Key Points
UBE2T is frequently amplified and/or overexpressed and is required for homologous recombination activity in multiple myeloma cells.
UBE2T is a potential therapeutic target to increase chemosensitivity in multiple myeloma cells.
Introduction
Multiple myeloma (MM) is a plasma cell malignancy, with malignant cells displaying diverse genomic alterations at diagnosis and acquiring additional changes over time.1 This underscores a significant role for genomic instability in MM oncogenesis.2-5 We have shown that dysregulated or spontaneously elevated homologous recombination (HR) contributes to genomic evolution and development of drug resistance in MM.3 However, the mechanisms mediating aberrant HR activity in MM are largely unclear. In this study, we investigated the role of UBE2T, a protein involved in ubiquitin signaling, in HR in MM. It has been shown that UBE2T is the E2 ubiquitin conjugase that monoubiquitinates FANCD2 during repair of DNA interstrand crosslinks through the Fanconi anemia (FA) pathway.6-9 We found that UBE2T gene locus is frequently amplified and the gene is highly expressed in MM cells, and increased copy number or expression of this gene is associated with poor patient survival. We also show that UBE2T is required for efficient DNA repair by HR in MM and its knockdown increases MM sensitivity to chemotherapeutic agents.
Methods
Cell lines and primary cells
Human MM cell lines (RPMI-8226, H929, KMS-12PE, MM.1S, OCI-MY7, OPM-1, OPM-2, and U266) were cultured in RPMI 1640 growth medium supplemented with l-glutamine and NaHCO3, 10% fetal bovine serum, and 1% penicillin-streptomycin (Thermo Fisher Scientific, Waltham, MA). For peripheral blood mononuclear cells (PBMCs), blood samples were collected from healthy voluntary donors and processed by Ficoll-Paque gradient (GE Healthcare, Boston, MA). Informed consent was obtained in accordance with the Helsinki Declaration and the review board of Dana Farber Cancer Institute.
Chemicals
Camptothecin (CPT; Selleckchem, Houston, TX), mitomycin C (MMC; Santa Cruz Biotechnology, Dallas, TX), and melphalan (MilliporeSigma, Burlington, MA) were dissolved in dimethyl sulfoxide and diluted in cell culture medium prior to use.
Evaluation of UBE2T expression
UBE2T expression was evaluated either by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) or western blotting. The forward and reverse primers for quantitative reverse transcription polymerase chain reation (qRT-PCR) included UBE2T (forward, 5′-GATGACCTGCGAGCTCAAATA-3′; reverse, 5′-GGATCTGAGGAGGTTCAAATGG-3′), GAPDH (forward, 5′-GTCCACTGGCGTCTTCACCA-3′; reverse, 5′-GGATCTGAGGAGGTTCAAATGG-3′)
Antibodies used in western blotting included UBE2T (Proteintech), β-actin (Santa Cruz Biotechnology), γ-H2AX (Cell Signaling Technology, Danvers, MA), phospho-RPA32 (S4/8) (Cell Signaling Technology), and RPA32 (Bethyl Laboratories, Montgomery, TX).
Lentiviral particles and infections
Cells were infected with the indicated short hairpin RNA (shRNA) lentiviral particles (MilliporeSigma) and selected in 2 μg/mL puromycin. Before experiments, dead cells were eliminated using Ficoll-Paque gradient (GE Healthcare) and live cells allowed to recover for 24 hours.
HR repair assays
Immunofluorescence staining and microscopy and cell cycle
Immunofluorescence staining was done as previously described.14 Briefly, cells treated with primary antibodies (mouse monoclonal anti-γH2AX and rabbit polyclonal anti-RAD51 from Santa Cruz Biotechnology) were washed and incubated with the appropriate secondary antibodies (Alexa Fluor 594–labeled goat anti-mouse immunoglobulin G from Abcam (Cambridge, MA) for γ-H2AX; Alexa Fluor 488–labeled goat anti-rabbit immunoglobulin G from Abcam for RAD51). Images were acquired with Yokogawa Spinning Disk Confocal/TIRF System with 63× oil objective. Bromodeoxyuridine incorporation and propidium iodide staining were carried out as previously described.14
Cell viability assays
Cells were treated as indicated for 72 hours and viability assessed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega, Chicago, IL) or Cell Counting Kit-8 (CCK-8) assay (MilliporeSigma) according to the manufacturer’s protocol.
Results and discussion
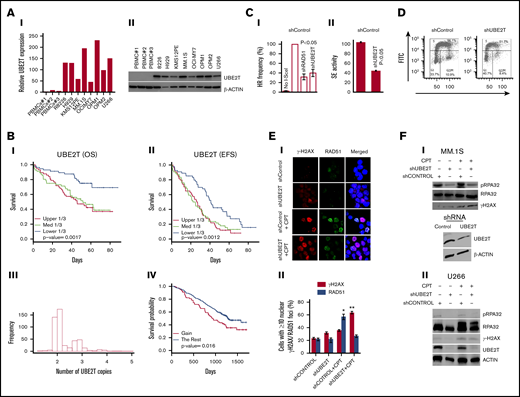
UBE2T expression was undetectable at both the messenger RNA and protein levels in normal PBMCs but highly expressed in all MM cell lines examined (Figure 1A, I-II). Consistent with a recent report in the GSE24080 MM dataset,15 we observed that elevated expression of UBE2T (GSE39754 dataset) is associated with poor overall as well as event-free survival in myeloma patients (Figure 1B, I-II). Moreover, 25% of these patients had >2 copies of the UBE2T gene, and UBE2T amplifications were also associated with poor patient survival (Figure 1B, III-IV). The MM database from the Arizona Translational Genomics Research Institute also indicated that UBE2T is highly expressed in the majority of MM cell lines, with increased copy number in several patients (not shown).
Increased UBE2T expression regulates HR activity in myeloma cells. (A) UBE2T is overexpressed in MM. UBE2T expression evaluated in normal PBMC samples (n = 3) and 8 MM cell lines by real-time qRT-PCR (I) or western blotting (II). (B) UBE2T expression and copy-number correlate with survival in a myeloma dataset (GSE39754; n = 170). Panels I and II show the association of UBE2T expression with overall survival (OS) (I) and event-free survival (EFS) (II). Panels III and IV show UBE2T copy-number alterations (III) and their correlation with overall survival (IV). (C) UBE2T regulates HR in MM cells. (I) HR monitored by the DRGFP assay. MM1S cells chromosomally integrated with the HR repair reporter substrate (DRGFP) were infected with lentiviral shRNA targeting UBE2T, RAD51, or control shRNA. The cells were then infected with a plasmid (AdGNU24i) that expresses the I-SceI endonuclease to initiate HR that is detected by flow cytometry as described in “Methods.” The fraction of GFP-positive cells, representing HR events, for control shRNA was taken as 100%, and other treatments were normalized to that as a percentage. No I-SceI, negative control with no HR; shCONTROL, cells transduced with control shRNA; shRAD51, cells transduced with RAD51 shRNA; shUBE2T, cells transduced with UBE2T shRNA. (II) HR monitored by strand exchange (SE) assay. Control and UBE2T knockdown MM.1S cells were analyzed for homologous strand exchange activity, as described in “Methods.” (D) Impact on cell cycle. Control and UBE2T knockdown MM.1S cells were analyzed for cell cycle using flow cytometry. (E) UBE2T is required for RAD51 focus formation. MM.1S cells transduced with lentivirus expressing the indicated shRNAs were treated with or without CPT (1 μM) for 1 hour and drug washed off. Cells were further cultured in drug-free medium for 6 hours and stained with antibodies to γH2AX and RAD51; DAPI staining was performed to define nuclei. Representative images of γH2AX and RAD51 foci (I) and bar graph showing percentage of cells with ≥5 nuclear γH2AX or RAD51 foci ± standard error of the mean (II) are shown. *P < .05; **P < .01. (F) UBE2T is required for DNA end resection. Myeloma cells, MM.1S (I) or U266 (II) were transduced with control shRNA (shCONTROL) or one targeting UBE2T (shUBE2T) and treated with CPT for 1 hour and evaluated for RPA32 and its phosphorylated form (pRPA S4/8; a marker of DNA end resection). Bottom figure in panel I shows the western blot confirming UBE2T knockdown in MM.1S cells. FITC, fluorescein isothiocyanate; PI, propidium iodide.
Increased UBE2T expression regulates HR activity in myeloma cells. (A) UBE2T is overexpressed in MM. UBE2T expression evaluated in normal PBMC samples (n = 3) and 8 MM cell lines by real-time qRT-PCR (I) or western blotting (II). (B) UBE2T expression and copy-number correlate with survival in a myeloma dataset (GSE39754; n = 170). Panels I and II show the association of UBE2T expression with overall survival (OS) (I) and event-free survival (EFS) (II). Panels III and IV show UBE2T copy-number alterations (III) and their correlation with overall survival (IV). (C) UBE2T regulates HR in MM cells. (I) HR monitored by the DRGFP assay. MM1S cells chromosomally integrated with the HR repair reporter substrate (DRGFP) were infected with lentiviral shRNA targeting UBE2T, RAD51, or control shRNA. The cells were then infected with a plasmid (AdGNU24i) that expresses the I-SceI endonuclease to initiate HR that is detected by flow cytometry as described in “Methods.” The fraction of GFP-positive cells, representing HR events, for control shRNA was taken as 100%, and other treatments were normalized to that as a percentage. No I-SceI, negative control with no HR; shCONTROL, cells transduced with control shRNA; shRAD51, cells transduced with RAD51 shRNA; shUBE2T, cells transduced with UBE2T shRNA. (II) HR monitored by strand exchange (SE) assay. Control and UBE2T knockdown MM.1S cells were analyzed for homologous strand exchange activity, as described in “Methods.” (D) Impact on cell cycle. Control and UBE2T knockdown MM.1S cells were analyzed for cell cycle using flow cytometry. (E) UBE2T is required for RAD51 focus formation. MM.1S cells transduced with lentivirus expressing the indicated shRNAs were treated with or without CPT (1 μM) for 1 hour and drug washed off. Cells were further cultured in drug-free medium for 6 hours and stained with antibodies to γH2AX and RAD51; DAPI staining was performed to define nuclei. Representative images of γH2AX and RAD51 foci (I) and bar graph showing percentage of cells with ≥5 nuclear γH2AX or RAD51 foci ± standard error of the mean (II) are shown. *P < .05; **P < .01. (F) UBE2T is required for DNA end resection. Myeloma cells, MM.1S (I) or U266 (II) were transduced with control shRNA (shCONTROL) or one targeting UBE2T (shUBE2T) and treated with CPT for 1 hour and evaluated for RPA32 and its phosphorylated form (pRPA S4/8; a marker of DNA end resection). Bottom figure in panel I shows the western blot confirming UBE2T knockdown in MM.1S cells. FITC, fluorescein isothiocyanate; PI, propidium iodide.
Since UBE2T has a known role in the FA pathway for DNA interstrand crosslink repair,6 which is upstream of HR,16 we evaluated whether high UBE2T expression contributes to elevated HR in MM. UBE2T knockdown in myeloma MM.1S cells reduced HR activity by ∼60% (P < .05; Figure 2C, I), as assessed by the pDRGFP assay.10,11,14,17,18 The inhibition of HR by UBE2T knockdown was comparable to the inhibition (65%; P < .01) observed following knockdown of RAD51, a central player in the HR process19 (Figure 2C, I). To further confirm the role of UBE2T in HR, we evaluated control and UBE2T-knockdown cells for homologous strand exchange activity, a distinct step involved in the initiation of HR by RAD51, as reported previously.12,13 As shown in Figure 2C, II, UBE2T knockdown led to ∼57% reduction (P < .05) in strand exchange activity in MM.1S cells. Furthermore, knockdown of UBE2T in the U2OS osteosarcoma cell line, which has been widely used to study DNA repair, also led to ∼60% reduction of HR repair (data not shown). Cell cycle analysis using propidium iodide staining indicated that the distribution of MM.1S cells following UBE2T knockdown was similar to that for control shRNA treatment (Figure 2D), suggesting that decreased HR observed with UBE2T knockdown was not related to cell cycle alteration. Moreover, pulse labeling with 5-bromo-2′-deoxyuridine followed by assessment of its incorporation into DNA also indicated that the overall levels of DNA replication in S-phase cells were not significantly altered by UBE2T knockdown (data not shown).
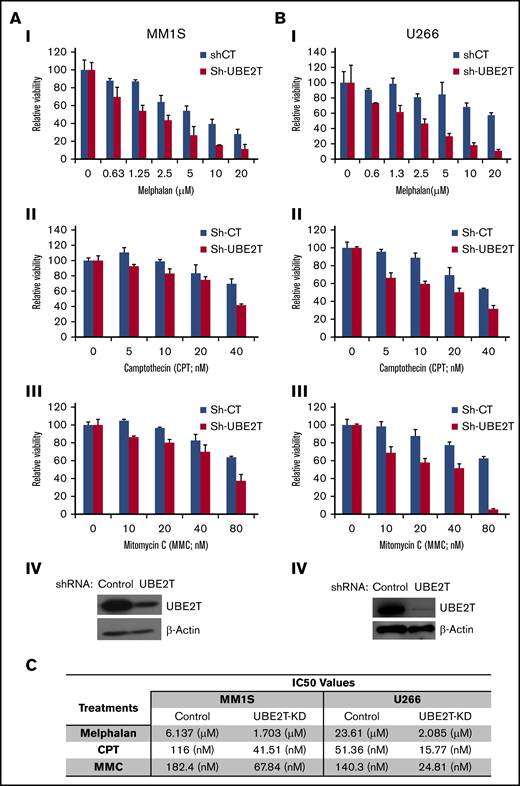
UBE2T-deficient MM cells are sensitive to DNA-damaging agents. (A-B) The MM cell lines MM1S (A) and U266 (B) were transduced with control shRNA (Sh-CT) or those targeting UBE2T (Sh-UBE2T) and cultured in the presence of indicated agents at the indicated doses. Cell viability was measured as described in “Methods.” Mean viability ± standard error of the mean is shown for 3 replicate experiments normalized to untreated controls. Western blots confirming UBE2T knockdown in MM1S and U266 cells are shown as panel IV in each figure. (C) Table showing 50% inhibitory concentration (IC50) values for the cytotoxicity of DNA-breaking agents in control and UBE2T-knockdown (UBE2T-KD) myeloma cells.
UBE2T-deficient MM cells are sensitive to DNA-damaging agents. (A-B) The MM cell lines MM1S (A) and U266 (B) were transduced with control shRNA (Sh-CT) or those targeting UBE2T (Sh-UBE2T) and cultured in the presence of indicated agents at the indicated doses. Cell viability was measured as described in “Methods.” Mean viability ± standard error of the mean is shown for 3 replicate experiments normalized to untreated controls. Western blots confirming UBE2T knockdown in MM1S and U266 cells are shown as panel IV in each figure. (C) Table showing 50% inhibitory concentration (IC50) values for the cytotoxicity of DNA-breaking agents in control and UBE2T-knockdown (UBE2T-KD) myeloma cells.
Nuclear foci of H2AX phosphorylated on serine 139 (γ-H2AX S139) indicate sites of double-stranded breaks (DSBs),20 while RAD51 foci designate RAD51 monomers bound to resected DSB ends engaged in HR repair.21 We therefore examined the effect of UBE2T depletion on the formation of RAD51 and γ-H2AX foci in the presence or absence of CPT, a topoisomerase I inhibitor that induces replication-dependent DSBs that are mainly repaired by HR.22 Cells were treated with or without CPT for 1 hour, the drug was removed, and cells were cultured in drug-free medium for 6 hours before immunofluorescence staining. Without CPT treatment, UBE2T-knockdown caused a small (8%) increase in basal levels of nuclear γ-H2AX foci without any apparent change in RAD51 foci (Figure 1E). CPT treatment caused a significant increase (35%; P < .05) in nuclear RAD51 foci in control shRNA–treated MM.1S cells, but not in UBE2T-knockdown MM.1S cells. In converse to nuclear RAD51 foci, nuclear γ-H2AX foci were significantly increased by CPT (>40%; P < .01) in cells depleted of UBE2T (Figure 1E). These results demonstrate that UBE2T knockdown is associated with impaired RAD51 focus formation at DSB sites, a key step in the HR repair process.
We also investigated the role of UBE2T in DSB end resection, a decisive and initiating step in HR that requires phosphorylation of RPA32 on S4/8.23,24 Remarkably, suppression of UBE2T led to reduction in CPT-induced levels of phospho-RPA32 (S4/8) relative to control cells (Figure 1F). As already indicated in Figure 1D, UBE2T knockdown did not impact cell cycle distribution. Moreover, the overall levels of DNA replication in S-phase cells were also not significantly altered by UBE2T knockdown (data not shown). These findings imply that the reduced RPA phosphorylation could not be due to changes in the cell cycle or DNA replication. Collectively, these results suggest that UBE2T may promote DSB end resection during HR repair.
With reduction in HR activity in UBE2T-knockdown cells, we tested their sensitivity to DNA-damaging agents (CPT, MMC, and melphalan) whose lesions require HR repair. UBE2T knockdown not only reduced MM-cell viability but also significantly increased the cytotoxicity of the DNA-damaging agents tested at all doses (Figure 2A-B). Overall, the 50% inhibitory concentration values for all 3 DNA-damaging agents (melphalan, CPT, and MMC) in UBE2T-knockdown myeloma cell lines were 2.7- to 11-fold lower than those of control cells (Figure 2C). We also observed that treatment of MM cells with agents that induce replication stress and DSBs, including CPT and hydroxyurea, induces UBE2T expression (data not shown).
In summary, we have identified UBE2T, a ubiquitin pathway gene, as a novel regulator of HR activity in MM. Its location on 1q32 may provide a possible basis for the development of high-risk disease that requires further investigation. The increased sensitivity of MM cells to chemotherapeutic agents following inhibition of UBE2T highlights its importance as a novel target to inhibit or reduce HR and genomic instability as well as overcome chemotolerance in myeloma. Although UBE2T inhibitors are currently not available, such inhibitors can be identified by screening for small molecules targeting this protein.
Acknowledgments
The authors thank N. Bahlis (University of Calgary, Calgary, AB, Canada) for providing MM.1S cells stably transfected with HR reporter plasmid (pDRGFP) and N. Bahlis (University of Calgary) and M. Nepveu (McGill University, Montreal, QC, Canada) for providing adenoviral vector AdNGUS24i that expresses I-SceI.
This work was supported by National Institutes of Health Research Program grants P01-155258 and P50 CA100707 (National Cancer Institute) (M.A.S., M.K.S., and N.C.M.), Department of Veterans Affairs Merit Review Award I01BX001584-01 (N.C.M.), and Leukemia and Lymphoma Society Translational Research grant (N.C.M.).
Authorship
Contribution: D.A.A. performed research and prepared the manuscript; S.K. and P.N. performed part of the research; S.T. performed part of the research and assisted in data analyses; L.B., C.C., M.K.S., and R.S. contributed to statistical and bioinformatics analyses; M.A.S. identified the target and contributed to experimental plan, data analyses/interpretations, and writing of the manuscript; and N.C.M. envisioned the study and contributed to experimental design, data interpretation, and final preparation of manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nikhil C. Munshi, Dana-Farber Cancer Institute, 450 Brookline Ave, Boston, MA 02215; e-mail: nikhil_munshi@dfci.harvard.edu; and Masood A. Shammas, Dana-Farber Cancer Institute, 44 Binney St, D1B25, Boston, MA 02215; e-mail: masood_shammas@dfci.harvard.edu.
References
Author notes
The clinical data sets used are publicly available at the Gene Expression Omnibus under accession number GSE39754 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39754) and the Arizona Translational Genomics Research Institute (TGen database; https://myelomagenomics.tgen.org/results_v3.php).
For original data, contact M.A.S. (masood_shammas@dfci.harvard.edu).