Key Points
The kindlin-3 FERM domain forms a cloverleaf-like structure, which is required for supporting integrin αIIbβ3 activation.
The kindlin-3 FERM domain exhibits a monomeric property, but dimerization is required for kindlin-3 to support integrin αIIbβ3 activation.
Abstract
Kindlin-3, a protein 4.1, ezrin, radixin, and moesin (FERM) domain–containing adaptor in hematopoietic cells, is essentially required for supporting the bidirectional integrin αIIbβ3 signaling in platelets by binding to the integrin β3 cytoplasmic tail. However, the structural details of kindlin-3’s FERM domain remain unknown. In this study, we crystalized the kindlin-3’s FERM domain protein and successfully solved its 3-dimensional structure. The structure shows that the 3 kindlin-3’s FERM subdomains (F1, F2, and F3) compact together and form a cloverleaf-shaped conformation, which is stabilized by the binding interface between the F1 and F3 subdomains. Interestingly, the FERM domain of kindlin-3 exists as a monomer in both crystal and solution, which is different from its counterpart in kindlin-2 that is able to form a F2 subdomain-swapped dimer; nonetheless, dimerization is required for kindlin-3 to support integrin αIIbβ3 activation, indicating that kindlin-3 may use alternative mechanisms for formation of a functional dimer in cells. To evaluate the functional importance of the cloverleaf-like FERM structure in kindlin-3, structure-based mutations were introduced into kindlin-3 to disrupt the F1/F3 interface. The results show that integrin αIIbβ3 activation is significantly suppressed in platelets expressing the kindlin-3 mutant compared with those expressing wild-type kindlin-3. In addition, introduction of equivalent mutations into kindlin-1 and kindlin-2 also significantly compromises their ability to support integrin αIIbβ3 activation in CHO cells. Together, our findings suggest that the cloverleaf-like FERM domain in kindlins is structurally important for supporting integrin αIIbβ3 activation.
Introduction
The kindlin family members (kindlin-1, -2, and -3) play an essential role in supporting the bidirectional integrin signaling.1,2 Among them, kindlin-3 is primarily expressed in hematopoietic cells, and its deficiency in humans leads to leukocyte adhesion deficiency type III, symptomatically associated with severe bleeding and recurrent infections due to disabled platelet aggregation and leukocyte recruitment.3-6 All kindlins are protein 4.1, ezrin, radixin, and moesin (FERM) domain–containing proteins with the same subdomain organization.7,8 In addition to 3 typical FERM subdomains (F1, F2, and F3), kindlins contain an N-terminal F0 subdomain, a loop region (F1L) in the F1, and a pleckstrin homology (PH) domain inserted in the F2. Although the overall full-length structure of any of the kindlin family members is currently unavailable, the structures of individual subdomains in kindlins have been solved. The F0 in kindlins adopts a ubiquitin-like fold and is functionally required for supporting integrin activation.9,10 The F1L is predominantly unfolded with a highly conserved polylysine motif, which may contribute to membrane attachment.9,11,12 The PH domain in kindlins possesses a putative phospholipid-binding pocket for membrane association.13-15 In particular, Li et al8 solved the structure of kindlin-2’s FERM domain using a truncated kindlin-2 protein that lacks the F1L and the PH domain, and disclosed a compact and cloverleaf-like conformation. Notably, the FERM domain of kindlin-2 can form an F2-swapped dimer, which is functionally important for supporting integrin activation.
Due to high homology between the kindlin family members, the structural feature of kindlin-2’s FERM domain may expectedly be shared by the other 2 kindlin members. However, a previous study revealed that the full-length kindlin-3 protein in solution was predominantly monomeric and mostly likely presented an extended conformation.7 Noticeably, unlike kindlin-1 and kindlin-2, kindlin-3 fails to cooperate with talin head domain to coactivate integrin αIIbβ3 in non-hematopoietic model cells.16 These observations indicate that the FERM domain of kindlin-3 may possess specific features compared with kindlin-1 and kindlin-2. In the current study, we crystalized the FERM domain of kindlin-3 and revealed a monomeric and cloverleaf-like structure. In addition, we show that the compact FERM domain in kindlin-3 is structurally important in supporting integrin αIIbβ3 signaling in platelets.
Materials and methods
Protein crystallization, data collection, and structure determination
To crystalize kindlin-3’s FERM domain, 2 human kindlin-3 mutant constructs (kindlin-3Δ and kindlin-3Δ’) were prepared by truncating the F1L and the PH domain, which share the same truncation in the F1L domain (Δ166-194) but have different truncating boundaries for the PH domain (Δ314-493 for kindlin-3Δ and Δ344-493 for kindlin-3Δ’). These 2 kindlin-3 constructs are equivalent to previously crystalized kindlin-2 mutants.8 Kindlin-3Δ and kindlin-3Δ’ were cloned into vector pET31b (Novagen) and expressed in Escherichia coli with a C-terminally fused 6×his tag. Proteins were purified by nickel-affinity chromatography followed by heparin cation exchange and size-exclusion chromatography, and concentrated to 10 mg/mL in 20 mM Tris-HCl (pH 8.0), 150 mM NaCl. Initial crystal hits for kindlin-3Δ were obtained by using hanging-drop vapor diffusion over reservoirs with 20% polyethylene glycol-3350 (PEG-3350) and 0.2 M sodium thiocyanate. The diffraction-quality crystals were obtained by microseeding of the initial microcrystals with 11% PEG-3350 and 0.2 M sodium thiocyanate. Crystals were cryoprotected by 20% ethylene glycol and flash frozen in liquid nitrogen. Kindlin-3Δ’ were crystalized against 0.2 M potassium nitrate, 12% PEG-3350, and 30% glycerol. Diffraction data were collected from a single crystal at Shanghai Synchrotron Radiation Facility Beamline 19U1 and then indexed, integrated, and scaled by using HKL2000.17 Molecular replacement trials in Phaser using previously solved kindlin-2 structure (Protein Data Bank [PDB], 5XPY) were successful. Model rebuilding was performed by Coot with high-quality electron density, and refinement was done with Phenix. Structure analysis was performed with CCP4. Molecular graphics were generated by using PyMOL (http://www.pymol.org). The structural files have been deposited to the PDB, and the PDB identifiers are 6V97 and 6V9G for kindlin-3Δ and kindlin-3Δ’, respectively.
Analytical gel filtration chromatography
Gel filtration chromatography (GFC) was performed by using a Superdex 200 Increase 10/300 GL column on an ÄKTA pure system (Cytiva). One aliquot of freshly purified kindlin-3Δ protein sample (∼20 µM) was analyzed according to analytic GFC. Two aliquots of the same batch of protein were placed in a cold room (∼4°C) for 3 days and 10 days followed by the same analytic GFC analysis.
Integrin αIIbβ3 activation in CHO cells and integrin αIIbβ3–mediated CHO cell spreading
Integrin αIIbβ3 activation in transfected CHO cells was evaluated by fluorescence-activated cell sorting, as previously described.18-20 In brief, DsRed-talin head and EGFP-kindlin were transiently coexpressed in CHO cells stably expressing integrin αIIbβ3 (CHO-αIIbβ3). The binding of activation-dependent PAC-1 antibody was measured on transfected cells by gating DsRed and EGFP double-positive cells; meanwhile, total expression of αIIbβ3 on the cell surface was also evaluated by 2G12 antibody (a gift of Ed Plow, Cleveland Clinic, Cleveland, OH) and used to normalize the PAC-1 binding. Dead cells were excluded by 4′,6-diamidino-2-phenylindole staining. The median fluorescence intensity of PAC-1 binding to control cells transfected with empty vectors was defined as 1, which serves as a basal line for all other samples. To evaluate the spreading of transfected cells, double-positive cells were sorted and allowed to spread on immobilized fibrinogen for 60 minutes followed by fixation and quantification.
Pull-down and coimmunoprecipitation assays
For pull-down assays, control EGFP, EGFP-fused kindlins, and the mutants were expressed in CHO cells and then enriched on protein A/G beads precoated with an anti-EGFP monoclonal antibody (homemade). After extensive washing, the beads were used to incubate with purified GST-fused integrin β3 CT proteins, which were constructed and expressed as previously described.18 The bound integrin β3 CT proteins and the coated EGFP proteins on the beads were then analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting. For coimmunoprecipitation assays, washed mouse platelets expressing EGFP–kindlin-3 or the designed mutant were lysed and incubated with protein A/G beads precoated with an anti-EGFP monoclonal antibody (homemade) for overnight in a cold room. After washing, the coprecipitants were evaluated by SDS-PAGE and western blotting.
Disuccinimidyl suberate crosslinking assay
GST–EGFP–kindlin-3 was constructed by fusion of the GST tag (obtained from pGEX-4T1) to the N terminus of EGFP–kindlin-3. To verify if GST–EGFP–kindlin-3 can form a dimer in CHO-αIIbβ3 cells, a disuccinimidyl suberate (DSS) crosslinking assay was used. In brief, cells transiently expressing EGFP–kindlin-3 or GST–EGFP–kindlin-3 were washed with phosphate-buffered saline and then lysed with cold phosphate-buffered saline with 1% Triton-X 100 plus protease inhibitors. Each lysate sample was split into 2 aliquots. One aliquot was used to incubate with DSS at 1 mM for 1 hour on ice followed by adding the quench solution, and the other aliquot was used as a non-crosslinking control. Finally, the molecular weights of EGFP–kindlin-3 and GST–EGFP–kindlin-3 in these lysates were determined by SDS-PAGE under reducing conditions followed by western blotting with an anti-EGFP antibody.
Lentiviral transduction and bone marrow transplantation
Kindlin-3fl/flMx1-Cre mice were intraperitoneally injected with poly(I:C) at 6 to 8 weeks age to induce deletion of kindlin-3 in hematopoietic cells. Deficiency of kindlin-3 in platelets in these mice was routinely verified by a homemade polyclonal antibody against mouse kindlin-3 as previously described.16 Sca1+ bone marrow cells were isolated from these mice by using an Sca1+ Selection Kit (Stemcell Technologies) and cultured in Dulbecco’s modified Eagle medium supplemented with 15% fetal bovine serum, 20 ng/mL interleukin-3, 50 ng/mL interleukin-6, and 50 ng/mL stem cell factor. EGFP-fused kindlin-3 was constructed into lentiviral vector pLeGo-G2 for generating lentiviral particles, which were used to transduce bone marrow cells on the next day after isolation at a multiplicity of infection of 5. EGFP-positive cells were sorted out 2 days after transduction and transplanted to lethally irradiated wild-type C57BL/6 recipient mice (∼1 × 106 cells/mouse); these mice were used for functional analysis of platelets after 6 to 8 weeks.
Functional analysis of mouse platelets expressing EGFP–kindlin-3 and the mutant
As previously described,16 platelets were isolated from peripheral blood of bone marrow–transplanted mice and washed. Integrin αIIbβ3 activation was tested by using the fibrinogen-binding assay with stimulations of collagen-related peptide (5 µg/mL) and protease-activated receptor 4 (PAR4)-agonist peptide (150 µM), both of which were synthesized by the Molecular Core at Versiti Blood Research Institute. To perform a platelet-spreading assay, washed platelets from bone marrow–transplanted mice were allowed to spread on immobilized fibrinogen in the present to PAR4 agonist peptide (150 µM) for 60 minutes, followed by fixation, permeabilization, and staining with Alexa Fluor 568 phalloidin. Spreading areas of EGFP-positive platelets were quantified.
All the animal studies were approved by the Institutional Animal Care and Use Committee.
Results and discussion
Structural features of the FERM domain of kindlin-3
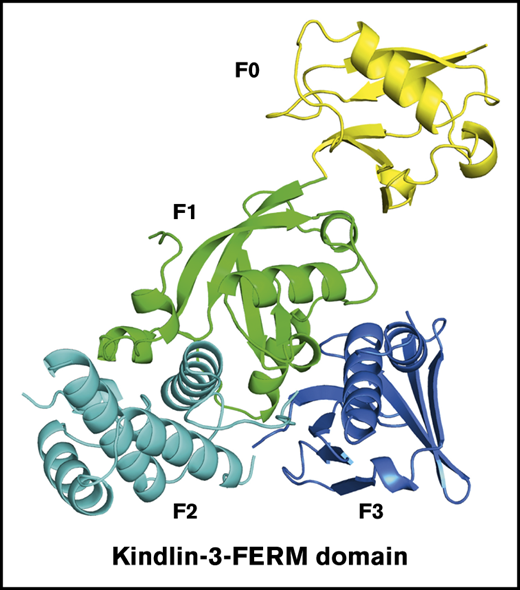
To determine the structure of the FERM domain of kindlin-3, kindlin-3Δ and kindlin-3Δ’ were constructed, as shown in Figure 1A, and proteins were expressed and purified. After crystallization screening and optimization, both kindlin-3Δ and kindlin-3Δ’ proteins were successfully crystalized in space group P212121 with 2 molecules in 1 asymmetric unit, and their structures were determined at 2.4 Å by molecular replacement (supplemental Table 1). In both FERM domain structures, 3 subdomains (F1, F2, and F3) pack against each other and form a cloverleaf-like conformation (Figure 1B-C), and the N-terminal F0 protrudes from the compact FERM domain by packing against the F1 subdomain. Superimposition of these 2 structures presents an identical conformation (Figure 1D). This finding suggests that the boundary variation in kindlin-3Δ and kindlin-3Δ’ does not affect the FERM domain packing, and it also indicates that formation of the compact FERM domain structure is likely independent of the other domains in kindlin-3.
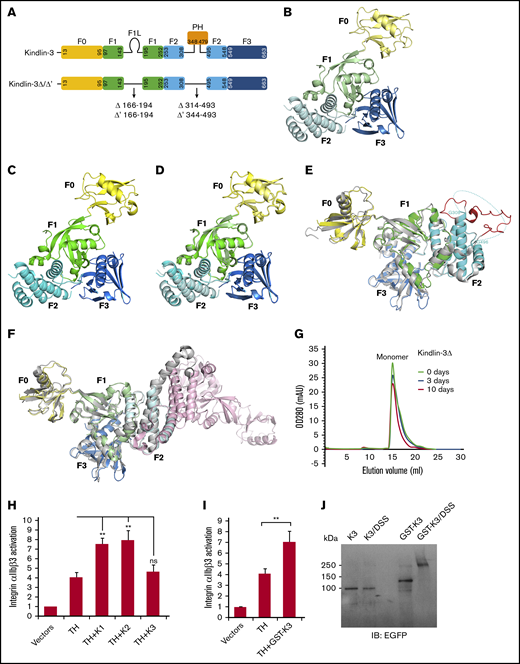
The FERM domain of kindlin-3 exhibits a monomeric and cloverleaf-like structure. (A) The organization of kindlin-3’s subdomains. The truncated regions in kindlin-3Δ and kindlin-3Δ’ were indicated. Ribbon representations of kindlin-3Δ (B) and kindlin-3Δ’ (C), in which the subdomains are labeled and colored using the similar or same color pattern as shown in panel A. (D) Superimposition of the structures of kindlin-3Δ and kindlin-3Δ’. (E) Structural comparison of kindlin-3Δ’ (color coded) and kindlin-2Δ’ (gray). The missing loop region in the F2 of kindlin-3Δ’ is indicated by hypothetical dotted line while the visible and fixed loop of the same region in kindlin-2Δ’ is highlighted in red. (F) Structural comparison of kindlin-3Δ (color coded) and the kindlin-2Δ dimer (gray and pink). The F2 subdomain-swapped dimer formed in kindlin-2Δ is not observed in kindlin-3Δ. (G) Analytic gel filtration analysis to show the monomeric property of kindlin-3Δ protein in solution at different storage time, either freshly purified protein (0 day) or the same batch of protein placed at 4°C for 3 or 10 days. (H) Integrin αIIbβ3 activation induced by coexpression of DsRed-fused talin head (TH) with EGFP-kindlins (K1, K2, and K3) was evaluated by using the PAC-1 binding assay in transfected CHO-αIIbβ3 cells. (I) The effect of GST-fused EGFP–kindlin-3 (GST-K3) on integrin αIIbβ3 activation was evaluated using the PAC-1 binding assay. (J) Dimerization of GST-fused EGFP–kindlin-3 (GST-K3) was confirmed by the DSS crosslinking assay and western blotting. EGFP–kindlin-3 (K3) was used as a control. Results present the mean ± standard deviation of 3 experiments. **P < .01. ns, nonsignificant.
The FERM domain of kindlin-3 exhibits a monomeric and cloverleaf-like structure. (A) The organization of kindlin-3’s subdomains. The truncated regions in kindlin-3Δ and kindlin-3Δ’ were indicated. Ribbon representations of kindlin-3Δ (B) and kindlin-3Δ’ (C), in which the subdomains are labeled and colored using the similar or same color pattern as shown in panel A. (D) Superimposition of the structures of kindlin-3Δ and kindlin-3Δ’. (E) Structural comparison of kindlin-3Δ’ (color coded) and kindlin-2Δ’ (gray). The missing loop region in the F2 of kindlin-3Δ’ is indicated by hypothetical dotted line while the visible and fixed loop of the same region in kindlin-2Δ’ is highlighted in red. (F) Structural comparison of kindlin-3Δ (color coded) and the kindlin-2Δ dimer (gray and pink). The F2 subdomain-swapped dimer formed in kindlin-2Δ is not observed in kindlin-3Δ. (G) Analytic gel filtration analysis to show the monomeric property of kindlin-3Δ protein in solution at different storage time, either freshly purified protein (0 day) or the same batch of protein placed at 4°C for 3 or 10 days. (H) Integrin αIIbβ3 activation induced by coexpression of DsRed-fused talin head (TH) with EGFP-kindlins (K1, K2, and K3) was evaluated by using the PAC-1 binding assay in transfected CHO-αIIbβ3 cells. (I) The effect of GST-fused EGFP–kindlin-3 (GST-K3) on integrin αIIbβ3 activation was evaluated using the PAC-1 binding assay. (J) Dimerization of GST-fused EGFP–kindlin-3 (GST-K3) was confirmed by the DSS crosslinking assay and western blotting. EGFP–kindlin-3 (K3) was used as a control. Results present the mean ± standard deviation of 3 experiments. **P < .01. ns, nonsignificant.
As exhibited in Figure 1A, kindlin-3Δ’ comprises a linker region preceding the PH domain (LF2-PH). However, amino acid residues of the LF2-PH region in kindlin-3Δ’ fail to show electron density, suggesting that this linker is structurally flexible in kindlin-3 (Figure 1E). In comparison, the equivalent linker in kindlin-2Δ’ presents appreciable electron density and can be traced as a fixed loop structure (PDB, 5XPY). Such a structural difference of the LF2-PH in kindlin-2 and kindlin-3 may reflect their functional discrepancy in model cells, in which kindlin-2 is able to bind integrin-linked kinase (ILK) via its LF2-PH to promote focal adhesion targeting, whereas kindlin-3 poorly binds ILK, consequently resulting in insufficient focal adhesion targeting.21-23
Strikingly, kindlin-3Δ does not structurally resemble kindlin-2Δ that can form an F2 subdomain-swapped dimer (Figure 1F). Although 2 kindlin-3Δ molecules are in one asymmetric unit (supplemental Figure 1A), a domain swapping between them does not occur. The buried surface area between these 2 kindlin-3Δ molecules in one asymmetric unit is ∼660 Å2 and only covers 3.1% of the overall surface (supplemental Figure 1C), which can be accounted for by general crystal packing contacts.24 Symmetry operation of 2 kindlin-3Δ molecules in one asymmetric unit exhibits 8 copies that form interfaces with them (supplemental Figure 1B). These interfaces were analyzed by using the Protein Interfaces, Surfaces, and Assemblies program,25 among which the largest interface is 1383 Å2 and covers 6.5% of the overall surface, which also falls into the category of crystal packing contacts.24 These structural features indicate that the FERM domain–swapping mediated dimerization does not occur for kindlin-3. Furthermore, analytic gel filtration chromatography was used to compare freshly purified kindlin-3Δ protein and the protein samples placed at 4°C for 3 days and 10 days, and found that all these protein samples presented as a monomer in solution (Figure 1G). Together, these results suggest that kindlin-3 may primarily exist as a monomer, which is consistent with previous observations in the literature.7,26
Because it was reported that kindlins contribute to promote integrin clustering,26 the difference between kindlin-2 and kindlin-3 on forming a dimer may be reflected by their ability to support talin head domain–induced integrin αIIbβ3 activation in non-hematopoietic CHO cells; this is because we detected that kindlin-2 (and kindlin-1 as well) but not kindlin-3 was able to enhance the binding of PAC-1 antibody (Figure 1H), a pentameric immunoglobulin M antibody specifically recognizing activated human integrin αIIbβ3. The functional difference between kindlin-3 and kindlin-1/kindlin-2 is not due to their expression level variation, as we performed the analysis on DsRed/EGFP double-positive CHO cells after transfection (see "Materials and methods"). In addition, the overall expression levels of talin head domain and kindlins in different transfectants were comparable, as evaluated by using western blotting (supplemental Figure 2). To test if dimerization of kindlin-3 is functionally required for supporting integrin αIIbβ3 activation, EGFP-kindlin-3 was fused with GST, a commonly used dimeric tag. Importantly, we found that expression GST-fused kindlin-3 was able to coactivate integrin αIIbβ3 in CHO cells (Figure 1I). Meanwhile, GST-mediated kindlin-3’s dimerization in transfected CHO cells was confirmed by using the DSS crosslinking assay (Figure 1J) and native PAGE (supplemental Figure 3). Collectively, the intrinsic property of kindlin-3 as a monomer and the functional requirement of kindlin-3’s dimerization (or polymerization) in supporting integrin activation raise the possibility that kindlin-3 may undergo dimerization in hematopoietic cells via a binding partner that either is a dimer or has more than 1 kindlin-3–binding site. If so, the expression specificity of such a binding partner could possibly determine the competency of kindlin-3 in supporting integrin signaling in different cells.
The compact FERM domain of kindlins in supporting integrin αIIbβ3 signaling
We next sought to evaluate the functional relevance of the compact FERM domain in kindlins. As shown in Figure 2A and B, the F1 and F3 subdomains in kindlin-2 and kindlin-3 contact with each other via multiple salt bridges, thereby stabilizing the cloverleaf-like FERM domain. The involved contacting residues in the interface are highly conserved in kindlins (Figure 2C). The salt bridges formed by F1-E137E138/F3-R640 and F1-K252/F3-E629 in kindlin-3 are also observed at equivalent positions in kindlin-2 (F1-E136E137/F3-R659 and F1-K273/F3-E648). Interestingly, the salt bridge of F1-D231/F3-R625 observed in kindlin-3 is not detected in kindlin-2, even though these residues are also conserved in kindlins; this finding indicates that the F1/F3 interface in different kindlins may not be exactly identical.
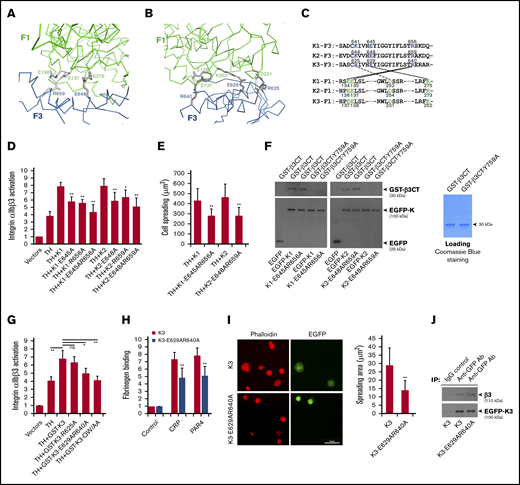
The compact structure of FERM domain in kindlins is required for supporting integrin αIIbβ3 activation. The salt bridges formed at the F1/F3 interface in kindlin-2 (A) and kindlin-3 (B) are shown, in which the side-chain of involved residues are shown in gray and the salt bridges are indicated with dotted lines. (C) The conserved residues mediating the F1/F3 interface in kindlins are positioned in both F1 and F3 subdomains. (D) Integrin αIIbβ3 activation induced by coexpression of the talin head domain (TH) and kindlins, including kindlin-1 (K1) and kindlin-2 (K2) and their mutants, was evaluated by using the PAC-1 binding assay in transfected CHO-αIIbβ3 cells. (E) Integrin αIIbβ3–mediated CHO cell spreading on immobilized fibrinogen was quantified. (F) EGFP-kindlin-1 (K1) and EGFP-kindlin-2 (K2) and their mutants expressed in CHO cells were enriched on protein A/G beads precoated with an anti-EGFP antibody. The beads were then used to incubate with purified GST-β3CT protein and the Y759A mutant, and after extensive washing, interaction between kindlin and GST-β3CT protein was evaluated by using SDS-PAGE and western blotting. (G) The effect of GST-fused EGFP-kindlin-3 (GST-K3) and the indicated mutants on integrin αIIbβ3 activation in CHO-αIIbβ3 cells was evaluated by using the PAC-1 binding assay. (H) Washed platelets from bone marrow–transplanted mice were stimulated with either collagen-related peptide (CRP; 5 µg/mL) or PAR4 agonist peptide (PAR4; 150 µM) and soluble fibrinogen binding to EGFP-positive platelets expressing either EGFP–kindlin-3 (K3) or the designed mutant (K3-E629AR640A) was quantified by fluorescence-activated cell sorting. (I) Washed platelets from bone marrow–transplanted mice were allowed to spread on immobilized fibrinogen in the present to PAR4 agonist peptide (150 µM) for 60 minutes followed by phalloidin staining. Spreading of EGFP-positive platelets expressing either EGFP-kindlin-3 (K3) or the designed mutant (K3-E629AR640A) was calculated. (J) Lysates of platelets expressing EGFP–kindlin-3 (K3) and the designed mutant (K3-E629AR640A) were used to incubate with protein A/G beads precoated with an anti-EGFP antibody. The coprecipitated EGFP–kindlin-3 and integrin αIIbβ3 were then evaluated by using western blotting. Results present the mean ± standard deviation of 3 experiments. *P < .05; **P < .01. IgG, immunoglobulin G.
The compact structure of FERM domain in kindlins is required for supporting integrin αIIbβ3 activation. The salt bridges formed at the F1/F3 interface in kindlin-2 (A) and kindlin-3 (B) are shown, in which the side-chain of involved residues are shown in gray and the salt bridges are indicated with dotted lines. (C) The conserved residues mediating the F1/F3 interface in kindlins are positioned in both F1 and F3 subdomains. (D) Integrin αIIbβ3 activation induced by coexpression of the talin head domain (TH) and kindlins, including kindlin-1 (K1) and kindlin-2 (K2) and their mutants, was evaluated by using the PAC-1 binding assay in transfected CHO-αIIbβ3 cells. (E) Integrin αIIbβ3–mediated CHO cell spreading on immobilized fibrinogen was quantified. (F) EGFP-kindlin-1 (K1) and EGFP-kindlin-2 (K2) and their mutants expressed in CHO cells were enriched on protein A/G beads precoated with an anti-EGFP antibody. The beads were then used to incubate with purified GST-β3CT protein and the Y759A mutant, and after extensive washing, interaction between kindlin and GST-β3CT protein was evaluated by using SDS-PAGE and western blotting. (G) The effect of GST-fused EGFP-kindlin-3 (GST-K3) and the indicated mutants on integrin αIIbβ3 activation in CHO-αIIbβ3 cells was evaluated by using the PAC-1 binding assay. (H) Washed platelets from bone marrow–transplanted mice were stimulated with either collagen-related peptide (CRP; 5 µg/mL) or PAR4 agonist peptide (PAR4; 150 µM) and soluble fibrinogen binding to EGFP-positive platelets expressing either EGFP–kindlin-3 (K3) or the designed mutant (K3-E629AR640A) was quantified by fluorescence-activated cell sorting. (I) Washed platelets from bone marrow–transplanted mice were allowed to spread on immobilized fibrinogen in the present to PAR4 agonist peptide (150 µM) for 60 minutes followed by phalloidin staining. Spreading of EGFP-positive platelets expressing either EGFP-kindlin-3 (K3) or the designed mutant (K3-E629AR640A) was calculated. (J) Lysates of platelets expressing EGFP–kindlin-3 (K3) and the designed mutant (K3-E629AR640A) were used to incubate with protein A/G beads precoated with an anti-EGFP antibody. The coprecipitated EGFP–kindlin-3 and integrin αIIbβ3 were then evaluated by using western blotting. Results present the mean ± standard deviation of 3 experiments. *P < .05; **P < .01. IgG, immunoglobulin G.
To evaluate the functional relevance, we destabilized the F1/F3 interface in kindlins by mutating the involved interface residues in the F3 subdomain, individually or in combination, and found that these mutations significantly suppressed the ability of kindlin-1 and kindlin-2 to support integrin αIIbβ3 activation in transfected CHO cells (Figure 2D). In addition, disruption of the F1/F3 interface in kindlin-1 and kindlin-2 also compromised their ability to support αIIbβ3-mediated CHO spreading (Figure 2E). To rule out the possible contribution of the F1/F3 interface in kindlins to integrin binding, pull-down assays were performed, and we found that wild-type kindlin-1 and kindlin-2 and their mutants bound equally to the integrin β3 CT (Figure 2F). As a control, neither of them interacted with the integrin β3 CT with a Y759A mutation that is known to be able to disable kindlin binding. Furthermore, we introduced the F1/F3 interface mutations into GST-fused EGFP–kindlin-3 and tested their functional effects on integrin αIIbβ3 activation in CHO-αIIbβ3 cells. As expected, the double mutation of E629AR640A in kindlin-3 significantly suppressed integrin αIIbβ3 activation (Figure 2G). However, the R625A mutation in kindlin-3 negligibly affected integrin activation, suggesting that the salt bridge formed by F1-D231/F3-R625 in kindlin-3 may not be the dominant force in mediating the F1/F3 interface formation. Here, we used the QW/AA mutation in kindlin-3 to disable its binding to integrin as a control. Together, these results suggest that the compact FERM domain in kindlins is structurally important in supporting integrin αIIbβ3 activation.
To further evaluate the platelet functions, we rescued kindlin-3’s expression in kindlin-3–deficient megakaryocytes/platelets in mice, as previously described.16 In brief, bone marrow cells isolated from Kindlin-3−/− mice were first transduced with lentiviral particles expressing either EGFP–kindlin-3 or the mutant bearing the F1/F3 interface–disrupting mutations (E629AR640A) and further transplanted into lethally irradiated wild-type C57BL/6 recipient mice. Expression of EGFP–kindlin-3 and the mutant in platelets isolated from recipient mice was confirmed by use of western blotting (supplemental Figure 4). As shown in Figure 2H, upon stimulation with either collagen-related peptide or PAR4 agonist peptide, platelets expressing the kindlin-3 mutant showed a compromised soluble fibrinogen binding compared with platelets expressing wild-type kindlin-3. In addition, the kindlin-3 mutant poorly supported platelet spreading on immobilized fibrinogen while platelets expressing wild-type kindlin-3 could spread well (Figure 2I); both exhibited a similar interaction, however, with the integrin β3 subunit (Figure 2J). These results show that the compact kindlin-3’s FERM domain is structurally important in supporting the integrin αIIbβ3 signaling in platelets.
The finding of the F1/F3 interface in kindlins in supporting integrin activation shows that the compact FERM domain structure in kindlins is functionally important, which is distinct from its coactivator talin head that has a linearized FERM domain structure.27 The difference of FERM structure in these 2 coactivators may reflect their functional properties in integrin signaling. The extended structure of the talin head can facilitate its association with flat membrane through multiple membrane-binding sites distributed in each of the subdomains, by which the talin head can efficiently interact with the integrin β CTs and unclasp the membrane-proximal integrin α/β CT complex, thus triggering integrin activation.28 Comparatively, the cloverleaf-like FERM domain structure of kindlins may act as a multifaceted adaptor essentially for recruiting and positioning cell adhesion complex molecules. In fact, it has been identified that kindlins can bind several cell adhesion molecules, such as migfilin,29 paxillin,16,30,31 ADAP,32 and ILK.21,22 However, the precise mechanisms by which kindlins regulate the integrin signaling by communicating with these binding partners are still poorly understood and need to be further elucidated.
Data-sharing requests should be submitted to the corresponding author (Yan-Qing Ma; e-mail, yma@versiti.org).
Acknowledgments
This work was supported by grants from the National Heart, Lung, and Blood Institute, National Institutes of Health (HL131654), and the National Natural Science Foundation of China (31370748 and 31770967).
Authorship
Contribution: J.S., D.X., Y.N., T.Z., Z.C., Z.X., H.N., and J.Z. contributed to acquisition of the data and reviewed the manuscript; G.C.W. and J.D. contributed to interpretation of the data and reviewed the manuscript; Y.-Q.M. and Z.X. contributed to design of the experiments, acquisition and analysis of the data, and wrote the manuscript; and all authors approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yan-Qing Ma, Versiti Blood Research Institute, 8727 Watertown Plank Rd, Milwaukee, WI 53226; e-mail: yma@versiti.org; or Zhen Xu, Versiti Blood Research Institute, 8727 Watertown Plank Rd, Milwaukee, WI 53226; e-mail: zxu@versiti.org.
References
Author notes
The full-text version of this article contains a data supplement.