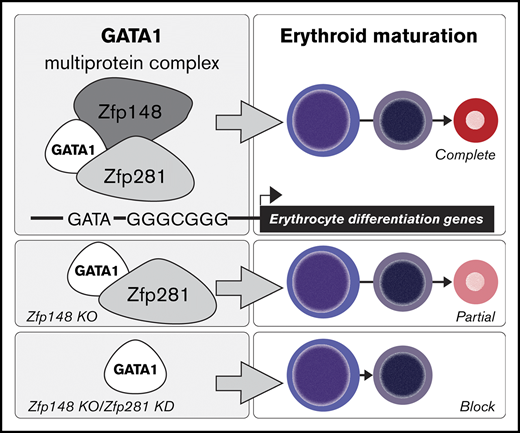
Key Points
The transcription factor Zfp281 (ZBP-99) plays a functional role during erythropoiesis that overlaps with its family member Zfp148 (ZBP-89).
Both Zfp281 and Zfp148 associate with GATA1.
Abstract
Erythroid maturation requires the concerted action of a core set of transcription factors. We previously identified the Krüppel-type zinc finger transcription factor Zfp148 (also called ZBP-89) as an interacting partner of the master erythroid transcription factor GATA1. Here we report the conditional knockout of Zfp148 in mice. Global loss of Zfp148 results in perinatal lethality from nonhematologic causes. Selective Zfp148 loss within the hematopoietic system results in a mild microcytic and hypochromic anemia, mildly impaired erythroid maturation, and delayed recovery from phenylhydrazine-induced hemolysis. Based on the mild erythroid phenotype of these mice compared with GATA1-deficient mice, we hypothesized that additional factor(s) may complement Zfp148 function during erythropoiesis. We show that Zfp281 (also called ZBP-99), another member of the Zfp148 transcription factor family, is highly expressed in murine and human erythroid cells. Zfp281 knockdown by itself results in partial erythroid defects. However, combined deficiency of Zfp148 and Zfp281 causes a marked erythroid maturation block. Zfp281 physically associates with GATA1, occupies many common chromatin sites with GATA1 and Zfp148, and regulates a common set of genes required for erythroid cell differentiation. These findings uncover a previously unknown role for Zfp281 in erythroid development and suggest that it functionally overlaps with that of Zfp148 during erythropoiesis.
Introduction
A complete understanding of hematopoietic transcriptional control requires that all functional trans-activating factors and cis-regulatory elements be identified. Within the erythroid system, the zinc finger transcription factor GATA1 acts as a master regulator controlling the expression of most, if not all, erythroid-specific genes.1-3 Targeted deletion of GATA1 blocks erythroid cell maturation and causes severe anemia in mice, leading to death by embryonic day 10.5 (e10.5) to e11.5.4,5 Enforced GATA1 expression reprograms alternate hematopoietic lineages into erythroid fates,6,7 and in combination with Tal1, Lmo2, and c-MYC, GATA1 directly converts fibroblasts into erythroid progenitor cells.8 These findings collectively highlight the dominant role that GATA1 plays in orchestrating erythroid cell fate decision and differentiation.
We previously performed a proteomic screen of GATA1-interacting proteins and identified the Krüppel-type zinc finger transcription factor Zfp148 (also called ZBP-89) as a GATA1 binding partner.9 Tetraploid complementation of Zfp148 gene-trap embryonic stem (ES) cells and chimeric mice showed that Zfp148 deficient cells have reduced contribution to definitive erythroid cells in vivo. Partial depletion of Zfp148 in primary human CD34+ (hCD34+) cells causes a mild impairment of erythroid maturation.10
The Zfp148 locus has been targeted previously in mice by disrupting exon 9, which removes 60% of the protein coding sequence but leaves a portion of the zinc finger DNA binding domain intact.11 This heterozygous allele was reported to cause defects in primordial germ cell development, and the allele was not able to be passed through the germline.11 A conditional knockout (cKO) mouse model targeting exons 8 and 9 deletion was subsequently generated,12,13 and Mx1-Cre–mediated Zfp148 deletion in the hematopoietic system showed acute, but transient anemia and thrombocytopenia in adult mice.12
The transcription factor Zfp281 (also called ZBP-99) was originally discovered as a guanine cytosine (GC)-rich DNA sequence binding Krüppel-like zinc finger protein that shares amino acid sequence homology with Zfp148 and binds to similar DNA sequences in vitro.14 Zfp281 has been extensively studied in ES cells where it is highly expressed and physically interacts with key stem cell transcription factors, including Nanog, Oct4, and Sox2, to regulate pluripotency genes.15-19 Zfp281 is dispensable for the establishment and maintenance of ES cells, but required for proper ES cell differentiation and embryo survival during the preimplantation blastocyst stage.20-22
Here, we report a new cKO mouse model for Zfp148 that deletes >80% of the protein coding domains, including the entire DNA binding zinc finger region. Hematopoietic selective loss causes mild anemia and delayed recovery from phenylhydrazine-induced hemolysis. We provide evidence that Zfp281 plays an overlapping role with Zfp148 in erythropoiesis, accounting for the mild erythroid phenotype associated with Zfp148 loss alone.
Methods
Animal studies
The Zfp148 locus spans ∼125 kb in mice and contains 9 exons (GRCm38, Ensembl). A targeting strategy, using pFlexible vector,23 was designed to allow Cre-LoxP–mediated deletion of exons 6 and 7, which introduces a premature translation termination codon “TGA” in exon 8. Homologous recombination of the targeting construct within Zfp148 locus was achieved in CJ9 (129/Sv) ES cells. A validated ES cell clone was injected into C57BL/6 blastocysts to generate chimeric mice (supplemental Methods). Germline transmission of the Zfp148 loxP allele was confirmed in F1 mice. To obtain hematopoietic-specific deletion of Zfp148, the Zfp148fl/fl mice were crossbred with Vav1-Cre24 transgenic mice. To induce stress erythropoiesis, mice were injected intraperitoneally with 60 mg/kg of phenylhydrazine (Sigma) in sterile phosphate-buffered saline (pH 7.4) on 2 consecutive days, as previously described.25 Zfp148 germline knockout mice were generated by interbreeding with CD-1 mice expressing Cre under the control of the GATA-1 promoter (GATA-1 Cre).26 The resulting Zfp148 heterozygous knockout (Zfp148+/−) F1 hybrids were mixed strains of 129/Sv, C57BL/6, and CD-1. All mice were backcrossed at least 5 to 6 generations with C57BL/6 mice. All experiments involving mice were approved by the Animal Care and Use Committee at Boston Children’s Hospital.
Antibodies and reagents
Generation of rabbit polyclonal Zfp148 N14 antibody has previously been described.9 Zfp148 N1-500 antibody was a gift from Juanita Merchant (University of Michigan Medical School). Additional antibodies were purchased from commercial sources (supplemental Methods), and all chemicals were purchased from Sigma-Aldrich, unless noted otherwise.
Blood counts and flow cytometry
Mouse peripheral blood was collected via retroorbital plexus and analyzed on a Hemavet HV950 multispecies hematology system (Drew Scientific, Inc). All flow cytometry was performed on FACSCalibur, LSR II flow cytometry, and Accuri instruments (BD Biosciences), and data were analyzed with FACSDiva and FlowJo software.
Complementary DNAs (cDNAs) and oligonucleotides
The human Zfp148 cDNA was amplified from MGC clone 4423572 (GE Healthcare Dharmacon, Inc); Zfp281 cDNA was synthesized (supplemental Methods), and oligonucleotides used for polymerase chain reactions (PCRs), reverse transcription–quantitative polymerase chain reactions (RT-qPCRs), chromatin immunoprecipitation–quantitative polymerase chain reactions (ChIP-qPCRs) and Southern blots are listed in supplemental Table 1.
Cell culture
The murine erythroid leukemic (MEL) BirA, MelBioGATA-1, K562 BirA, K562BioZfp148, and K562BioZfp281 cell lines were generated in house, and primary hCD34+ cells were obtained from the Yale Center of Excellence. Cells were cultured as previously described (supplemental Methods).9,10
Coimmunoprecipitation assays
Coimmunoprecipitation assays were performed by standard protocols as previously described,9,10 using 1 to 5 mg of nuclear extract protein (supplemental Methods).
ChIP and sequencing analysis
K562 cell lines expressing recombinant BioZfp148 and BioZfp281 were induced with 50 μM hemin for 48 hours. ChIP assays and ChIP-seq analysis were performed using standard methods (supplemental Methods).
RNA interference, CRISPR/Cas9, and lentiviruses
Lentiviral short hairpin RNA (shRNA) knockdown was performed as previously described.10,21 shRNAs seed sequences against Zfp281 are shown in supplemental Table 2. CRISPR/Cas9 genomic RNA (gRNA) expressing plasmid targeting exon 4 of Zfp148 (Target ID: HS0000451815) was obtained from Sigma-Aldrich.
Fetal liver erythroid differentiation
Fetal liver erythroid precursor cells were differentiated ex vivo as previously described.27 In brief, fetal livers were isolated from e12.5 to e13.5 C57BL/6 wild-type (WT) and Zfp148−/− embryos. These were made into a single-cell suspension and grown in an expansion medium. WT and knockout cells were infected with lentiviruses harboring shLuc control or shZfp281 for 24 hours, allowed to recover for 48 hours in expansion medium, and then selected for 48 hours in puromycin (0.3 µg/mL). Surviving cells were switched to differentiation media for 48 hours (supplemental Methods).
Histocytochemistry
Cytocentrifuged cells were stained with o-dianisidine (benzidine) for the presence of hemoglobin according to standard procedures.28 Counts were performed in a blinded fashion.
Results
Generation of Zfp148 cKO mice
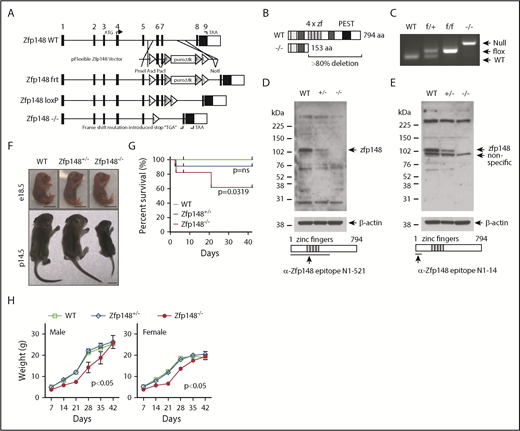
Targeted deletions of Zfp148 in mice have yielded varying phenotypes, suggesting that different strategies and/or background strains can influence the in vivo outcome.9,11-13 Previously reported gene deletion strategies are predicted to retain a portion of the zinc finger DNA binding domain.11-13 We generated a new Cre-LoxP cKO mouse that deletes >80% of the Zfp148 coding region, including the entire zinc finger DNA binding domain, making it the most complete knockout model to our knowledge (Figure 1A-B). Correct gene targeting in 129/Sv ES cells was confirmed by Southern blot and PCR analysis (supplemental Figure 1A-B). Zfp148fl/+ ES clones were karyotyped and injected into C57BL/6 blastocysts. High-level chimeric animals were obtained, and germline transmission of the floxed allele (Zfp148fl/+) was achieved.
Gene targeting of Zfp148 locus. (A) Schematic diagram of the Zfp148 locus and gene targeting strategy, showing the Zfp148 allele with short Flippase recognition target (frt, F) and Cre recombinase recognition target (LoxP, L). (B) Diagram of the Zfp148 protein in WT and null allele showing >80% deletion, including all 4 zinc finger domains. (C) PCR-based genotyping of WT, Zfp148fl/+, Zfp148fl/fl, and Zfp148−/− from tail DNA. (D-E) Western blot analysis of protein extracts from e14.5 WT, Zfp148+/−, and Zfp148−/− whole embryos on mixed genetic background strains using 2 independent anti-Zfp148 antibodies from Woo et al9 in panel D and Taniuchi et al52 in panel E. The epitope used to generate the antibodies is indicated on the schematic diagrams. (F) Photographs of representative Zfp148 conventional KO e18.5 embryos (top) and p14.5 (bottom) neonates on mixed C57BL/6, CD-1, and 129/Sv genetic background showing the runted postnatal phenotype. Scale bars, 1 cm. (G) Survival curves of Zfp148 WT (n = 14), Zfp148+/− (n = 21), and Zfp148−/− (n = 11) mice over the first 6 weeks of life. A significant difference in survival curve is observed between WT and Zfp148−/− (Mantel-Cox Log-rank P = .0319), whereas the WT and Zfp148+/− are not significant (Log-rank P = ns). (H) Growth curves of male WT (n = 5), Zfp148+/− (n = 7), and Zfp148−/− (n = 3) (left) panel, and female WT (n = 8), Zfp148+/− (n = 11), and Zfp148−/− (n = 6) mice (right) over the first 6 weeks of life. Growth of both female and male Zfp148−/− mice is significantly delayed compared with WT (2-tailed paired Student t test, P < .05). PEST, peptide domain rich in proline, glutamic acid, serine, and threonine.
Gene targeting of Zfp148 locus. (A) Schematic diagram of the Zfp148 locus and gene targeting strategy, showing the Zfp148 allele with short Flippase recognition target (frt, F) and Cre recombinase recognition target (LoxP, L). (B) Diagram of the Zfp148 protein in WT and null allele showing >80% deletion, including all 4 zinc finger domains. (C) PCR-based genotyping of WT, Zfp148fl/+, Zfp148fl/fl, and Zfp148−/− from tail DNA. (D-E) Western blot analysis of protein extracts from e14.5 WT, Zfp148+/−, and Zfp148−/− whole embryos on mixed genetic background strains using 2 independent anti-Zfp148 antibodies from Woo et al9 in panel D and Taniuchi et al52 in panel E. The epitope used to generate the antibodies is indicated on the schematic diagrams. (F) Photographs of representative Zfp148 conventional KO e18.5 embryos (top) and p14.5 (bottom) neonates on mixed C57BL/6, CD-1, and 129/Sv genetic background showing the runted postnatal phenotype. Scale bars, 1 cm. (G) Survival curves of Zfp148 WT (n = 14), Zfp148+/− (n = 21), and Zfp148−/− (n = 11) mice over the first 6 weeks of life. A significant difference in survival curve is observed between WT and Zfp148−/− (Mantel-Cox Log-rank P = .0319), whereas the WT and Zfp148+/− are not significant (Log-rank P = ns). (H) Growth curves of male WT (n = 5), Zfp148+/− (n = 7), and Zfp148−/− (n = 3) (left) panel, and female WT (n = 8), Zfp148+/− (n = 11), and Zfp148−/− (n = 6) mice (right) over the first 6 weeks of life. Growth of both female and male Zfp148−/− mice is significantly delayed compared with WT (2-tailed paired Student t test, P < .05). PEST, peptide domain rich in proline, glutamic acid, serine, and threonine.
Fertility of Zfp148 heterozygous mice and survival of null mice on a mixed strain background
We attempted generation of Zfp148−/+ mice to validate the previously reported germ cell defect associated with disrupting exon 9.11 The Zfp148 floxed allele was first excised in the germline by breeding with GATA1-Cre mice, which express Cre in sperm cells.26 In contrast to the previous report,11 we obtained live heterozygous Zfp148 knockout (Zfp148−/+) pups. Interbreeding of the heterozygous mice produced homozygous null Zfp148 (Zfp148−/−) mice at the expected Mendelian ratio (Figure 1C; supplemental Table 3). Sequencing of the Zfp148 transcripts from the knockout animals showed the expected fusion of exons 5 to 8 (supplemental Figure 1C-D). Western blot analysis of whole embryos using 2 independent antibodies raised against aminoterminal regions of Zfp148 demonstrated an allele dose-dependent loss of the protein (Figure 1D-E).
Severe growth retardation of Zfp148−/− neonatal mice on mixed genetic background
The Zfp148−/− pups, on a mixed C57BL/6, CD-1, and 129/Sv genetic background, were severely runted compared with their littermate controls (Figure 1F). They appeared to have normal growth up until at least e18.5 estimated days postcoitus (e18.5 d.p.c.). The pups had poor weight gain, low plasma glucose levels, and increased mortality over the first few weeks of life (Figure 1G; supplemental Figure 2A-B). The early lethality did not appear to be related to hematologic causes. The Zfp148−/− animals had milk in their stomach indicating an ability to suckle (supplemental Figure 2B). The Zfp148−/− pups continued to exhibit poor growth even when control littermates were removed. The animals that survived achieved equivalent body weights and glucose levels compared with their heterozygous and WT littermates by ∼6 weeks of age (supplemental Figure 2A) and had normal long-term survival.
Neonatal lethality of Zfp148−/− mice on a pure C57BL/6 background
Zfp148−/+ mice backcrossed onto a pure C57BL/6 background remained fertile but produced only rare live Zfp148−/− mice (≤2%) beyond 2 days of life (Table 1). Most of the homozygous fetuses were lost between e18.5 d.p.c. and birth. The etiology of their death remains unclear. The embryos did not have severe anemia, hemorrhage, or gross structural defects. No defects were observed upon histological analysis of lung tissues at postnatal day 1 (p1.5) (supplemental Figure 3).
Microcytic, hypochromic anemia of Vav1-Cre; Zfp148fl/fl mice
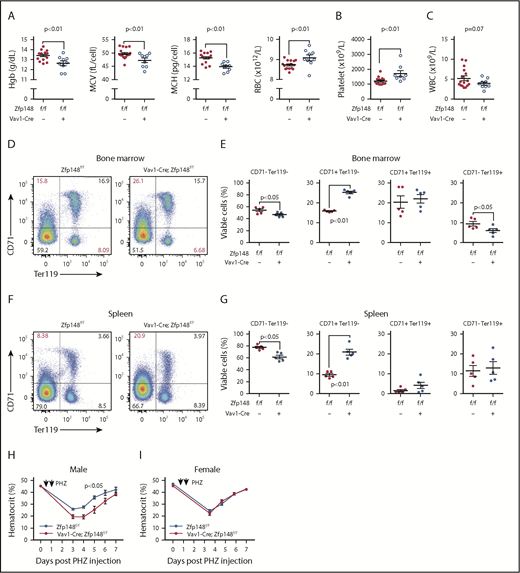
Zfp148fl/fl mice on a pure C57BL/6 background were then bred to pan-hematopoietic Cre expressing Vav1-Cre mice.24 The Vav1-Cre; Zfp148fl/fl mice were viable and had a normal lifespan. The mice had decreased hemoglobin levels, mean cell volume, mean cell hemoglobin, and increased red blood cell (RBC) number consistent with a mild thalassemia-like phenotype (Figure 2A). There was no statistically significant difference in plasma iron levels or reticulocyte counts between the knockout and control mice (supplemental Figure 4A-B). The platelet counts were mildly elevated, and the mean platelet volumes were not significantly different (Figure 2B). The total white blood cell count showed a lower trend in the Vav1-Cre; Zfp148fl/fl mice, but this was not statistically significant (Figure 2C). There was mildly impaired erythroid maturation based on flow cytometry for CD71 and Ter119, with an accumulation of immature CD71+Ter119− cells and reduction of mature CD71+Ter119+ cells in the bone marrow (Figure 2D-E). Splenic erythropoiesis was similar but only accumulation of CD71+Ter119− cells was observed (Figure 2F-G). We examined recovery following phenylhydrazine-induced transient hemolytic anemia. This showed a modestly delayed recovery of peripheral RBCs in male Vav1-Cre; Zfp148fl/fl mice, but not in female mice (Figure 2H-I).
Defective erythropoiesis in mice with pan-hematopoietic Zfp148 deletion. (A-C) Peripheral blood counts and indices from Zfp148fl/fl and Vav1-Cre; Zfp148fl/fl mice at 15 to 26 weeks of age. (D) Representative flow cytometric analysis profiles of bone marrow for erythroid progenitor maturation using anti-CD71 and anti-Ter119 antibodies in Zfp148fl/fl and Vav1-Cre; Zfp148fl/fl mice. (E) Dot plots depicting the percentage of live BM cells in non- and early erythroid cells (CD71−Ter119−), late proerythroblasts (CD71+Ter119−), early erythroid cells (CD71+Ter119+) and RBCs (CD71−Ter119+) from the flow analysis in panel D (n = 5). (F-G) Representative flow cytometric analysis profile for spleen as in panels D and E. (H-I) Recovery from phenylhydrazine (PHZ)-induced hemolytic anemia in male (n = 8) and female (n = 12) mice. Spun hematocrit levels from zfp148fl/fl, Vav1-Cre, and Zfp148fl/fl control littermates preceding and following a 2-day course of phenylhydrazine intraperitoneal injection. A baseline spun hematocrit was obtained prior to phenylhydrazine injection and then repeated on days 3, 4, 5, 6, and/or 7 following initial injection. Hgb, hemoglobin; MCH, mean cell hemoglobin; MCV, mean cell volume; WBC, white blood cell.
Defective erythropoiesis in mice with pan-hematopoietic Zfp148 deletion. (A-C) Peripheral blood counts and indices from Zfp148fl/fl and Vav1-Cre; Zfp148fl/fl mice at 15 to 26 weeks of age. (D) Representative flow cytometric analysis profiles of bone marrow for erythroid progenitor maturation using anti-CD71 and anti-Ter119 antibodies in Zfp148fl/fl and Vav1-Cre; Zfp148fl/fl mice. (E) Dot plots depicting the percentage of live BM cells in non- and early erythroid cells (CD71−Ter119−), late proerythroblasts (CD71+Ter119−), early erythroid cells (CD71+Ter119+) and RBCs (CD71−Ter119+) from the flow analysis in panel D (n = 5). (F-G) Representative flow cytometric analysis profile for spleen as in panels D and E. (H-I) Recovery from phenylhydrazine (PHZ)-induced hemolytic anemia in male (n = 8) and female (n = 12) mice. Spun hematocrit levels from zfp148fl/fl, Vav1-Cre, and Zfp148fl/fl control littermates preceding and following a 2-day course of phenylhydrazine intraperitoneal injection. A baseline spun hematocrit was obtained prior to phenylhydrazine injection and then repeated on days 3, 4, 5, 6, and/or 7 following initial injection. Hgb, hemoglobin; MCH, mean cell hemoglobin; MCV, mean cell volume; WBC, white blood cell.
Expression of Zfp281 in erythroid cells and interaction with GATA1
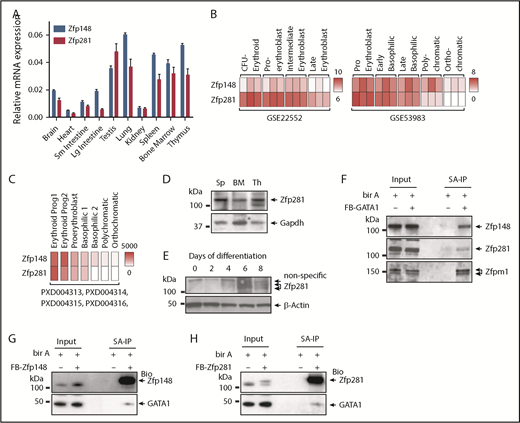
The relatively mild hematopoietic phenotype of the Vav1-Cre; Zfp148fl/fl mice led us to hypothesize that Zfp281 may play a functionally redundant role with Zfp148 during erythroid development. Zfp281 shares 79% and 71% amino acid sequence similarity and identity, respectively, within the zinc finger DNA binding region of Zfp148.29 In ES cells, Zfp281 binds to consecutive guanine (G)-rich DNA motifs30 that are similar to Zfp148 motifs found in human primary erythroid cells.10 We began investigating a role for Zfp281 in erythroid cells by first examining its expression in major hematopoietic tissues in mice. Both Zfp148 and Zfp281 are expressed in a variety of tissues, but with higher levels in spleen, bone marrow, and thymus (Figure 3A,D). In humans, Zfp281 is expressed concomitantly with Zfp148 at both the messenger RNA (mRNA) and the protein level in purified erythroid populations from ex vivo differentiated from CD34+ peripheral blood mononuclear cells (PBMCs)31 and cord blood cells,32,33 with higher levels at immature vs late stages (Figure 3B-C). We confirmed the emergence of Zfp281 protein by western blot on day 4 of erythroid ex vivo differentiation of PBMC-derived hCD34+ cells (Figure 3E). Zfp281 protein was not detected in undifferentiated primary human PBMC-derived CD34+ cells. This expression pattern mirrors that of Zfp148.10
Zfp281 expression and physical interaction with GATA1 in erythroid cells. (A) qRT-PCR analysis showing Zfp148 and Zfp281 mRNA levels relative to Gapdh (glyceraldehyde-3-phosphate dehydrogenase) in C57BL/6 mouse tissues. (B) Heat map of Zfp148 and Zfp281 mRNA levels in flow cytometric sorted erythroid progenitor cells from ex vivo differentiated PBMC (GSE22552)31 (left) and CD34+ cord blood cells (GSE53983)32 (right). (C) Heat map showing mean copy number of Zfp148 and Zfp281 protein per erythroid progenitor cell, determined by mass spectrometry–based absolute quantification approach (PXD004313, PXD004314, PXD004315, and PXD004316).33 Following the expansion of cord blood CD34+ cells, CD36+ progenitors were flow sorted and differentiated ex vivo under erythroid conditions. Erythroid Prog1 and 2 equates to burst-forming unit-erythroid (BFU-E) and colony-forming unit-erythroid (CFU-E) cells, respectably.33 (D) Western blot analysis of Zfp281 protein in major hematopoietic organs, spleen (Sp), bone marrow (BM), and thymus (Th) in mice. (E) Western blot showing Zfp281 protein levels during erythroid ex vivo differentiation of hCD34+ cells. (F) Western blot analysis following streptavidin affinity purification (SA-IP) from nuclear extracts of MEL cells stably expressing Bir A alone or Bir A and recombinant GATA1 containing an amino-terminal FLAG and BirA recognition motif (FB-GATA1). Two percent of the input material is shown. (G) Western blot analysis following SA-IP from nuclear extracts of K562 cells stably expressing Bir A alone or Bir A and recombinant FB-Zfp148. Two percent of the input material is shown. (H) Western blot analysis following SA-IP from nuclear extracts of K562 cells stably expressing Bir A or Bir A and recombinant FB-Zfp281. Two percent of the input material is shown. Lg, large; Sm, small.
Zfp281 expression and physical interaction with GATA1 in erythroid cells. (A) qRT-PCR analysis showing Zfp148 and Zfp281 mRNA levels relative to Gapdh (glyceraldehyde-3-phosphate dehydrogenase) in C57BL/6 mouse tissues. (B) Heat map of Zfp148 and Zfp281 mRNA levels in flow cytometric sorted erythroid progenitor cells from ex vivo differentiated PBMC (GSE22552)31 (left) and CD34+ cord blood cells (GSE53983)32 (right). (C) Heat map showing mean copy number of Zfp148 and Zfp281 protein per erythroid progenitor cell, determined by mass spectrometry–based absolute quantification approach (PXD004313, PXD004314, PXD004315, and PXD004316).33 Following the expansion of cord blood CD34+ cells, CD36+ progenitors were flow sorted and differentiated ex vivo under erythroid conditions. Erythroid Prog1 and 2 equates to burst-forming unit-erythroid (BFU-E) and colony-forming unit-erythroid (CFU-E) cells, respectably.33 (D) Western blot analysis of Zfp281 protein in major hematopoietic organs, spleen (Sp), bone marrow (BM), and thymus (Th) in mice. (E) Western blot showing Zfp281 protein levels during erythroid ex vivo differentiation of hCD34+ cells. (F) Western blot analysis following streptavidin affinity purification (SA-IP) from nuclear extracts of MEL cells stably expressing Bir A alone or Bir A and recombinant GATA1 containing an amino-terminal FLAG and BirA recognition motif (FB-GATA1). Two percent of the input material is shown. (G) Western blot analysis following SA-IP from nuclear extracts of K562 cells stably expressing Bir A alone or Bir A and recombinant FB-Zfp148. Two percent of the input material is shown. (H) Western blot analysis following SA-IP from nuclear extracts of K562 cells stably expressing Bir A or Bir A and recombinant FB-Zfp281. Two percent of the input material is shown. Lg, large; Sm, small.
We previously showed that Zfp148 physically and functionally interacts with GATA1 to regulate erythroid cell differentiation.9,10 To examine if Zfp281 also associates with GATA1, we performed a streptavidin pull-down of multiprotein complexes containing metabolically biotinylated GATA1 (BioGATA1) in MEL cells.9 Western blot analysis showed that Zfp281 coprecipitated with these2 complexes, similar to Zfp148 and Zfpm1 (friend of GATA protein-1), a known GATA1 cofactor (Figure 3F). We validated their association in the reverse direction by expressing BioZfp148 and BioZfp281 in human K562 erythroid leukemia cells followed by streptavidin pull-down and GATA1 western blot (Figure 3G-H).
Common chromatin occupancy of Zfp148, Zfp281, and GATA1 in erythroid cells
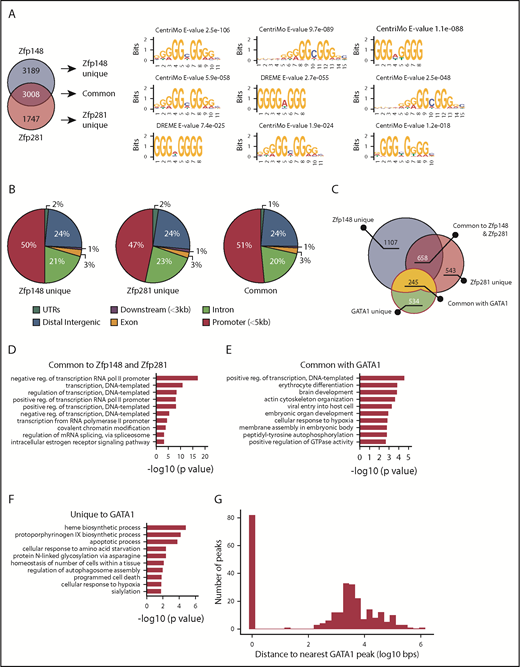
ChIP-seq was performed next to assess the degree of chromatin occupancy overlap between Zfp148 and Zfp281. To avoid potential antibody crossreactivity, we used a streptavidin BioChIP-seq approach34 using K562 BioZfp148 and BioZfp281 clones that express metabolically biotinylated Zfp148 or Zfp281 at or below the endogenous protein levels (supplemental Figure 5A-B). Parental cells expressing the biotin ligase Bir A alone were used as controls. Remarkably, close to half of the Zfp148 peaks (3008 of 6197, 49%) and two-thirds of the Zfp281 peaks (3008 of 4755, 63%) corresponded to each other’s occupancy sites (hypergeometric test of overlap, P < 10500) (Figure 4A; supplemental Figure 6). Consecutive stretches of G-rich DNA motifs were highly enriched in all Zfp281/Zfp148 occupancy sites, consistent with prior reports of their DNA binding sequence preferences.10,14,21,29,30 The distributions of their peaks were also similar, with about half occurring at the promoter (Figure 4B). These occupancy peaks were then assigned to nearest target genes. More than half of Zfp281 (728 of 1339, 54%) and a third of Zfp148 target genes (728 of 1942, 37%) were in common with each other (supplemental Figure 7A). Gene ontology (GO) term analysis revealed that the common target genes were enriched for transcriptional processes, nucleosome disassembly, and covalent chromatin modification (supplemental Figure 7D).
Common gene targets of Zfp148 and Zfp281 in erythroid cells. (A) Venn diagram showing the overlap of Zfp148 and Zfp281 chromatin occupancy peaks (left). MEME-ChIP motif analysis53 result showing 3 representative motifs for unique and common peaks (right). (B) Pie graphs showing the distribution of chromatin occupancy peak location. UTRs, untranslated regions. (C) Venn diagram showing the number of genes commonly occupied by Zfp148, Zfp281, and/or GATA1. (D) GO term analysis of the 658 target genes common to Zfp148 and Zfp281, but devoid of GATA1. (E) GO term analysis of the 245 target genes common to GATA1 and Zfp148 and/or Zfp281. (F) GO term analysis of the 534 genes unique to GATA1. GO term analysis of unique Zfp148 and Zfp281 target genes are shown in supplemental Figure 8. (G) Distribution of Zfp148/Zfp281 peaks with respect to GATA1 peaks at the 245 commonly occupied genes. reg., regulation.
Common gene targets of Zfp148 and Zfp281 in erythroid cells. (A) Venn diagram showing the overlap of Zfp148 and Zfp281 chromatin occupancy peaks (left). MEME-ChIP motif analysis53 result showing 3 representative motifs for unique and common peaks (right). (B) Pie graphs showing the distribution of chromatin occupancy peak location. UTRs, untranslated regions. (C) Venn diagram showing the number of genes commonly occupied by Zfp148, Zfp281, and/or GATA1. (D) GO term analysis of the 658 target genes common to Zfp148 and Zfp281, but devoid of GATA1. (E) GO term analysis of the 245 target genes common to GATA1 and Zfp148 and/or Zfp281. (F) GO term analysis of the 534 genes unique to GATA1. GO term analysis of unique Zfp148 and Zfp281 target genes are shown in supplemental Figure 8. (G) Distribution of Zfp148/Zfp281 peaks with respect to GATA1 peaks at the 245 commonly occupied genes. reg., regulation.
Zfp148 and Zfp281 target genes were next compared with those of GATA1 in K562 cells (GEO accession: GSM1003608)35 (Figure 4C). About 245 GATA target genes also contain occupancy by Zfp148 and/or Zfp281 (supplemental Table 4). These genes are enriched for functions associated with positive regulation of transcription (GO:0045893) and erythrocyte differentiation (GO:0030218; Figure 4E). GATA1 unique genes showed enrichment for heme biosynthetic processes (GO:0006783; Figure 4F). There was a bimodal chromatin occupancy peak distribution among genes bound by both GATA1 and Zfp148/Zfp281with 83 showing exact overlap of the GATA1 and Zfp148/Zfp281 peaks and the remainder being separated by several kilobases (Figure 4G). No statistically significant DNA binding motif enrichment was identified that distinguished these 2 classes of binding configurations. Together with the physical interaction findings, the overall chromatin occupancy data suggest a cooperative role for both Zfp281 and Zfp148 in driving a subset of the GATA1-mediated erythroid gene program.
Zfp281 plays a redundant role with Zfp148 in γ-globin gene regulation
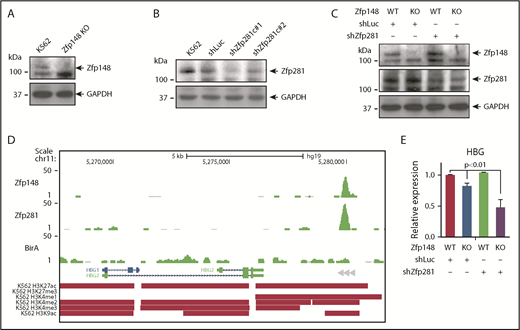
We previously showed that Zfp148 occupies both the α- and β-globin loci in adult erythroid cells and positively regulate their gene expression.10 To examine the functional consequence of depleting both Zfp148 and Zfp281 in globin gene regulation, we first deleted Zfp148 in K562 cells using CRISPR-Cas9 and confirmed the protein depletion by western blot (Figure 5A). Despite trying various guide RNA sequences to disrupt the Zfp281 locus, we were unable to obtain knockout clones. As an alternative approach, lentiviral delivered shRNAs were used to knockdown Zfp281 expression (Figure 5B). Zfp148 knockout cells were then infected with shZfp281c#1 lentivirus to establish K562 cells depleted for both Zfp148 and Zfp281 (Figure 5C). K562 cells express predominantly γ-globin, and both Zfp148 and Zfp281 occupy a chromatin site ∼3 kb upstream of the HBG2 transcriptional start site (Figure 5D). Zfp148 knockout alone led to a modest decrease in γ-globin mRNA levels (Figure 5E), whereas Zfp281 knockdown by itself had no significant effect (Figure 5E). However, the double depletion of Zfp148 and Zfp281 reduced levels to ∼50% lower than Zfp148 knockout by itself, supporting a redundant role for Zfp281 with Zfp148 in regulating γ-globin gene expression in this cell system.
Redundant roles of Zfp281 and Zfp148 in γ-globin gene expression in K562 cells. (A) Western blot showing knockout (KO) of Zfp148 in K562 cells by CRISPR/Cas9. (B) Western blot showing knockdown of Zfp281 protein levels in K562 cells using lentiviral transduction of shRNA constructs c#1 and c#2. A lentiviral shRNA against luciferase (shLuc) is shown as a control. (C) Western blot showing Zfp148 and Zfp281 protein levels in Zfp148 KO cells or control cells transduced with shZfp281c#1 or shLuc. (D) Representative BioChIP-seq signals at the γ-globin locus in hemin-induced K562 cells expressing Bir A alone or Bir A and BioZfp148 or BioZfp281. Peak calls for H3K27ac, H3K27me3, H3K4me1, H3K4me2, H3K4me3, and H3K9ac from ENCODE35 are indicated. (E) Quantitative RT-PCR analysis of γ-globin mRNA transcripts from cells in panel C. The levels were normalized to GAPDH mRNA transcript levels and are shown relative to the Zfp148 WT and shLuc control. HBG, hemoglobin subunit gamma.
Redundant roles of Zfp281 and Zfp148 in γ-globin gene expression in K562 cells. (A) Western blot showing knockout (KO) of Zfp148 in K562 cells by CRISPR/Cas9. (B) Western blot showing knockdown of Zfp281 protein levels in K562 cells using lentiviral transduction of shRNA constructs c#1 and c#2. A lentiviral shRNA against luciferase (shLuc) is shown as a control. (C) Western blot showing Zfp148 and Zfp281 protein levels in Zfp148 KO cells or control cells transduced with shZfp281c#1 or shLuc. (D) Representative BioChIP-seq signals at the γ-globin locus in hemin-induced K562 cells expressing Bir A alone or Bir A and BioZfp148 or BioZfp281. Peak calls for H3K27ac, H3K27me3, H3K4me1, H3K4me2, H3K4me3, and H3K9ac from ENCODE35 are indicated. (E) Quantitative RT-PCR analysis of γ-globin mRNA transcripts from cells in panel C. The levels were normalized to GAPDH mRNA transcript levels and are shown relative to the Zfp148 WT and shLuc control. HBG, hemoglobin subunit gamma.
Cross examination of histone modification marks deposited in ENCODE35 showed an overlap of active histone marks (H3K27ac, H3K4me1, K3K4me2, H3K9ac) and depletion of a repressive mark (H3K27me3) at the Zfp148/Zfp281 occupied peak (Figure 5D), suggesting that both factors act predominantly as mediators of gene activation in this context.
Both Zfp281 and Zfp148 are required for erythroid development
To examine the consequences of combined Zfp148 and Zfp281 loss in vivo, we attempted to generate Zfp281 cKO mice. However, extensive trials failed to produce a clone with the properly targeted allele. This may be related to the high GC content of this locus and its unusual exonic structure (2 exons with a 216 base pair GC-rich intron). Thus, we resorted to using ex vivo differentiation27 of murine fetal liver progenitor cells from Zfp148−/− mice in combination with Zfp281 lentiviral shRNA knockdown.
shRNAs that efficiently knock down murine Zfp281 in MEL cells were first identified (Figure 6A). Fetal liver cells from WT and Zfp148−/− mice (e12.5-e13.5 d.p.c.) were harvested, cultured in expansion medium for 8 days during which time they were transduced with Zfp281c#3 shRNA or control (shLuc) lentiviral vectors, and selected (Figure 6B). The cells were then switched to differentiation medium and analyzed (see “Methods”). Benzidine staining showed >60% of fetal liver cells transduced with shLuc became hemoglobinized at 48 hours (Figure 6C-D). Zfp148 knockout led to a slightly increased percentage of benzidine-positive cells in this system and no statistically significant alteration in erythroid maturation based on CD71 and Ter119 surface expression. Fetal liver cells transduced with the Zfp281 shRNA had a marginally lower percentage of benzidine-positive cells and trends toward higher R2 (CD71highTer119low) and lower R3 (CD71highTer119high) cell populations, indicating a mild perturbation of erythropoiesis (Figure 6C-F). In contrast, when Zfp281 was depleted in Zfp148−/− fetal liver cells, the erythroid maturation became significantly more impaired compared with either knockout or knockdown alone with <40% benzidine-positive cells, and higher R2 and a significantly lower R3 population (Figure 6C-F). These data support the model that Zfp148 and Zfp281 play functionally redundant roles in erythroid maturation.
Redundant roles of Zfp148 and Zfp281 in primary murine fetal liver erythroid cells. (A) Western blot showing knockdown of murine Zfp281 protein levels in MEL cells transduced with lentiviral shRNA constructs shZfp281c#3, zhZfp281c#4, and shLuc. (B) Schematic diagram showing an experimental timeline of ex vivo culture, lentiviral transduction, and puromycin selection of fetal liver cells. (C) Representative benzidine stains with May-Grünwald counterstain of cytospun cells from murine e12.5-e13.5 d.p.c. Zfp148−/− or WT (littermates) fetal liver cells transduced with the indicated shRNA lentiviruses, selected with puromycin, and ex vivo differentiated into erythroid cells (8 days in expansion medium followed by 48 hours in differentiation medium; see “Methods”). Magnification ×600. (D) Bar graph shows blinded benzidine counts from 5 biologic replicates of cultures in panel C. (E-F) Representative CD71/Ter119 flow cytometric plots and quantitation of the cultures in panel C. Each plot shows the percentage of R1 to R5 populations in 5 biological replicates. *P < .05; **P < .01.
Redundant roles of Zfp148 and Zfp281 in primary murine fetal liver erythroid cells. (A) Western blot showing knockdown of murine Zfp281 protein levels in MEL cells transduced with lentiviral shRNA constructs shZfp281c#3, zhZfp281c#4, and shLuc. (B) Schematic diagram showing an experimental timeline of ex vivo culture, lentiviral transduction, and puromycin selection of fetal liver cells. (C) Representative benzidine stains with May-Grünwald counterstain of cytospun cells from murine e12.5-e13.5 d.p.c. Zfp148−/− or WT (littermates) fetal liver cells transduced with the indicated shRNA lentiviruses, selected with puromycin, and ex vivo differentiated into erythroid cells (8 days in expansion medium followed by 48 hours in differentiation medium; see “Methods”). Magnification ×600. (D) Bar graph shows blinded benzidine counts from 5 biologic replicates of cultures in panel C. (E-F) Representative CD71/Ter119 flow cytometric plots and quantitation of the cultures in panel C. Each plot shows the percentage of R1 to R5 populations in 5 biological replicates. *P < .05; **P < .01.
Discussion
In this study, we uncover a novel role for the Krüppel-type zinc finger transcription factor Zfp281 in erythroid development and demonstrate that it functionally overlaps with that of its family member Zfp148. We also show that these 2 factors, likely in combination with other coregulators, associate with GATA1 to regulate genes during erythroid maturation.
Although Zfp281 was initially identified during a screen for Zfp148 gene family members,29 recent studies have focused on its activity in ES cell biology.16,17,20-22,36 Our study demonstrates that Zfp281 also has functional roles in adult tissues, such as the hematopoietic system. To the best of our knowledge, this is the first study to demonstrate that Zfp281 and Zfp148 occupy common chromatin sites and functionally compensate for one another. We also show that both Zfp148 and Zfp281 physically associate with and occupy a common set of chromatin sites with GATA1, although we have not yet established whether these interactions are direct. Interestingly, Zfp281 was recently shown to bind with the GATA family member GATA4 and enhance GATA4 activity in reprogramming of cardiac cells.37 We previously showed that Zfp148 interacts with GATA2 and GATA3, in addition to GATA1.9 These findings suggest that Zfp148/Zfp281 and GATA family transcription factors may cooperate broadly in diverse tissues and cellular contexts.
We previously showed a high correlation between Zfp148 chromatin occupancy and activating histone marks in human erythroid cells.10 In addition, we and others showed that Zfp148 physically associates with the p30010,38 and the GCN5/Trrap (GNAP) histone acetyltransferases, and that Zfp148 deficiency reduces GCN5 recruitment at selected activated gene loci.10 Our new ChIP-seq data show a predominance of Zfp148 and Zfp281 chromatin occupancy at gene promoters. Analysis of GATA1 occupancy sites of activated vs repressed genes in an estradiol-induced GATA1-ER cell system39,40 also shows high enrichment for Zfp148/Zfp281 consensus binding motifs within the first intron of GATA1 activated genes (A.G., G.P., E.F., and A.B.C., unpublished observation, 2 February 2017). Collectively, these data suggest that Zfp148 and Zfp281 functionally synergize with GATA1 and act predominantly in gene activation during erythroid maturation, likely through mechanisms operating near gene transcription start sites.
Although Zfp148 and Zfp281 are present in a wide array of tissues, their expression was not detected in hCD34+ cells, which represent stem and early multipotent progenitor cells. Both proteins are upregulated during hCD34+ cell erythroid ex vivo differentiation, first becoming detectable at the protein level in early erythroid progenitors (Figure 3C,E; Woo et al10 ). This developmental regulation is consistent with them playing stage-specific roles during erythroid commitment and maturation.
Hematopoietic-specific deletion of Zfp148 causes a hypochromic and microcytic anemia with elevated RBC number. This likely represents a mild thalassemia-like phenotype given our previous data showing that Zfp148 occupies key cis-regulatory elements within the globin locus in primary human erythroid progenitor cells, that Zfp148 shRNA knockdown reduces globin mRNA levels in primary erythroid progenitors, and that the new data in this study showing Zfp148/Zfp281 occupancy upstream of the γ-globin gene in K562 cells and reduced γ-globin mRNA transcript levels upon double Zfp148/Zfp281 depletion. This supports a direct role for Zfp148/Zfp281 family transcription factors in globin gene expression. However, although Zfp281 clearly compensates for Zfp148 loss in general, the fact that Zfp148 knockout mice have an erythroid phenotype indicates that Zfp281 by itself is insufficient to completely replace Zfp148 deficiency. Whether this is because there is a total combined level of Zfp148/Zfp281 protein necessary for normal erythropoiesis or that Zfp148 and Zfp281 have unique biologic properties will require additional study.
Delayed recovery from phenylhydrazine-induced anemia of Vav1-Cre; Zfp148fl/fl mice was observed only in male mice. The etiology of this remains unclear. Testosterone facilitates erythropoiesis by inducing erythropoietin expression; however, glucocorticoids counteract this effect.41-43 The glucocorticoid receptor synergizes with hypoxia response genes to rapidly expand erythroid progenitor cell populations during stress erythropoiesis.44,45 Zfp281 occupied target genes are enriched for hypoxia response (GO:0001666; supplemental Figure 7C). Thus, it is possible that in the absence of Zfp148, an altered Zfp281-mediated hypoxia response impacts the testosterone-erythropoietin axis. Because GATA1 is X-linked and Zfp148 controls GATA1 expression,46 another possibility is that female mice are able to epigenetically compensate for Zfp148 loss in ways not possible in male mice. Future in vivo studies will be needed to clarify these mechanisms.
Our study also reports a novel Zfp148 cKO mouse model. Although our targeting strategy deletes more coding region than the model previously described by Takeuchi et al,11 including the entire DNA binding region, we did not observe the primordial germ cell development defects and infertility described in heterozygous mice in that report. One possible explanation for this discrepancy is that the residual Zfp148 protein containing DNA binding generated in the earlier report may have neomorphic effects. Introduction of nonsense mutations or premature termination codons in the last exon of genes increases the chance of nonsense-mediated mRNA decay escape, generating a C-terminal truncated protein.47,48 In support of this notion, various truncating de novo heterozygous mutations in exon 9 of Zfp148 have been described recently in humans.49 These are associated with pleiotropic developmental abnormalities, suggesting possible dominant-negative or gain-of-function effects of the truncated Zfp148 protein.
The cause of death in Zfp148−/− neonates remains unclear. Pulmonary defects in some neonates from a gene-trap mouse strain have been previously described50 ; however, these were not observed in our Zfp148−/− neonates (supplemental Figure 3). We did find a requirement for Zfp148 in regulating glucose metabolism and growth, consistent with previous reports of Zfp148 controlling the growth hormone gene.51 However, the observed phenotype was transient. Recently described symptoms commonly associated with de novo mutations in exon 9 of Zfp148 in humans include mild to moderate developmental delay, short stature, and feeding problems. Further study will be required to fully understand the mechanisms of perinatal lethality of Zfp148−/− mice. It is of interest to note the strong genetic background effect on the Zfp148 knockout phenotype. This implies that genetic modifiers significantly influence Zfp148 activity.
In summary, this study adds Zfp281 to the network of known transcription factors involved in erythroid differentiation and globin regulation. It also provides evidence for the potential cooperative role of Zfp148/Zfp281 family transcription factors in GATA1-mediated gene control.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE121133).
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank Yuko Fujiwara for assistance in generating Zfp148 gene–targeted mice, Rod Bronson for rodent pathology, and Jonathan Snow and Ronald Mathieu for flow cytometry assistance.
This work is supported by the Royal Perth Hospital Medical Research Foundation, Cancer Council of Western Australia, Department of Health, Government of WA, the Ride to Conquer Cancer, Perth (A.J.W.), and the National Institutes of Health, National Heart, Lung, and Blood Institute grant P01 HL32262-30 (A.B.C.).
Authorship
Contribution: A.J.W., C.-A.A.P., A.G., G.P., D.Y., K.Z., T.P., M.H., J.L., and J.-H.L. performed the experiments and analyzed the data; J.Y.W., M.F., P.J.L., J.W., and E.F. provided key materials and analyzed the data; J.W. edited the manuscript; and A.J.W. and A.B.C. designed the experiments, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alan B. Cantor, Division of Pediatric Hematology/Oncology, Boston Children's Hospital and Dana-Farber Cancer Institute, 300 Longwood Ave, Mailstop 3142, Boston, MA 02115; e-mail: alan.cantor@childrens.harvard.edu; and Andrew J. Woo, Harry Perkins Institute of Medical Research and UWA Centre for Medical Research, Level 7, Room 729, 6 Verdun St, Nedlands, WA 6009, Australia; e-mail: andrew.j.woo@uwa.edu.au.