GC-induced apoptosis in human ALL cells in vitro. Human ALL cell lines were treated with vehicle (DMSO), dexamethasone (1 μM), DY131 (10 μM), or dexamethasone+DY131 for 72 hours, and apoptosis was assayed by annexin V-FITC/7-AAD staining followed by flow cytometry. Dexamethasone and DY131 dual treatment increase dexamethasone-induced gene expression in the human T-ALL cell line KOPTK1 in vitro. (B) Cells were treated with vehicle, dexamethasone (1 μM), DY131 (10 μM), or dexamethasone+DY131 for 24 hours, and NR3C1, BCL2L11, and TSC23D3 expression was analyzed by qRT-PCR. The copy number was normalized to β-actin by the ΔΔCT method. T-ALL patient samples are sensitive to dexamethasone and DY131 dual treatment. (C) Cells were treated with vehicle or dexamethasone (50 nM) and/or DY131 (50 μM), and cell viability was assayed by CellTiterGlo. Treatment with dexamethasone and GSK4716 increased GC-induced apoptosis in human ALL cells in vitro. (D) Human ALL cell lines were treated with vehicle (DMSO), dexamethasone (1 μM), GSK4716 (10 μM), or dexamethasone+GSK4716 for 72 hours, and apoptosis was assayed by annexin V-FITC/7-AAD staining followed by flow cytometry. The ESRRB agonists GSK4716 and DY131 require ESRRB to induce leukemic cell death. (E) NS or ESRRB-deficient mouse T-ALL cells were treated with DY131 or GSK4716 (10 μM) for 48 hours, and apoptosis was assayed by annexin V-FITC/7-AAD staining followed by flow cytometry. GSK4716 synergized with dexamethasone to induce human leukemic cell death. The human T-ALL cell lines KOPTK1 and DND-41 were treated with increasing doses of GSK4716 and dexamethasone, and cell viability was determined by CellTiterGlo. (F) The CI was calculated by using the formula a/A + b/B. Synergisms, additive effect, and antagonism of combined treatment assays were defined as CI < 1, CI = 1, and CI > 1, respectively. The results are averages of 3 independent experiments; error bars represent SEM. *P < .05; **P < .01; ***P < .001.
Discussion
Despite increases in overall survival, relapse remains a critical challenge in the care of pediatric patients with ALL. Although failure to respond to GCs can predict a patient’s outcome,3,4 GC resensitizing agents have not entered the clinic. Our functional screen for GC-resistance genes identified ESRRB as a dexamethasone-inducible transcription factor that cooperates with the GR to maximize the GC-regulated gene expression signature. We provide evidence that ESRRB directly contributes to dexamethasone-induced gene expression in human T-ALL cells through direct binding to ERREs in the regulatory regions of the GR target genes examined.
It is likely that ESRRB cooperates with the GR to regulate gene expression through DNA binding, followed by coactivator recruitment in leukemic cells, similar to findings in ES cells where ESRRB interacts with NCOA3 to mediate gene transcription.49 The coactivator(s) that function with ESRRB remain unclear; however, EP300, NCOA4, and NCOA5 were identified in this screening (supplemental Table 3), suggesting that they may collaborate with ESRRB in leukemic cells. Through transcriptome analyses, we established that ESRRB and GR had overlapping gene regulatory functions in the GC response in leukemic cells. However, only 66% of the dexamethasone-induced genes depended on ESRRB (Figure 2), indicating that other transcription factors remain to be discovered that coregulate GR transcriptional activity in lymphoid cells.
ESRRB functions as a pioneer factor in the reprogramming of epiblast stem cells, leading us to speculate that ESRRB may increase chromatin accessibility at select GR target genes.50 Our Hi-ChIP analysis of GR- and ESRRB-dependent genes showed that enhancer elements known to bind the GR in human T-ALL cells39 contain ESRRB binding sites and that dexamethasone increases the frequency of these enhancer/promoter interactions (Figure 4). These data lead us to speculate that ESRRB and GR potentiate GC-induced gene expression by supporting these chromatin interactions. Our Hi-ChIP data also detected preexisting chromatin interactions at dexamethasone-responsive loci in mouse and human T-ALL cells (Figures 3 and 4), indicating that lymphoid cells may be poised for a rapid transcriptional GC response.
Our work establishes ESRRB repression as a mechanism of GC resistance and relapse in ALL. Treatment of human T- or B-ALL cell lines and samples with the ESRRB agonists DY131 or GSK4716 enhanced dexamethasone-induced gene expression and cell death. These in vitro data led us to administer DY131 in vivo to assess its effects on leukemic burden. The solubility resulting from the hydrophobic nature of the agonist51 prevented assessment of its cooperating/synergizing with dexamethasone to prevent leukemic growth in vivo. Future collaborative efforts will be directed at improving agonist pharmacokinetics.
This work reveals novel functions of ESRRB as a GR regulator. We showed that ESRRB is a GC-responsive gene and is coenriched, along with GR at select GC-regulated target genes in human ALL cells. Our study provides mechanistic insight into GR transcription and establishes ESRRB as a potential therapeutic target to enhance GC action in acute leukemia and potentially other lymphoid malignancies.
The sequencing data reported in this article have been deposited in the Gene Expression Omnibus (accession number GSE115363).
Acknowledgments
The authors thank members of the Kelliher laboratory for helpful discussions; UMMS Flow Cytometry, Deep Sequencing Core and Ai Cores for support; and Véronique Azuara, Imperial College London Faculty of Medicine for supplying the ESRRB-FLAG plasmid.
This work was supported by National Institutes of Health, National Cancer Institute (NCI) grant (CA096899), an Alex’s Lemonade Stand Innovator Award and Hyundai Hope on Wheels Award (M.A.K.); an NCI Cancer Biology grant (T32 CA130807) (K.M.G. and K.W.O.); Medical Science Training Program, National Institute of General Medical Sciences grant (T32 GM107000) (K.W.O.); and an American Cancer Society Postdoctoral Fellowship Award (125087-PF-13-247-01-LIB) (J.E.R.).
Authorship
Contribution: K.M.G. conceived and performed the experiments and wrote the manuscript; J.E.R. and M.R.G. designed the shRNA screen; J.E.R. executed the screening, interpreted the data, and helped prepare the manuscript; L.M. contributed to the experimental work; K.W.O. contributed to the experimental work, data interpretation, and manuscript preparation; S.H.T. performed H3K27Ac Hi-ChIP and T.K.T. and T.S. analyzed the data; J.Y., J.Z., and R.L. performed key bioinformatic analyses; and M.A.K. conceived and supervised the study and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michelle A. Kelliher, Department of Molecular, Cell and Cancer Biology, University of Massachusetts Medical School, 364 Plantation St, Lazare Research Building, Floor 4, Worcester, MA 01605; e-mail: michelle.kelliher@umassmed.edu.
References
Author notes
The full-text version of this article contains a data supplement.