Key Points
Genome-wide CRISPR/Cas9 screens identify novel mediators of resistance to lenalidomide, pomalidomide, and CC-122 in PEL cells.
SENP8 and UBE2G1 are modulators of CRL4CRBN, and their inactivation drives resistance to CMs in PEL-derived cell lines.
Abstract
Genome-wide CRISPR/Cas9 screens represent a powerful approach to studying mechanisms of drug action and resistance. Cereblon modulating agents (CMs) have recently emerged as candidates for therapeutic intervention in primary effusion lymphoma (PEL), a highly aggressive cancer caused by Kaposi’s sarcoma-associated herpesvirus. CMs bind to cereblon (CRBN), the substrate receptor of the cullin-RING type E3 ubiquitin ligase CRL4CRBN, and thereby trigger the acquisition and proteasomal degradation of neosubstrates. Downstream mechanisms of CM toxicity are incompletely understood, however. To identify novel CM effectors and mechanisms of CM resistance, we performed positive selection CRISPR screens using 3 CMs with increasing toxicity in PEL: lenalidomide (LEN), pomalidomide (POM), and CC-122. Results identified several novel modulators of the activity of CRL4CRBN. The number of genes whose inactivation confers resistance decreases with increasing CM efficacy. Only inactivation of CRBN conferred complete resistance to CC-122. Inactivation of the E2 ubiquitin conjugating enzyme UBE2G1 also conferred robust resistance against LEN and POM. Inactivation of additional genes, including the Nedd8-specific protease SENP8, conferred resistance to only LEN. SENP8 inactivation indirectly increased levels of unneddylated CUL4A/B, which limits CRL4CRBN activity in a dominant negative manner. Accordingly, sensitivity of SENP8-inactivated cells to LEN is restored by overexpression of CRBN. In sum, our screens identify several novel players in CRL4CRBN function and define pathways to CM resistance in PEL. These results provide rationale for increasing CM efficacy on patient relapse from a less-efficient CM. Identified genes could finally be developed as biomarkers to predict CM efficacy in PEL and other cancers.
Introduction
Primary effusion lymphoma (PEL) is a non-Hodgkin B cell lymphoma caused by Kaposi’s Sarcoma-associated Herpesvirus.1,2 PEL most commonly arises in HIV-infected individuals, where it comprises ∼4% of HIV-related non-Hodgkin B cell lymphomas.3 PEL carries a poor prognosis, despite chemotherapy and antiretrovirals.4,5 Recent work by us and others suggests that cereblon modulators (CMs), including the immunomodulatory drugs lenalidomide (LEN) and pomalidomide (POM), may present a promising treatment strategy in PEL.6,7 LEN and POM are used in multiple myeloma (MM), as frontline therapy or on relapse from LEN, respectively. Despite substantial efficacy of immunomodulatory drugs in MM, patients commonly relapse. LEN is also effective in myelodysplastic syndrome with deletion of chromosome 5q [del(5q) MDS]. CC-122 is a fourth-generation CM currently under preclinical investigation in diffuse large B cell-lymphoma (DLBCL).8 The molecular target of CMs is cereblon (encoded by CRBN), a substrate receptor of the cullin-RING type E3 ubiquitin (Ub) ligase CRL4CRBN.9
The covalent conjugation of Ub to target proteins is mediated by an enzymatic cascade that involves an E1 Ub activating enzyme, E2 Ub conjugating enzyme, and E3 Ub ligase, which attaches Ub to lysine (K) residues of protein substrates or Ub itself.10 K48-linked Ub chains of 4 or more moieties canonically target substrates for degradation by the 26S proteasome. The modular cullin-RING E3 Ub ligases (CRLs) are composed of 1 of 7 cullin family scaffold proteins (CUL1, 2, 3, 4A, 4B, 5, or 7), which recruits a RING family protein with E3 Ub ligase function and a substrate recognition module.11,12 The CRL4 complex specifically consists of cullin4A (CUL4A) or cullin4B (CUL4B), the RING protein RBX1, the adaptor protein DDB1 (damaged DNA binding 1), and 1 of several substrate receptors, including CRBN (Figure 1A).9,13,14 CRLs are regulated by dynamic modification of the cullin subunit with the Ub-like modifier Nedd8.11,12,15,16 Similar to Ub, Nedd8 requires the sequential action of Nedd8-specific E1, E2, and E3 enzymes. Nedd8-modification of cullins is required for E3 ligase activity by exposing RBX1 and thereby positioning the associated E2 enzyme in proximity of substrates.16 Conversely, deneddylation of cullins is required for the exchange of the substrate receptor module.17,18 Cullin deneddylation is mediated by the multisubunit COP9 signalosome (CSN).19,20 Thus, cullin Nedd8-modification is highly dynamic, with neddylation required for E3 ligase activity and deneddylation required for substrate receptor exchange.
CRISPR-Cas9 inactivation-based CM resistance screens in the PEL cell line BC-3. (A) Schematic of the CM-bound CRL4CRBN E3 Ub ligase complex. (B) Experimental outline of CM resistance screens. (C) Cumulative live cell counts of BC-3/Cas9 during the duration of the screens. Cell numbers were normalized to those from dimethyl sulfoxide (DMSO)–treated control cells. Arrows indicate early and late points at which a subset of cells were taken from the pool for analysis of sgRNA distribution. (D) Absolute live cell counts of LEN, POM, or CC-122–treated BC-3/Cas9 cells at late times in the screen shows emergence of proliferating cell pools under continued drug treatment.
CRISPR-Cas9 inactivation-based CM resistance screens in the PEL cell line BC-3. (A) Schematic of the CM-bound CRL4CRBN E3 Ub ligase complex. (B) Experimental outline of CM resistance screens. (C) Cumulative live cell counts of BC-3/Cas9 during the duration of the screens. Cell numbers were normalized to those from dimethyl sulfoxide (DMSO)–treated control cells. Arrows indicate early and late points at which a subset of cells were taken from the pool for analysis of sgRNA distribution. (D) Absolute live cell counts of LEN, POM, or CC-122–treated BC-3/Cas9 cells at late times in the screen shows emergence of proliferating cell pools under continued drug treatment.
CMs bind to CRBN within the substrate recognition surface, and thereby enable acquisition of neosubstrates, resulting in their polyubiquitination and proteasomal degradation.21-26 In MM, CM-induced degradation of neosubstrates IKZF1 and IKZF3 and the subsequent downregulation of their transcriptional target IRF4 is thought be a major trigger of CM toxicity.21,22 Degradation of the neosubstrate CK1α, which is preferentially targeted by LEN, is thought to explain the efficacy of LEN in del(5q) MDS.27 Recently, CMs have been shown to induce efficient cell death in PEL cell lines,6,7 and LEN is currently part of a clinical trial in PEL (clinicaltrials.gov identifier: NCT02911142). As in MM,28 the transcription factor IRF4 is a highly essential survival gene in PEL cells, and its expression is reduced on treatment with CMs.6,7,29 Surprisingly, however, in this setting, downregulation of IRF4 and CM toxicity were independent of IKZF1 and/or IKZF3.7 In PEL cell lines, CK1α is furthermore commonly essential, and its degradation represents an IRF4-independent arm of CM toxicity.7 In contrast to the reported lack of CK1α degradation in POM-treated del(5q) MDS cells,27 we consistently observe degradation of CK1α on treatment of PEL cells with POM, at only slightly lower efficiency than with LEN.7 Degradation of CK1α in POM-treated cells is in agreement with recently reported in vitro data.25 The individual or combined reexpression of CK1α and IRF4 significantly, but incompletely, rescued PEL cell lines from CM-induced cell death,7 suggesting that additional mechanisms of CM action exist in PEL. Thus, further mechanistic investigation of CM toxicity in this and other cell types is warranted.
Genome-wide CRISPR screens represent a powerful tool to probe for genes whose inactivation confers drug resistance.30,31 To identify CM effectors, we applied this unbiased screening approach to identify genes whose inactivation confers resistance to LEN, POM, or CC-122 in PEL cells. Our results suggest that decreased CRL4CRBN activity is the dominant pathway to resistance and identify several novel regulators of CRL4CRBN. We validate the E2 enzyme UBE2G1 and the Nedd8-specific protease SENP8 as genes whose inactivation can confer resistance to LEN and/or POM. Inactivation of SENP8 indirectly increased the levels of unneddylated CUL4A/B, which interferes with LEN toxicity in a dominant-negative manner.
Methods
Cell lines, reagents, and CRISPR screens
Cell lines were maintained and CRISPR knockout cell pools were generated as described recently and further detailed in the supplemental Methods.29 Lenalidomide was from Cayman Chemicals (Ann Arbor, MI), and pomalidomide and CC-122 were from Selleck Chemicals (Houston, TX). All compounds were dissolved in DMSO (Sigma-Aldrich, St. Louis, MO). The Brunello sgRNA library was obtained from Addgene (Cambridge, MA).32 CRISPR screens were performed in clonal BC-3/Cas9 cells, as described and detailed in the supplemental Methods.29 Raw sgRNA read counts are in supplemental Table 1. sgRNA read counts after exclusion of those in the bottom 5% are in supplemental Table 2. Raw data are available under GEO accession number GSE122040.
Cloning
Lentiviral sgRNA vectors were based on pLenti-guide puro30,33 (Addgene #52963; see supplemental Table 5 for sgRNA sequences and primers). For lentiviral protein expression, cDNA sequences were placed into pLC/P2Ahygro (CRBN, UBE2G1, SENP8) or pLC/P2Apuro (CUL4A WT/K705R). Detailed cloning procedures are described in the supplemental Methods. Primer sequences are listed in supplemental Table 6, and cDNA nucleotide sequences are listed in supplemental Table 7. Vectors are available through Addgene.
Cell counting, western blotting, and CM dose-response assays
Growth curve and CM dose-response and growth curve assays were performed essentially as described,7 with minor modifications (supplemental Methods). Quantitative western blot analysis (LI-COR) was performed as reported,7,29 and primary antibodies are listed in supplemental Table 8. The CK1α antibody detects a nonspecific band of a molecular weight that was only sometimes present on the imaged piece of membrane, depending on how the membrane was cut for various Western analyses.
Results
Genome-wide CRISPR/Cas9-based positive selection screens against CMs in PEL
We conducted genome-wide CRISPR/Cas9 resistance screens against LEN, POM, and CC-122 in the commonly used PEL cell line BC-3 (Figure 1B) to identify effectors of CMs in an unbiased approach. Half maximal inhibitory concentrations (IC50) of these drugs in BC-3 are ∼2 µM for LEN, 213 nM for POM, and 117 nM for CC-122 (supplemental Figure 1). Cas9-expressing BC-3 cells were transduced with the Brunello genome-wide sgRNA library and cultured for 2 weeks after antibiotic selection to allow for depletion of cells with sgRNAs targeting essential genes.29 The resulting cell pool was divided and treated with DMSO or lethal concentrations of CMs (ie, 5 µM LEN, 1 µM POM, or 1 µM CC-122). High concentrations were chosen to assay for robust CM resistance under stringent conditions. A first sample was collected when a minimum of live cells remained: on day 14 for LEN, or day 9 for POM or CC-122 (Figure 1C). A second sample was harvested after a proliferating cell pool emerged under drug treatment (Figure 1D). The later point was intended to assay for complete resistance, whereas the early point was included to detect more subtle delays in CM toxicity. sgRNA composition over 2 replicate screens per drug was assessed by next-generation sequencing and analyzed with the MAGeCK-VISPR algorithm.34,35
For each drug, sgRNAs targeting fewer genes were significantly enriched in the live cell population at late points vs at early points (Figure 2A-F). The number of hits furthermore decreased with increasing toxicity of the CM, such that only inactivation of CRBN appeared to confer complete resistance to CC-122 (Figure 2F). An enrichment for guides against CRBN was expected, because its inactivation has previously been shown to confer CM resistance in PEL6,7 and MM cell lines.21,22 Other genes with strong sgRNA enrichments in at least a subset of settings include: the CRL4 cullin subunit CUL4B; GLMN, a previously identified binding partner of RBX136 ; and the E2 Ub conjugating enzyme UBE2G1, which has very recently been implicated as the preferred E2 enzyme for Ub chain elongation by CRL4.37,38 sgRNAs targeting 4 other genes (ILF2, ILF3, YPEL5, SENP8) were highly and specifically enriched in LEN-resistant cells (supplemental Table 4; Figure 2G), and therefore represent novel candidates for modulators of LEN toxicity. Of these, ILF2 and ILF3 have previously been identified as candidate interacting partners of several CRL complexes,39 but have not been functionally investigated as potential regulators of CRL activity. YPEL5 has been identified as a candidate binding partner of the COP9 signalosome subunit COPS5,40 but also participates in an unrelated E3 Ub ligase complex,41 among other roles. SENP8 (also known as DEN1 or NEDP1) encodes a Nedd8-specific cysteine protease that has recently been shown to remove Nedd8 modifications from proteins other than cullins, and specifically the Nedd8 conjugation machinery, including Nedd8-specific E1 and E2 enzymes.42 Although SENP8 inactivation has recently been shown to cause decreased neddylation of CUL1 and CUL5,42 SENP8 has not previously been implicated in the regulation of CRL4 activity or CM toxicity.
CRISPR screens identify candidates for genes whose inactivation confers CM resistance. (A-F) Gene level analysis of changes in sgRNA distribution in cell pools harvested at early times (day 13 for LEN or day 9 for POM and CC-122) (A,C,E) or late times (day 21) (B,D,F). The most relevant genes are highlighted and labeled using identical colors in all panels. (G) Venn diagram depicting overlap of most confident candidates for genes whose inactivation confers outright resistance against LEN, POM, or CC-122. Cutoffs used were FDR adjusted (adj). P < .05 and a median sgRNA fold change >2 on day 21.
CRISPR screens identify candidates for genes whose inactivation confers CM resistance. (A-F) Gene level analysis of changes in sgRNA distribution in cell pools harvested at early times (day 13 for LEN or day 9 for POM and CC-122) (A,C,E) or late times (day 21) (B,D,F). The most relevant genes are highlighted and labeled using identical colors in all panels. (G) Venn diagram depicting overlap of most confident candidates for genes whose inactivation confers outright resistance against LEN, POM, or CC-122. Cutoffs used were FDR adjusted (adj). P < .05 and a median sgRNA fold change >2 on day 21.
Both the experimental settings for our screens and cutoffs for data analyses presented in Figure 2 were selected for high stringency. Lowering statistical cutoffs identifies additional genes as candidates for modulators of CM toxicity, among known components or regulators of CRL4, such as RBX1 and CSN subunits (supplemental Tables 3 and 4). We note that DDB1, RBX1, and several CSN subunits are potentially essential in PEL based on our reported essentiality screens29 (supplemental Figure 2), which would prevent enrichments of sgRNAs targeting these genes in the CM-resistant cell population. On the basis of the prominent enrichments for CUL4B, UBE2G1, ILF2/3, YPEL5, and SENP8-specific sgRNAs, these genes were selected for further validation.
Inactivation of CUL4A or CUL4B confers resistance to lenalidomide and delays toxicity of pomalidomide
Our detection of enrichments for CUL4B sgRNAs is interesting, because it suggests that overall CUL4A and CUL4B expression levels in BC-3 cells is limiting for the efficacy of even high concentrations of CMs. To confirm this result, we transduced Cas9-expressing BC-3 cells with lentiviruses carrying 2 independent sgRNAs, each targeting CUL4B or CUL4A, or an established negative control guide targeting the noncoding locus AAVS1. A previously reported CRBN knockout BC-3 clone served as a positive control for CM resistance.7 Indeed, individual CRISPR/Cas9-mediated inactivation of CUL4B substantially delayed the toxicity of POM and conferred robust resistance to LEN (supplemental Figure 3A-C). Inactivation of CUL4A similarly conferred significant resistance to LEN, although CUL4A-specific sgRNAs were not enriched in our screens (supplemental Figure 3D-E). Inactivation of CUL4A had a smaller effect on the response to POM in BC-3 cells (supplemental Figure 3F). Overall, these data confirm that CUL4B and CUL4A are both expressed in PEL and contribute to CRL4CRBN-dependent CM toxicity in BC-3. The failure for CUL4A sgRNAs to meet our statistical cutoff (Figure 2) may reflect a relative inefficiency of these guides in the pooled approach and/or, possibly, the high stringency of the experimental setup and cutoffs. Similar results were obtained in the PEL cell lines BCBL-1 and BC-1, which are EBV-negative and EBV-positive, respectively (supplemental Figure 3G-R). Together, these data suggest that low CUL4A/B expression levels may indeed limit CM toxicity in PEL cell lines.
Inactivation of UBE2G1 confers resistance to lenalidomide and pomalidomide
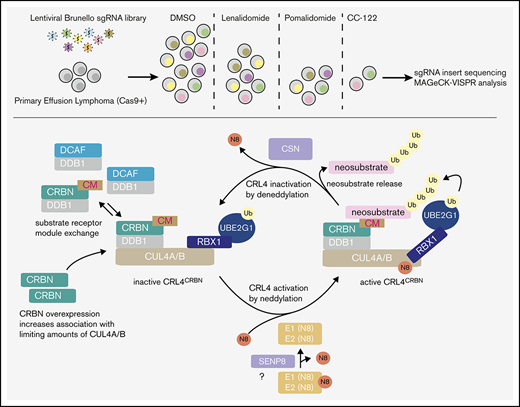
Remarkably, UBE2G1 was the only E2 Ub conjugating enzyme out of 40 human E2 enzymes with enriched sgRNAs at early (LEN, POM, CC-122) and late (LEN, POM) times (Figure 2; supplemental Figure 4). To test the hypothesis that inactivation of UBE2G1 confers CM resistance, we infected Cas9-expressing BC-3 cells with lentiviruses carrying 2 independent sgRNAs targeting UBE2G1 or the sgAAVS1-negative control guide. Inactivation of UBE2G1 was confirmed by western blot analysis (Figure 3A). Inactivation of UBE2G1 indeed conferred outright resistance against either LEN or POM (Figure 3B-C). Although live cell numbers of UBE2G1 knockout cells under CM treatment were lower than those of CBRN-KO cells, these cells continued to proliferate under drug treatment. Reexpression of an sgRNA-resistant UBE2G1 cDNA in the context of UBE2G1 inactivation restored sensitivity to CMs, confirming the specificity of sgUBE2G1-mediated CM resistance (Figure 3D-E). Western blot analyses of UBE2G1-inactivated cells demonstrate substantial impairment of CM-induced neosubstrate degradation in LEN or POM-treated UBE2G1-inactivated cell pools for IKZF1, IKZF3, and CK1α (Figure 3F-G), which were analyzed as indicators of overall CRL4CRBN activity. The LEN/POM-induced indirect downregulation of IRF4 was similarly rescued in UBE2G1-inactivated cells compared with the negative control cells. The rescue of LEN- and POM-induced phenotypes strongly suggest that UBE2G1 acts at the level of the CM-bound CRL4CRBN complex, most likely as the preferred E2 Ub conjugation enzyme participating in the polyubiquitination of CM neosubstrates in PEL. Validation experiments furthermore confirmed that inactivation of UBE2G1 modestly delays toxicity of the more potent CC-122 but does not confer resistance against this drug (Figure 3H-I). Thus, other E2 enzymes must be able to compensate for loss of UBE2G1, albeit at lower efficiency. These results parallel 2 recent reports,37,43 which similarly identified UBE2G1 as hits in genome-wide CRISPR/Cas9 screens for LEN or POM resistance in MM. UBE2G1 was also identified as the preferred E2 in CM-induced CRL4CRBN activity in an E2 enzyme-focused CRISPR screen in MM.38 Sievers et al37 and Lu et al38 confirmed a role for UBE2G1 in CM-induced toxicity in MM and proposed a “prime-extend” mechanism, whereby the E2 enzyme UBE2D3 mediates substrate mono-ubiquitination, whereas UBE2G1 extends the Ub chain via addition of further K48-linked Ub moieties. Inactivation of UBE2D3 alone, which did not score in our screens, did not confer significant CM resistance in PEL, suggesting the existence of redundant E2 enzymes that can fulfill this function in our system (supplemental Figure 5). Overall, our data suggest a preference for CRL4CRBN for UBE2G1, similar to that seen in MM.37,38,43
Inactivation of UBE2G1 confers resistance to LEN and POM, but not CC-122. (A) Representative western blot analysis confirms efficient CRISPR-induced inactivation of UBE2G1 in BC-3/Cas9 cells by 2 independent UBE2G1-specific sgRNAs (sg1, sg2). sgAAVS1 is a negative control guide. GAPDH served as loading control. (B-C) Growth curve analyses of UBE2G1 inactivated BC-3/Cas9 cells after treatment with 5 µM LEN (B) or 1 µM POM (C). BC-3/Cas9 cells transduced with sgAAVS1 were included as a CM-sensitive negative control, whereas previously described clonal CRBN-inactivated BC-3 cells (CRBN KO) served as a positive control for complete CM resistance. Absolute live cell numbers were normalized to corresponding DMSO vehicle-treated cells at each passage (represented by the dotted line). These experiments were performed in parallel with SENP8-inactivated cells shown in Figure 4B-C, and thus share common negative and positive controls for several or all replicates. (n = 3; error bars represent standard error of the mean [SEM]). (D) Representative western blot analysis (anti-Flag) confirms lentiviral expression of sg2-resistant, Flag-tagged UBE2G1 in UBE2G1-inactivated BC-3/Cas9 cells (UBE2G1 sgRNA sg2). GAPDH served as loading control. (E) Growth curve analysis of UBE2G1-inactivated BC-3/Cas9 cells (UBE2G1 sgRNA sg2) expressing Flag tagged UBE2G1 (sg2-resistant; UBE2G1/sg2R) or control ZsGreen after treatment with 5 µM LEN confirms restoration of CM sensitivity on UBE2G1 reexpression. BC3/Cas9/sg AAVS1 was included as a LEN-sensitive positive control. Live cell counts were normalized to corresponding DMSO vehicle-treated cells, which are represented by the dotted line. These assays were run in parallel with those in Figure 4E, and thus share common controls (n = 3; error bars represent SEM). (F-G) Representative time course western blot analyses of CM neosubstrates IKZF1, IKZF3, and CK1α, and IRF4 in WT or UBE2G1-inactivated BC-3/Cas9 cells upon treatment with 5 µM LEN (F) or 1 µM POM (G). Lysates are matched with growth curves shown in panels B and C, respectively. GAPDH served as loading control. The asterisk marks a nonspecific band in panel F. (H) Growth curve analyses of UBE2G1-inactivated BC-3/Cas9 cells treated with 1 µM CC-122. sgAAVS1 and CRBN KO cells were included as negative and positive controls, respectively. Live cell counts were normalized to corresponding DMSO vehicle-treated cells, represented by the dotted line (n = 3; error bars represent SEM). (I) Representative western blot analysis of IKZF1, IKZF3, CK1α, and IRF4 at indicated points confirms a delay in neosubstrate degradation and IRF4 downregulation in UBE2G1-inactivated BC-3/Cas9 cells compared with control pools as in panel H. GAPDH served as loading control. Statistical analyses throughout this figure were performed by unpaired Student t tests comparing specified conditions to corresponding sgAAVS1 or ZsGreen controls. *P <. 05; **P < .01; ***P < .001; ****P < .0001. n.s., not significant.
Inactivation of UBE2G1 confers resistance to LEN and POM, but not CC-122. (A) Representative western blot analysis confirms efficient CRISPR-induced inactivation of UBE2G1 in BC-3/Cas9 cells by 2 independent UBE2G1-specific sgRNAs (sg1, sg2). sgAAVS1 is a negative control guide. GAPDH served as loading control. (B-C) Growth curve analyses of UBE2G1 inactivated BC-3/Cas9 cells after treatment with 5 µM LEN (B) or 1 µM POM (C). BC-3/Cas9 cells transduced with sgAAVS1 were included as a CM-sensitive negative control, whereas previously described clonal CRBN-inactivated BC-3 cells (CRBN KO) served as a positive control for complete CM resistance. Absolute live cell numbers were normalized to corresponding DMSO vehicle-treated cells at each passage (represented by the dotted line). These experiments were performed in parallel with SENP8-inactivated cells shown in Figure 4B-C, and thus share common negative and positive controls for several or all replicates. (n = 3; error bars represent standard error of the mean [SEM]). (D) Representative western blot analysis (anti-Flag) confirms lentiviral expression of sg2-resistant, Flag-tagged UBE2G1 in UBE2G1-inactivated BC-3/Cas9 cells (UBE2G1 sgRNA sg2). GAPDH served as loading control. (E) Growth curve analysis of UBE2G1-inactivated BC-3/Cas9 cells (UBE2G1 sgRNA sg2) expressing Flag tagged UBE2G1 (sg2-resistant; UBE2G1/sg2R) or control ZsGreen after treatment with 5 µM LEN confirms restoration of CM sensitivity on UBE2G1 reexpression. BC3/Cas9/sg AAVS1 was included as a LEN-sensitive positive control. Live cell counts were normalized to corresponding DMSO vehicle-treated cells, which are represented by the dotted line. These assays were run in parallel with those in Figure 4E, and thus share common controls (n = 3; error bars represent SEM). (F-G) Representative time course western blot analyses of CM neosubstrates IKZF1, IKZF3, and CK1α, and IRF4 in WT or UBE2G1-inactivated BC-3/Cas9 cells upon treatment with 5 µM LEN (F) or 1 µM POM (G). Lysates are matched with growth curves shown in panels B and C, respectively. GAPDH served as loading control. The asterisk marks a nonspecific band in panel F. (H) Growth curve analyses of UBE2G1-inactivated BC-3/Cas9 cells treated with 1 µM CC-122. sgAAVS1 and CRBN KO cells were included as negative and positive controls, respectively. Live cell counts were normalized to corresponding DMSO vehicle-treated cells, represented by the dotted line (n = 3; error bars represent SEM). (I) Representative western blot analysis of IKZF1, IKZF3, CK1α, and IRF4 at indicated points confirms a delay in neosubstrate degradation and IRF4 downregulation in UBE2G1-inactivated BC-3/Cas9 cells compared with control pools as in panel H. GAPDH served as loading control. Statistical analyses throughout this figure were performed by unpaired Student t tests comparing specified conditions to corresponding sgAAVS1 or ZsGreen controls. *P <. 05; **P < .01; ***P < .001; ****P < .0001. n.s., not significant.
Inactivation of SENP8 specifically confers resistance to lenalidomide
SENP8, ILF2, ILF3, and YPEL5 scored specifically for LEN, but not POM or CC-122. To test effects of their inactivation on CM toxicity, we initially targeted each gene in BC-3 cells using 2 sgRNAs. Consequences of ILF2 inactivation could not be tested because of inefficient inactivation (supplemental Figure 6A). Western blot analyses confirmed robust target inactivation for ILF3, YPEL5, and SENP8 (supplemental Figure 6B,D; Figure 4A). Inactivation of ILF3 and YPEL5 only modestly delayed LEN toxicity in BC-3 cells (supplemental Figure 6C,E). For YPEL5, similarly modest effects were also seen in BCBL-1 cells (supplemental Figure 6D,F). Although these results suggest that ILF3 and YPEL5 may indeed modulate CM toxicity, their effect was minor, and was therefore not studied further. In contrast, inactivation of SENP8 in BC-3 conferred pronounced resistance to LEN (Figure 4B). Because SENP8 has not been implicated in CRL4 function or CM resistance to date, we decided to further investigate the role of SENP8. As predicted by the screens, inactivation of SENP8 delayed and attenuated toxicity of POM in BC-3 cells, but did not confer resistance at later points into drug treatment (Figure 4C). Observed effects were specific to SENP8 inactivation, as reexpression of sgRNA-resistant SENP8 restored LEN sensitivity (supplemental Figure 7A-B). Western blot analyses of SENP8-inactivated BC-3 cells demonstrate rescue of IRF4 and the neosubstrates IKZF1, IKZF3, and CK1α on treatment with LEN, but not with POM (Figure 4D-E). Similar results were obtained in the PEL cell line BCBL-1 (supplemental Figure 7C-E). SENP8 and the positive control CRBN were additionally targeted for inactivation in the PEL cell line BC-1 and in the MM cell lines KMS12-BM and MM.1S (supplemental Figure 7F-I). The inclusion of MM cell lines is of interest, as SENP8 did not score in reported CRISPR CM resistance screens in MM.37,43 Dose-response analyses for LEN and POM in each cell line confirmed that inactivation of SENP8 conferred significant resistance to LEN in each PEL cell line and in MM.1S (Figure 4F). Inactivation of SENP8 did not confer resistance to LEN in KMS12-BM. None of the cell lines exhibited strongly altered dose response to POM (supplemental Figure 7J). The range of concentrations tested includes physiologically achievable drug concentrations.44,45 Overall, these results show that inactivation of SENP8 confers significant resistance to LEN in PEL cell lines and at least the MM cell line MM.1S, but does not strongly affect the response to POM.
Inactivation of SENP8 confers resistance to LEN, but not POM. (A) Representative western blot analysis confirms efficient CRISPR-induced inactivation of SENP8 in BC-3/Cas9 cells by 2 independent SENP8-specific sgRNAs (sg1, sg2). sgAAVS1 served as negative control guide, GAPDH served as loading control. (B-C) Growth curve analyses of SENP8-inactivated BC-3/Cas9 cells after treatment with 5 µM LEN (B) or 1 µM POM (C). BC-3/Cas9 cells transduced with sgAAVS1 were included as a CM-sensitive negative control, whereas previously described clonal CRBN-inactivated BC-3 cells (CRBN KO) served as a positive control for complete CM resistance. Absolute live cell numbers were normalized to corresponding DMSO vehicle-treated cells at each passage (represented by the dotted line). These experiments were performed in parallel with UBE2G1-inactivated cells shown in Figure 3B-C, and thus share common negative and positive controls for several or all replicates (n = 3; error bars represent SEM). (D-E) Representative western blot analyses of IKZF1, IKZF3, CK1α, and IRF4 at indicated points confirm incomplete neosubstrate degradation and IRF4 downregulation in SENP8-inactivated BC-3/Cas9 cells compared with sgAAVS1 transduced BC-3/Cas9 control cells upon treatment with 5 µM LEN (D) or 1 µM POM (E). Lysates are matched with growth curves shown in panels B and C, respectively. GAPDH served as loading control. The asterisk marks a nonspecific band. (F) Dose-response analysis of LEN toxicity in Cas9-expressing PEL cell lines BC-3, BCBL-1, BC-1, and MM cell lines KMS12-BM and MM.1S on inactivation of SENP8. sgAAVS1- or sgCRBN-transduced cell lines served as negative or positive controls for drug resistance, respectively. Readout of the assay was on day 7 (BC-3, BCBL-1, and MM.1S) or day 9 (BC-1, KMS12-BM) into CM treatment. Statistical analyses in panels B and C were performed by unpaired Student t tests comparing specified conditions to corresponding sgAAVS1 controls. *P < .05; **P < .01; ***P < .001; ****P < .0001.
Inactivation of SENP8 confers resistance to LEN, but not POM. (A) Representative western blot analysis confirms efficient CRISPR-induced inactivation of SENP8 in BC-3/Cas9 cells by 2 independent SENP8-specific sgRNAs (sg1, sg2). sgAAVS1 served as negative control guide, GAPDH served as loading control. (B-C) Growth curve analyses of SENP8-inactivated BC-3/Cas9 cells after treatment with 5 µM LEN (B) or 1 µM POM (C). BC-3/Cas9 cells transduced with sgAAVS1 were included as a CM-sensitive negative control, whereas previously described clonal CRBN-inactivated BC-3 cells (CRBN KO) served as a positive control for complete CM resistance. Absolute live cell numbers were normalized to corresponding DMSO vehicle-treated cells at each passage (represented by the dotted line). These experiments were performed in parallel with UBE2G1-inactivated cells shown in Figure 3B-C, and thus share common negative and positive controls for several or all replicates (n = 3; error bars represent SEM). (D-E) Representative western blot analyses of IKZF1, IKZF3, CK1α, and IRF4 at indicated points confirm incomplete neosubstrate degradation and IRF4 downregulation in SENP8-inactivated BC-3/Cas9 cells compared with sgAAVS1 transduced BC-3/Cas9 control cells upon treatment with 5 µM LEN (D) or 1 µM POM (E). Lysates are matched with growth curves shown in panels B and C, respectively. GAPDH served as loading control. The asterisk marks a nonspecific band. (F) Dose-response analysis of LEN toxicity in Cas9-expressing PEL cell lines BC-3, BCBL-1, BC-1, and MM cell lines KMS12-BM and MM.1S on inactivation of SENP8. sgAAVS1- or sgCRBN-transduced cell lines served as negative or positive controls for drug resistance, respectively. Readout of the assay was on day 7 (BC-3, BCBL-1, and MM.1S) or day 9 (BC-1, KMS12-BM) into CM treatment. Statistical analyses in panels B and C were performed by unpaired Student t tests comparing specified conditions to corresponding sgAAVS1 controls. *P < .05; **P < .01; ***P < .001; ****P < .0001.
SENP8 inactivation results in accumulation of unneddylated CUL4A/B
A recent report showed that SENP8 cleaves Nedd8 from noncullin neddylation substrates, including the Nedd8-specific E2 enzyme UBE2M and other components of the neddylation machinery.42 Accumulation of aberrant neddylated forms of these proteins appears to render neddylation of cullins in SENP8 inactivated cells inefficient, as shown for CUL1 and CUL5 in HeLa and HEK293T cells.42 The same study reported no effect on the neddylation status of CUL4, however. To begin to address the role of SENP8 in PEL, we probed wild-type (WT) and SENP8-inactivated BC-3 cells for Nedd8 (Figure 5A). As reported for 293T,42 SENP8-inactivated BC-3 showed a depletion of unconjugated Nedd8 and an accumulation of several Nedd8-modified proteins, presumably substrates for Nedd8 deconjugation by SENP8. In both BC-3 and BCBL-1 cells, there was an increase in the levels of unneddylated CUL4A and CUL4B (Figure 5B-C). Importantly, the expression of CRBN was unchanged in SENP8-inactivated cell pools (Figure 5B). A similar increase in unneddylated CUL4A and CUL4B was observed in SENP8-inactivated MM.1S cells, where SENP8 inactivation conferred LEN resistance, but not in SENP8-inactivated KMS12-BM cells, where SENP8 inactivation was inconsequential (supplemental Figure 8A). We note that KMS12-BM expresses moderately higher levels of CUL4A than the other cell lines (supplemental Figure 8B). Overall, our data suggest that the effect of SENP8 on CM efficacy is most likely at the level of reduced CUL4A/B neddylation. CUL4A/B neddylation and LEN sensitivity in the context of SENP8 inactivation could not be restored by overexpression of either active or precursor Nedd8 (supplemental Figure 8C-E). These data suggest that the accumulation of unneddylated CUL4A/B in SENP8 inactivated cells is not a consequence of limiting pools of unconjugated Nedd8.
SENP8 inactivation leads to an increase of unneddylated CUL4A/B and depletion of free Nedd8. (A) Representative western blot analysis of SENP8-inactivated and control sgAAVS1-transduced BC-3/Cas9 cells indicates a strong depletion of unconjugated Nedd8 and an increase in Nedd8-conjugated proteins. GAPDH served as loading control. SENP8 sg1 and sg2 are 2 independent SENP8-specific sgRNAs used. (B) Representative western blot analyses of CUL4A, CUL4B, and CRBN in SENP8-inactivated BC-3/Cas9 and BCBL-1/Cas9 cells indicate an increase in unneddylated CUL4A/B and no significant change in CRBN protein levels. Neddylated (N8) and unneddylated forms of CUL4A/B are indicated by arrows. GAPDH served as loading control. (C) Quantitative analysis of the data shown in panel B over replicates. Protein levels were first normalized to corresponding GAPDH levels and then to sg AAVS1 levels (n = 2; error bars represent SEM).
SENP8 inactivation leads to an increase of unneddylated CUL4A/B and depletion of free Nedd8. (A) Representative western blot analysis of SENP8-inactivated and control sgAAVS1-transduced BC-3/Cas9 cells indicates a strong depletion of unconjugated Nedd8 and an increase in Nedd8-conjugated proteins. GAPDH served as loading control. SENP8 sg1 and sg2 are 2 independent SENP8-specific sgRNAs used. (B) Representative western blot analyses of CUL4A, CUL4B, and CRBN in SENP8-inactivated BC-3/Cas9 and BCBL-1/Cas9 cells indicate an increase in unneddylated CUL4A/B and no significant change in CRBN protein levels. Neddylated (N8) and unneddylated forms of CUL4A/B are indicated by arrows. GAPDH served as loading control. (C) Quantitative analysis of the data shown in panel B over replicates. Protein levels were first normalized to corresponding GAPDH levels and then to sg AAVS1 levels (n = 2; error bars represent SEM).
CRBN overexpression restores sensitivity to lenalidomide in the context of SENP8 inactivation
Because expression of Nedd8-modified CUL4A/B was not strongly reduced in the context of SENP8 inactivation, we reasoned that unneddylated CUL4A/B might dominant negatively interfere with CM toxicity. Interference could be through sequestration of CRBN on unneddylated complexes. Alternatively, increased substrate receptor exchange facilitated by CUL4 deneddylation could cause competition with alternative substrate adaptors. In either case, overexpression of CRBN would be expected to drive increased association of CRBN/DDB1 with active CRL4 complexes under conditions in which activation of CUL4 by neddylation is limiting because of the loss of SENP8. Indeed, lentiviral overexpression of Flag-tagged CRBN restored full sensitivity of BC-3 cells to LEN in the context of SENP8 inactivation (Figure 6A). Western blot analysis confirmed restoration of LEN-induced neosubstrate degradation and IRF4 downregulation in CRBN overexpressing cells (Figure 6B). We finally directly tested whether overexpression of a nonneddylatable CUL4A K705R mutant dominant negatively interferes with CM toxicity (Figure 6C). Although overexpression of WT CUL4A did not affect the toxicity of LEN or POM, K705R-mutant CUL4A expressing BC-3 were completely resistant to high concentrations of either LEN or POM, which formally shows that unneddylated CUL4 can drive resistance to CMs in a dominant-negative manner (Figure 6D-E; supplemental Figure 9).
Overexpression of CRBN restores LEN sensitivity in SENP8-inactivated cells. (A) Growth curve analyses of SENP8-inactivated BC-3/Cas9 cells (using SENP8-specific sg2) lentivirally transduced to overexpress Flag-tagged CRBN or ZsGreen and treated with 5 µM LEN. sgAAVS1-transduced cells serve as a control for LEN-sensitive BC-3 and CRBN-inactivated BC-3 cells (CRBN KO) were used as a control for LEN-resistant cells. Live cell counts were normalized to DMSO vehicle-treated cells, represented by the dotted line (n = 3; error bars represent SEM). (B) Representative western blot analysis of IKZF1, IKZF3, CK1α, IRF4, CRBN, and SENP8 at indicated points into the experiment shown in panel A. GAPDH served as loading control. Asterisks mark nonspecific bands. (C) Representative western blot analysis of BC-3 cells transduced with lentiviral vectors constitutively expressing WT CUL4A, K705R mutant CUL4A, or ZsGreen. Membranes were probed for CUL4A, CRBN, IRF4, or the loading control GAPDH. Neddylated (N8) and unneddylated forms of CUL4A are indicated by arrows. (D-E) Growth curve analyses of BC-3 cells from panel C and treated with 5 µM LEN (D) or 1 µM POM (E). CRBN-inactivated BC-3 cells (CRBN KO) were used as a control for LEN-resistant cells. Live cell counts were normalized to DMSO-treated control cells, as represented by the dotted line (n = 3; error bars represent SEM). For western blot analysis of this experiment, see supplemental Figure 8. Statistical analyses throughout this figure were performed by unpaired Student t tests comparing specified conditions to corresponding ZsGreen controls. *P < .05; **P < .01; ***P < .001; ****P < .0001.
Overexpression of CRBN restores LEN sensitivity in SENP8-inactivated cells. (A) Growth curve analyses of SENP8-inactivated BC-3/Cas9 cells (using SENP8-specific sg2) lentivirally transduced to overexpress Flag-tagged CRBN or ZsGreen and treated with 5 µM LEN. sgAAVS1-transduced cells serve as a control for LEN-sensitive BC-3 and CRBN-inactivated BC-3 cells (CRBN KO) were used as a control for LEN-resistant cells. Live cell counts were normalized to DMSO vehicle-treated cells, represented by the dotted line (n = 3; error bars represent SEM). (B) Representative western blot analysis of IKZF1, IKZF3, CK1α, IRF4, CRBN, and SENP8 at indicated points into the experiment shown in panel A. GAPDH served as loading control. Asterisks mark nonspecific bands. (C) Representative western blot analysis of BC-3 cells transduced with lentiviral vectors constitutively expressing WT CUL4A, K705R mutant CUL4A, or ZsGreen. Membranes were probed for CUL4A, CRBN, IRF4, or the loading control GAPDH. Neddylated (N8) and unneddylated forms of CUL4A are indicated by arrows. (D-E) Growth curve analyses of BC-3 cells from panel C and treated with 5 µM LEN (D) or 1 µM POM (E). CRBN-inactivated BC-3 cells (CRBN KO) were used as a control for LEN-resistant cells. Live cell counts were normalized to DMSO-treated control cells, as represented by the dotted line (n = 3; error bars represent SEM). For western blot analysis of this experiment, see supplemental Figure 8. Statistical analyses throughout this figure were performed by unpaired Student t tests comparing specified conditions to corresponding ZsGreen controls. *P < .05; **P < .01; ***P < .001; ****P < .0001.
Discussion
Here we report genome-wide CRISPR KO screens for genes whose inactivation confers resistance to 3 generations of CMs (ie, LEN, POM, and CC-122) in the PEL cell line BC-3. Our study confirms that CRBN is strictly required for CM toxicity. In addition, screens and validation experiments show that the E2 Ub conjugating enzyme UBE2G1 and the deneddylase SENP8 are required for optimal CM toxicity in PEL. While our work was in progress, similar screens against LEN37 or POM43 were reported in MM. These screens and an additional E2-targeted CRISPR screen38 similarly identified loss of UBE2G1 as a mechanism of LEN and POM resistance in MM. Our screens additionally identified SENP8 and other novel candidates as modulators of CM toxicity in PEL. Our validation experiments show that SENP8 expression is required for toxicity of LEN in at least a subset of MM cell lines. It is unclear why SENP8 did not score in the reported CRISPR-based LEN resistance screen in MM.1S cells.37
Our results suggest that genes that confer robust CM resistance are mostly, or possibly exclusively, modulators of CRL4CBRN activity. This can be explained by our previous finding that mechanisms of CM action in PEL are multifactorial.7 Interestingly, the number of genes whose inactivation confers resistance decreases with increased efficacy of the drug. Even relatively subtle impairments in CRL4CRBN activity (eg, after loss of SENP8) appear to limit LEN toxicity. In contrast, POM and especially CC-122 remain effective under conditions of suboptimal CRL4CRBN activity, such as reduced expression of overall CUL4 or loss of SENP8. Strikingly, the drug target CRBN was the only gene whose inactivation conferred complete resistance to CC-122 in our system.
Among the CRL4CRBN regulators identified in our study was the E2 Ub conjugating enzyme UBE2G1. We confirmed that inactivation of UBE2G1 in PEL cells confers resistance to LEN and to POM, but not CC-122. LEN and POM resistance of UBE2G1-inactivated cells was recently also reported in MM by Sievers et al37 and Lu et al.38 These groups proposed a “prime-extend” mechanism, in which the E2 conjugating enzyme UBE2D3 mediates neosubstrate mono-ubiquitination, whereas UBE2G1 adds additional Ub moieties for polyubiquitination. Our results extend this role of UBE2G1 to PEL, where CM toxicity is independent of CM-induced IKZF1/3 degradation.7 Future CRISPR screens in the context of UBE2D3 and/or UBE2G1 inactivation could be used to identify E2 enzymes that can compensate for these enzymes, especially during treatment with CC-122.
Inactivation of either CUL4A or CUL4B conferred significant resistance to LEN in several PEL cell lines, suggesting that total CUL4A/B levels are commonly limiting for CM toxicity. Limited CUL4 expression could sensitize cells to alterations in CUL4 neddylation status, such as those seen in SENP8-inactivated cells. It is conceivable that certain tumor types, including PEL, have low CUL4A/B expression, or that cells with low CUL4A/B expression exist within tumors for various cancers, which may affect the response to CM treatment and increase the likelihood of relapse.
Our screens and validation experiments identified a role for the Nedd8-specific protease SENP8 in the modulation of LEN toxicity and CRL4 activity. Specifically, inactivation of SENP8 leads to accumulation of deneddylated CUL4, perhaps a consequence of aberrant neddylation and reduced activity of the Nedd8 conjugation machinery.42 Increased amounts of deneddylated CUL4A/B may lead to the sequestration of DDB1/CRBN on inactive CRL4 complexes, thus limiting CM toxicity in SENP8-inactivated cells. Accordingly, overexpression of CRBN restored sensitivity of SENP8 KO cells to LEN. Conversely, overexpression of nonneddylatable CUL4A dominant negatively interfered with CM toxicity, and thereby phenocopied SENP8 knockout.
Finally, our study identified and validated ILF3 and YPEL5 as candidates for modulators of CM toxicity. ILF2 and ILF3 were recently identified as candidate binding partners of CRL complexes and their roles should be addressed in future studies.39 YPEL5 is a component of the unrelated GID/CTLH E3 ligase complex.41 The early point for LEN identified GID components WDR26 and MEMA, as well as the cognate E2 enzyme of this complex (UBE2H) as candidates for genes with delayed LEN toxicity, perhaps implicating the GID E3 ligase as a regulator of CM toxicity. However, YPEL5 has also been identified as a candidate interacting partner of the CSN subunit COPS540 and has unrelated functions.
In summary, we propose a model in which overall expression levels of CRBN, CUL4A, CUL4B, UBE2G1, and SENP8 dictate sensitivity of CMs in PEL (Figure 7). Future sequencing and gene expression studies will be required to identify roles of the expression or mutation of these genes in the response to CMs and the development of CM resistance in patients. The expression and/or mutational status of the genes identified in this study may be developed as biomarkers for whether treatment with a more potent CM is indicated on relapse from LEN or POM. Moving forward, it will be critical to identify the relevant neosubstrate(s) and mechanisms of CM toxicity in PEL. An improved understanding of the mechanisms of CM toxicity might also enable combination therapy with other drugs.
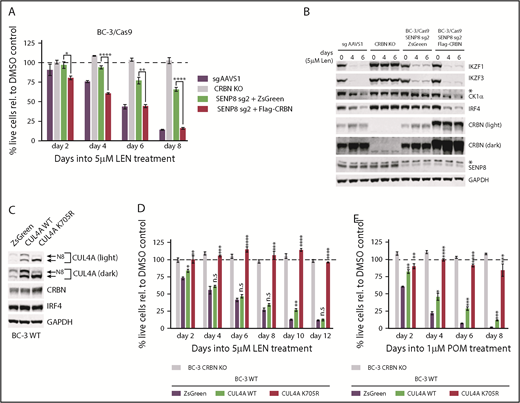
Model of the roles of UBE2G1 and SENP8 in CM toxicity. We hypothesize that UBE2G1 is the preferred E2 Ub conjugating enzyme for polyubiquitination by CM-bound CRL4CRBN. SENP8 affects the neddylation status of CUL4A and CUL4B, potentially by cleaving detrimental Nedd8 (N8)-conjugates from Nedd8-specific E1 [E1(N8)] and E2 [E2(N8)] enzymes, and thereby affects CRL4CRBN activity. CRBN overexpression promotes assembly of active CRL4CRBN complexes, likely by outcompeting other DDB1/DDB1-CUL4–associated factors (DCAF) substrate recognition modules for limiting pools of CUL4A/B. Conversely, increased expression of unneddylated CUL4A/B, either as a consequence of SENP8 inactivation or as a result of expression of nonneddylatable mutants, interferes with CRL4CRBN activity in a dominant-negative manner, potentially by sequestration of DDB1/CRBN on inactive complexes. Our data and this model suggest that CRBN and CUL4A/B expression can be limiting. CUL4A, CUL4B (together referred to as CUL4A/B), DDB1, UBE2G1, SENP8, RBX1, and CRBN represent gene symbols. CSN, COP9 signalosome.
Model of the roles of UBE2G1 and SENP8 in CM toxicity. We hypothesize that UBE2G1 is the preferred E2 Ub conjugating enzyme for polyubiquitination by CM-bound CRL4CRBN. SENP8 affects the neddylation status of CUL4A and CUL4B, potentially by cleaving detrimental Nedd8 (N8)-conjugates from Nedd8-specific E1 [E1(N8)] and E2 [E2(N8)] enzymes, and thereby affects CRL4CRBN activity. CRBN overexpression promotes assembly of active CRL4CRBN complexes, likely by outcompeting other DDB1/DDB1-CUL4–associated factors (DCAF) substrate recognition modules for limiting pools of CUL4A/B. Conversely, increased expression of unneddylated CUL4A/B, either as a consequence of SENP8 inactivation or as a result of expression of nonneddylatable mutants, interferes with CRL4CRBN activity in a dominant-negative manner, potentially by sequestration of DDB1/CRBN on inactive complexes. Our data and this model suggest that CRBN and CUL4A/B expression can be limiting. CUL4A, CUL4B (together referred to as CUL4A/B), DDB1, UBE2G1, SENP8, RBX1, and CRBN represent gene symbols. CSN, COP9 signalosome.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank the Northwestern University NUSeq Core Facility and the University of Chicago Genomics Facility for their Illumina sequencing services, and Kylee Morrison and Samuel Harvey for their feedback on this manuscript.
This study was supported by the National Institutes of Health, National Cancer Institute grant R21 CA210904, by Searle and Zell Scholar Awards from the Robert H. Lurie Comprehensive Cancer Center (E.G.), and by Chicago Biomedical Consortium Postdoctoral Award PDR-061 (M.M.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Authorship
Contribution: A.P. and E.G. designed the study, analyzed data, and wrote the manuscript; A.P. conducted all experiments; and M.M. prepared a subset of next generation sequencing libraries, analyzed data, and provided feedback on the study and manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Eva Gottwein, Department of Microbiology-Immunology, Northwestern University Feinberg School of Medicine, 320 E Superior St, Tarry Building, Room 6-735, Chicago, IL 60611; e-mail: e-gottwein@northwestern.edu.



![Figure 3. Inactivation of UBE2G1 confers resistance to LEN and POM, but not CC-122. (A) Representative western blot analysis confirms efficient CRISPR-induced inactivation of UBE2G1 in BC-3/Cas9 cells by 2 independent UBE2G1-specific sgRNAs (sg1, sg2). sgAAVS1 is a negative control guide. GAPDH served as loading control. (B-C) Growth curve analyses of UBE2G1 inactivated BC-3/Cas9 cells after treatment with 5 µM LEN (B) or 1 µM POM (C). BC-3/Cas9 cells transduced with sgAAVS1 were included as a CM-sensitive negative control, whereas previously described clonal CRBN-inactivated BC-3 cells (CRBN KO) served as a positive control for complete CM resistance. Absolute live cell numbers were normalized to corresponding DMSO vehicle-treated cells at each passage (represented by the dotted line). These experiments were performed in parallel with SENP8-inactivated cells shown in Figure 4B-C, and thus share common negative and positive controls for several or all replicates. (n = 3; error bars represent standard error of the mean [SEM]). (D) Representative western blot analysis (anti-Flag) confirms lentiviral expression of sg2-resistant, Flag-tagged UBE2G1 in UBE2G1-inactivated BC-3/Cas9 cells (UBE2G1 sgRNA sg2). GAPDH served as loading control. (E) Growth curve analysis of UBE2G1-inactivated BC-3/Cas9 cells (UBE2G1 sgRNA sg2) expressing Flag tagged UBE2G1 (sg2-resistant; UBE2G1/sg2R) or control ZsGreen after treatment with 5 µM LEN confirms restoration of CM sensitivity on UBE2G1 reexpression. BC3/Cas9/sg AAVS1 was included as a LEN-sensitive positive control. Live cell counts were normalized to corresponding DMSO vehicle-treated cells, which are represented by the dotted line. These assays were run in parallel with those in Figure 4E, and thus share common controls (n = 3; error bars represent SEM). (F-G) Representative time course western blot analyses of CM neosubstrates IKZF1, IKZF3, and CK1α, and IRF4 in WT or UBE2G1-inactivated BC-3/Cas9 cells upon treatment with 5 µM LEN (F) or 1 µM POM (G). Lysates are matched with growth curves shown in panels B and C, respectively. GAPDH served as loading control. The asterisk marks a nonspecific band in panel F. (H) Growth curve analyses of UBE2G1-inactivated BC-3/Cas9 cells treated with 1 µM CC-122. sgAAVS1 and CRBN KO cells were included as negative and positive controls, respectively. Live cell counts were normalized to corresponding DMSO vehicle-treated cells, represented by the dotted line (n = 3; error bars represent SEM). (I) Representative western blot analysis of IKZF1, IKZF3, CK1α, and IRF4 at indicated points confirms a delay in neosubstrate degradation and IRF4 downregulation in UBE2G1-inactivated BC-3/Cas9 cells compared with control pools as in panel H. GAPDH served as loading control. Statistical analyses throughout this figure were performed by unpaired Student t tests comparing specified conditions to corresponding sgAAVS1 or ZsGreen controls. *P <. 05; **P < .01; ***P < .001; ****P < .0001. n.s., not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/3/14/10.1182_bloodadvances.2019031732/2/m_advances031732f3.png?Expires=1765016619&Signature=DtzaEu05zY9nbUOt1YWYxq5iMMYCF~CbHrjWbTaN7KkKtFKMLrqMddA3wlPdu5GAowaC-J8rpnwHdBNed2ERJJo-Ps6~8zaZlUk-BUdMOhi3LIEqgk~2QwyQPtLDeYMpiNRknCTD07eIKL044cQigiZm0PCqpMIHmScFn4rQRQ~1~RGAQwTB~KEiP7Gcwq9ooQqbXziUImPSd4GTQ0eTTfquOiex3uNgVsL-xqtN82ekUiTys8ZwkKeNspXxNCuQi-tMMrlsauL0uziYMLzPuj2srQyTWTyIFukl6Cn5YDojNnj3B4dIjGP8H8Dq~Raf2YN70cyt7zzdUPty8LTE6Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 7. Model of the roles of UBE2G1 and SENP8 in CM toxicity. We hypothesize that UBE2G1 is the preferred E2 Ub conjugating enzyme for polyubiquitination by CM-bound CRL4CRBN. SENP8 affects the neddylation status of CUL4A and CUL4B, potentially by cleaving detrimental Nedd8 (N8)-conjugates from Nedd8-specific E1 [E1(N8)] and E2 [E2(N8)] enzymes, and thereby affects CRL4CRBN activity. CRBN overexpression promotes assembly of active CRL4CRBN complexes, likely by outcompeting other DDB1/DDB1-CUL4–associated factors (DCAF) substrate recognition modules for limiting pools of CUL4A/B. Conversely, increased expression of unneddylated CUL4A/B, either as a consequence of SENP8 inactivation or as a result of expression of nonneddylatable mutants, interferes with CRL4CRBN activity in a dominant-negative manner, potentially by sequestration of DDB1/CRBN on inactive complexes. Our data and this model suggest that CRBN and CUL4A/B expression can be limiting. CUL4A, CUL4B (together referred to as CUL4A/B), DDB1, UBE2G1, SENP8, RBX1, and CRBN represent gene symbols. CSN, COP9 signalosome.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/3/14/10.1182_bloodadvances.2019031732/2/m_advances031732f7.png?Expires=1765016619&Signature=yAEsaMfENvKE~6tnrO888DXHLKc~4Dx6HEYyID-olbx2M6NQ953W8cdkGBRT6kimMFx5W-hoiiqHhNMMWlsNkWPpPJxYv7SaCrVSJFgvcWFbOZ4XJhrnxJFRU7fgk1HJMO8VNzK7~jymGq6cv6hXs40FUaXXACs7bVtO-mxipfW9K5ASFJrcJIsVl~LFl3ZabStTl4wfUq8GPjD2ihn5iGDvrmt4CQ8z25woDdPNM~NlrYQR~eaSPTIeBXzuIO3y3nam2qrx4fgx83YzNvGNUqXkJ3V7pHm2pdeKyercFvEYN1huISNVr7dlVF86~c-SQwe33n5Dj7ZyQnYjTzxVgA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)