Key Points
Tet2 is a tumor suppressor in B cells.
Loss of Tet2 in B cells leads to age-dependent transformation that requires AID.
Abstract
The TET2 gene encodes an α-ketoglutarate–dependent dioxygenase able to oxidize 5-methylcytosine into 5-hydroxymethylcytosine, which is a step toward active DNA demethylation. TET2 is frequently mutated in myeloid malignancies but also in B- and T-cell malignancies. TET2 somatic mutations are also identified in healthy elderly individuals with clonal hematopoiesis. Tet2-deficient mouse models showed widespread hematological differentiation abnormalities, including myeloid, T-cell, and B-cell malignancies. We show here that, similar to what is observed with constitutive Tet2-deficient mice, B-cell–specific Tet2 knockout leads to abnormalities in the B1-cell subset and a development of B-cell malignancies after long latency. Aging Tet2-deficient mice accumulate clonal CD19+ B220low immunoglobulin M+ B-cell populations with transplantable ability showing similarities to human chronic lymphocytic leukemia, including CD5 expression and sensitivity to ibrutinib-mediated B-cell receptor (BCR) signaling inhibition. Exome sequencing of Tet2−/− malignant B cells reveals C-to-T and G-to-A mutations that lie within single-stranded DNA–specific activation-induced deaminase (AID)/APOBEC (apolipoprotein B messenger RNA editing enzyme, catalytic polypeptide-like) cytidine deaminases targeted motif, as confirmed by the lack of a B-cell tumor in compound Tet2-Aicda–deficient mice. Finally, we show that Tet2 deficiency accelerates and exacerbates T-cell leukemia/lymphoma 1A–induced leukemogenesis. Together, our data establish that Tet2 deficiency predisposes to mature B-cell malignancies, which development might be attributed in part to AID-mediated accumulating mutations and BCR-mediated signaling.
Introduction
The incidence and type of hematological malignancies vary with age. The incidence of mature lymphoid malignancies increases with age. This includes lymphoma but also chronic lymphocytic leukemia (CLL), the most frequent adult leukemia in Western countries with a median age at diagnosis of 72 years. The reasons are not yet fully understood, but they may be due to a modification of hematopoietic stem/progenitor cell (HSC/P) microenvironment1 or cell-intrinsic alterations of HSC,2,3 or both.4 The importance of cell-intrinsic defects is also supported by the age-related incidence of somatic aberrations detected in the blood cells5 and in the HSC6 compartment of individuals devoid of clinical signs of hematological disorder. The frequency of clonal hematopoiesis increases with age and is associated with a risk for developing hematological malignancy. Mutations of genes whose products are directly or indirectly involved in the control of DNA methylation, such as IDH1, IDH2, TET2, and DNMT3A, represent a large proportion of the mutations associated with clonal dominance.5
The TET proteins are α-ketoglutarate (α-KG)–dependent dioxygenases able to oxidize 5-methylated cytosine (mC) into hydroxymethylated C (hmC), which represent a step toward active or passive DNA demethylation, or both. The TET2 gene is mutated in 15% of all types of human myeloid neoplasms,7-9 2% to 10% of B-cell lymphomas,10,11 and 10% of T-cell lymphomas, particularly of the angioimmunoblastic subtype.8 However, no major member of the DNA methylation control pathway was recurrently found mutated in CLL and malignant B-cell differentiation.12-16
Another player in B-cell malignant development is the activation-induced cytidine deaminase (AID) gene, which encodes a cytidine deaminase and is known to initiate both class switch recombination and somatic hypermutation, 2 main mechanisms implicated in the maturation of the antibody response. AID expression is tightly regulated, and its aberrant activity has been shown to induce mutations in nonimmunoglobulin genes, thus contributing to cellular transformation.17
Mouse models have demonstrated that Tet2 deficiency endows the cell with growth advantage over wild-type cells and have suggested that the development of a full-blown malignancy depends on the occurrence of additional mutations.8,18,19
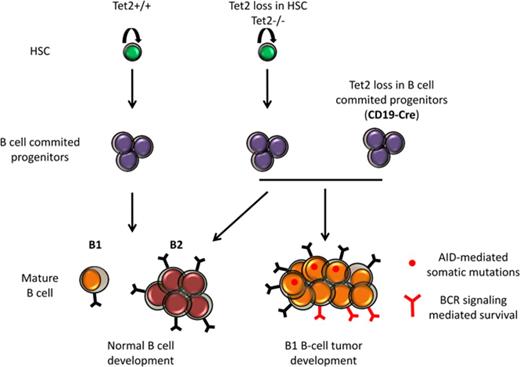
In our previously published Tet2-deficient models, a fraction (∼30%) of aging animals develops a chronic myelomonocytic leukemia-like disease, whereas another third shows the accumulation of an abnormal B-cell population characterized by a low level of B220 at the cell surface (B220low B cells).8 Here, we report the analyses of the B-cell differentiation and B-cell malignancy development in both constitutive and B-cell–specific (CD19-Cre) Tet2-deficient mice. Our results show that in addition to myeloid and T-cell malignancies, Tet2 deficiency predisposes to B-cell malignancies, which depend on AID-induced mutation for their development and on B-cell receptor (BCR) signaling for their survival.
Materials and methods
Additional information can be found in the supplemental Methods.
Mice
Mice carrying conditional inactivated Tet2 alleles have been previously described.8 Mice harboring Tet2flox alleles with exon 11 flanked by 2 loxP sites were intercrossed with CD19-Cre transgenic mice expressing specifically the Cre recombinase in the B-cell compartment that induces Tet2 inactivation specifically in the B-cell compartment.20 We used the following nomenclature: Tet2CD19-Cre+ for CD19-Cre+Tet2flox/flox mice and Tet2CD19-Cre− for CD19-Cre−Tet2flox/flox mice. We generated mice constitutively inactivated for Tet2 by crossing Tet2flox/flox mice with EIIA-Cre mice. Intercrossing heterozygous mice gave rise to mendelian ratios of the 3 genotypes: Tet2 wild-type (Tet2+/+), heterozygous (Tet2−/+), and homozygous deleted Tet2 (Tet2−/−). The Tet2−/− mice are fertile. Aicda-deficient mice were previously described.21 All mice used in this study, including wild-type C57BL/6 mice, were maintained in our breeding facility (SCEA, Gustave Roussy, Villejuif, France) and were sacrificed at the indicated times or when they became moribund or sick. Sick mice were identified on the basis of elevated white blood cell count of 20 × 109/L and higher (normal white blood counts are <10 × 109/L) or when they developed obvious splenomegaly. Animal experiments were conducted according to the Gustave Roussy institutional guidelines and authorized by the Direction Départementale des Services Vétérinaires du Val de Marne.
Genescan and CDR3 analysis
Complementary DNA (cDNA) from sorted Tet2-deficient abnormal B cells (B220lowCD19+) and wild-type (WT) B cells (B220+CD19+) was polymerase chain reaction (PCR) amplified using VH family primers (VH1, VH2, VH3, VH5, VH6, and VH7, as indicated) and a common JH fluorescein amidite–conjugated primer to assess immunoglobulin H (IgH) CDR3 diversity.22 PCR products were run on a 3130xl sequencer (Applied Biosystems), and data were analyzed using Peakscanner software (Applied Biosystems). PCR fragments were sequenced using the same VH family primer used for amplification. Sequences were analyzed using IgBlast23 and International ImMunoGeneTics (IMGT) V-Quest,24-26 and IgH V, D, and J genes with highest significance in both analyses are shown in Table 1.
IgH CDR3 of Tet2-deficient CD19+B220low B cells reveals CLL-like features
| Mouse . | Tag . | Tet2 genotype . | V . | D . | J . | CDR3 . | Length, nt . | D reading frame . | IP . |
|---|---|---|---|---|---|---|---|---|---|
| Tet2 ANO mice | C12 | ANO/WT | V1-9 | D-FL 16.2 | J1 | C AR SYYGYS WYFDV W | 13 | 3 | 6,14 |
| A | ANO/WT | V2-2 | D-FL 16.3 | J4 | C AR KTNEVNYYDMDY W | 14 | 1 | 4,72 | |
| Tet2 CD19-Cre mice | G212 | Flox/WT | V1-55 | D-FL 16.1 | J1 | C AR IYYGSSYWYFDV W | 14 | 3 | 6,14 |
| G203 | Flox/WT | V3-2 | D-SP2.2 | J4 | C ARYYDYDYYAMDY W | 13 | 3 | 4,15 | |
| G322 | Flox/WT | V5-2 | D-FL 16.1 | J4 | C ARLWYYVG YAMDY W | 13 | 3 | 6,14 | |
| G328 | Flox/WT | V3-6 | D-SP2.9 | J4 | C AR DDGYYGD YAMDY W | 14 | 3 | 3,99 | |
| G211 | Flox/flox | V3-6 | D-SP2.5 | J4 | C A NYGNYIS YAMDY W | 13 | 3 | 3,75 | |
| G200 | Flox/flox | V3-6 | D-SP2.2 | J4 | C S TCPYDYDGS YYAMDY W | 16 | 3 | 3,46 | |
| G635 | Flox/flox | V2-2 | D-FL 16.1 | J4 | C AR TPPYITTVVA PYAMDY W | 18 | 1 | 6,14 | |
| G630 | Flox/flox | V5-15 | D-FL 16.1 | J4 | C AR HGGS AMDY W | 10 | 3 | 7,17 | |
| G103 | Flox/flox | V10-1 | D-Q52 | J2 | C VR HDWDG FDY W | 10 | 3 | 4,71 | |
| G213 | Flox/flox | V5-9-1 | D-SP2.5 | J2 | C TR EYLGSTTVPTIL DY W | 16 | 1 | 4,44 |
| Mouse . | Tag . | Tet2 genotype . | V . | D . | J . | CDR3 . | Length, nt . | D reading frame . | IP . |
|---|---|---|---|---|---|---|---|---|---|
| Tet2 ANO mice | C12 | ANO/WT | V1-9 | D-FL 16.2 | J1 | C AR SYYGYS WYFDV W | 13 | 3 | 6,14 |
| A | ANO/WT | V2-2 | D-FL 16.3 | J4 | C AR KTNEVNYYDMDY W | 14 | 1 | 4,72 | |
| Tet2 CD19-Cre mice | G212 | Flox/WT | V1-55 | D-FL 16.1 | J1 | C AR IYYGSSYWYFDV W | 14 | 3 | 6,14 |
| G203 | Flox/WT | V3-2 | D-SP2.2 | J4 | C ARYYDYDYYAMDY W | 13 | 3 | 4,15 | |
| G322 | Flox/WT | V5-2 | D-FL 16.1 | J4 | C ARLWYYVG YAMDY W | 13 | 3 | 6,14 | |
| G328 | Flox/WT | V3-6 | D-SP2.9 | J4 | C AR DDGYYGD YAMDY W | 14 | 3 | 3,99 | |
| G211 | Flox/flox | V3-6 | D-SP2.5 | J4 | C A NYGNYIS YAMDY W | 13 | 3 | 3,75 | |
| G200 | Flox/flox | V3-6 | D-SP2.2 | J4 | C S TCPYDYDGS YYAMDY W | 16 | 3 | 3,46 | |
| G635 | Flox/flox | V2-2 | D-FL 16.1 | J4 | C AR TPPYITTVVA PYAMDY W | 18 | 1 | 6,14 | |
| G630 | Flox/flox | V5-15 | D-FL 16.1 | J4 | C AR HGGS AMDY W | 10 | 3 | 7,17 | |
| G103 | Flox/flox | V10-1 | D-Q52 | J2 | C VR HDWDG FDY W | 10 | 3 | 4,71 | |
| G213 | Flox/flox | V5-9-1 | D-SP2.5 | J2 | C TR EYLGSTTVPTIL DY W | 16 | 1 | 4,44 |
Sequence analysis of PCR products obtained from GeneScan analysis was down using the JH primer. Results obtained were subject to analysis using IMGT/Vquest and IgBlast software to define all V, D, and J genes, as well as the CDR3 sequence. Columns show the identified IgH-V, -D, and -J genes; the resulting CDR3 sequence, the CDR3 length; the D genes reading frame, and the isoelectric point (IP) of the CDR3. The background of the mice that developed the abnormal B-cell population is indicated. 2 abnormal populations heterozygous for Tet2 gene-trap model (Tet2LacZ ANO) are shown, as well as 4 heterozygous Tet2 floxed alleles (Tet2−/+) and 6 homozygous inactivated Tet2 (Tet2−/−) in CD19-Cre Tet2 background. The last 3 lines represent analysis of 3 abnormal Tet2 populations that have emerged in 3 secondary recipients engrafted from the same primary Tet2CD19-Cre+ mice. In the “CDR3” column, amino acid residues encoded by the V or J segments (underlined) are separated by spaces from those encoded by the D segment (in bold and underlined).
Statistical and quantification analyses
Results are expressed as mean values ± standard deviation or standard error of the mean, as is indicated in each of the figures. Statistical significance of differences between the results was assessed using a 2-tailed unpaired Student t test with Welch’s correction, performed using Prism (GraphPad software, version 5.03). Statistically significant P values are <.05, <.01, and <.005.
Retroviral infection and in vivo cell transfer
T-cell leukemia/lymphoma 1A (TCL1A) cDNA (NM_001098725) was subcloned into murine stem cell virus (MSCV)–green fluorescent protein (GFP) backbone. BRAF constructs, viral particles, and transduction procedures were described previously.27
Results
Cell autonomous effect of Tet2 deficiency in B220low B-cell population accumulation
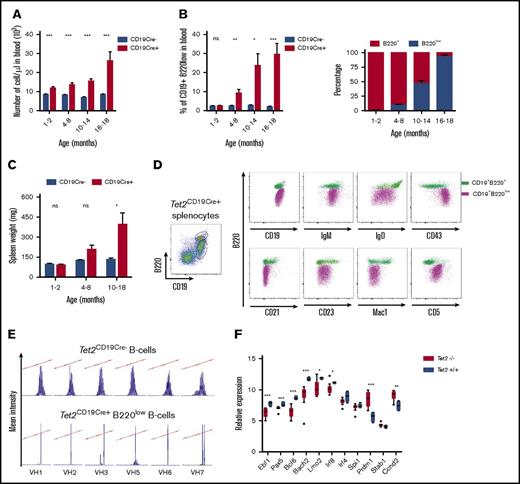
We previously reported a phenotypically abnormal B-cell population, characterized by low B220 cell surface expression in the 2 Tet2-deficient mouse models, a constitutive gene-trap Lac-z insertion model (Tet2LacZ ANO) and an inducible Mx1-Cre model (Tet2Mx1-Cre+) (supplemental Figure 1A and Quivoron et al8 ). To establish the cell-autonomous nature of the mature B-cell abnormality, we used the CD19-Cre transgenic mice to inactivate Tet2 specifically during B-cell differentiation.20 In those mice (Tet2CD19-Cre+), Tet2 deletion was restricted to the B-cell lineage (supplemental Figure 1B). We monitored the mice by monthly blood sampling and showed that the white blood cell numbers increased with age in Cre+ animals (Figure 1A). This correlates with the accumulation of the same abnormal CD19+ IgM+ B220low population and was comparable to what we detected in Tet2Mx1-Cre+, Tet2LacZ ANO, and Tet2−/− mice (supplemental Figure 1A,C). The accumulation started at approximately 5 months of age and invaded the entire B-cell compartment (Figure 1B). Interestingly, May-Grünwald Giemsa–stained blood smears of the Tet2-deficient mice showed a population of cells referred to as “smudge cells,” and 2 out of 3 mouse blood samples displayed abnormal lymphocyte cells (supplemental Figure 1C), which are routinely observed in blood samples from CLL patients. Sick mice were killed, and the spleen showed enlargement (Figure 1C) due to lymphoid cell infiltration observed in the hematoxylin eosin saffron staining of a representative spleen section (supplemental Figure 1C). This correlates with the increased B220low splenocytes showed by fluorescence-activated cell sorter (FACS) analyses (Figure 1D). Comparison of the immunophenotype of B220low splenocytes with B cells expressing a normal amount of B220 antigen (B220+) showed that they were IgM+IgD−/lowCD43+CD21−CD23−Mac1lowCD5+ cells (Figure 1D). Together, these results demonstrate that specific Tet2 deficiency in the B-cell lineage is sufficient to induce mature B-cell transformation, indicating that this development is cell autonomous. This result supports the cell-autonomous nature of the appearance of the abnormal B220low B-cell population in the previously published constitutive (Tet2LacZ ANO) and inducible (Tet2Mx1-Cre+) Tet2-deficient models and is linked to the Tet2 deficiency in the B-cell compartment (supplemental Figure 1A).
Tet2 deficiency leads to the accumulation of clonal B220lowB-cell population in a cell-autonomous fashion. (A) Monthly monitoring of blood cell count in Tet2CD19-Cre− and Tet2CD19-Cre+ mice. ***P < .001. (B) Monthly monitoring of CD19+ B220low abnormal cell population in the blood of Tet2CD19-Cre− and Tet2CD19-Cre+ mice (left panel). Significance was tested using an unpaired Student t test. Time-dependent representation of the proportion of the B220low population within the circulating B cells (CD19+) in the same mice as in panel A (right panel). Significance was tested using an unpaired Student t test. (C) Spleen weight of sacrificed mice at various time points was measured for both types of mice. Significance was tested using an unpaired Student t test. (D) Immunostaining of total splenocytes (polychromatic dot plot) from a representative sick Tet2CD19-Cre+ mouse (same genotypes as above). Splenocytes were first stained with B220 and CD19 to define the B220+ and B220low population of CD19+ cells (left polychromatic dot plot). Other dot plots show B220 expression together with the indicated antibody on the x-axis of the overlaid B220+CD19+ population (green) and B220lowCD19+ population (violet). (E) Analyses of BCR rearrangement clonality in abnormal Tet2CD19-Cre+ B cells. V-J junctions were PCR-amplified from cDNA of sorted B cells using VH family primers (VH1, VH2, VH3, VH5, VH6, and VH7, as indicated) and conjugated consensus JH primer22 to assess the IgH CDR3 diversity. Top panels show Results shown are obtained from sorted Tet2CD19Cre− B splenocytes (B220+CD19+) (top) and from age-matched Tet2CD19-Cre+ sorted abnormal B cells (B220lowCD19+) (bottom). (F) Transcription of selected genes important for B-cell differentiation differs between normal WT and Tet2-deficient tumor B cells. Histograms show quantitative reverse transcription (qRT)–PCR analysis results ΔΔCT-normalized by Abl1 expression levels. Results are an average of 3 independent duplicate experiments. The differences were validated in an extension cohort for 8 of the 11 genes. Significance was tested using the Mann-Whitney U test. *P < .05; **P < .01; ***P < .001. ns, not significant.
Tet2 deficiency leads to the accumulation of clonal B220lowB-cell population in a cell-autonomous fashion. (A) Monthly monitoring of blood cell count in Tet2CD19-Cre− and Tet2CD19-Cre+ mice. ***P < .001. (B) Monthly monitoring of CD19+ B220low abnormal cell population in the blood of Tet2CD19-Cre− and Tet2CD19-Cre+ mice (left panel). Significance was tested using an unpaired Student t test. Time-dependent representation of the proportion of the B220low population within the circulating B cells (CD19+) in the same mice as in panel A (right panel). Significance was tested using an unpaired Student t test. (C) Spleen weight of sacrificed mice at various time points was measured for both types of mice. Significance was tested using an unpaired Student t test. (D) Immunostaining of total splenocytes (polychromatic dot plot) from a representative sick Tet2CD19-Cre+ mouse (same genotypes as above). Splenocytes were first stained with B220 and CD19 to define the B220+ and B220low population of CD19+ cells (left polychromatic dot plot). Other dot plots show B220 expression together with the indicated antibody on the x-axis of the overlaid B220+CD19+ population (green) and B220lowCD19+ population (violet). (E) Analyses of BCR rearrangement clonality in abnormal Tet2CD19-Cre+ B cells. V-J junctions were PCR-amplified from cDNA of sorted B cells using VH family primers (VH1, VH2, VH3, VH5, VH6, and VH7, as indicated) and conjugated consensus JH primer22 to assess the IgH CDR3 diversity. Top panels show Results shown are obtained from sorted Tet2CD19Cre− B splenocytes (B220+CD19+) (top) and from age-matched Tet2CD19-Cre+ sorted abnormal B cells (B220lowCD19+) (bottom). (F) Transcription of selected genes important for B-cell differentiation differs between normal WT and Tet2-deficient tumor B cells. Histograms show quantitative reverse transcription (qRT)–PCR analysis results ΔΔCT-normalized by Abl1 expression levels. Results are an average of 3 independent duplicate experiments. The differences were validated in an extension cohort for 8 of the 11 genes. Significance was tested using the Mann-Whitney U test. *P < .05; **P < .01; ***P < .001. ns, not significant.
We next performed serial transplant experiments. In 3 independent experiments, we engrafted from a symptomatic Tet2CD19-Cre+ mouse 1 million splenic mononuclear cells per mouse into 5 sublethally (5 Gy) irradiated syngeneic wild-type mice. The recipient mice showed the accumulation of abnormal B cells with a phenotype similar to the donor sample after a 6- to 8-month latency. Transplantation from secondary recipients was then performed and resulted in a similar outcome, but with reduced latency (4-6 months; supplemental Figure 1E). Because we injected whole-cell mixtures during those experiments, we next looked at the cell-autonomous capacity of the abnormal B-cell population to engraft and develop. So we sorted CD19+CD5+ spleen abnormal B cells from a 14-month-old sick mouse (Tet2−/−) and engrafted 400 000 cells/recipient into 4 recipients; they all died from the accumulation of B220low CD19+CD5+ abnormal B cells before 7 months postengraftment (data not shown).
To investigate the clonal nature of this B-cell tumor, we analyzed the IgH gene rearrangement using Genescan analyses of FACS-sorted abnormal B cells.22 A representative analysis is shown in Figure 1E. The abnormal B-cell population showed oligo/monoclonality (Figure 1E, bottom panel), markedly different in comparison with the polyclonality observed in lanes corresponding to wild-type B-cell populations (Figure 1E, top panel). Nucleotide sequence analyses of the fragments amplified from the abnormal B cells confirmed the amplification of the 1-in-frame rearrangement. Prediction of the corresponding IgH protein (Table 1) revealed an enrichment of hydrophilic amino acids (Tyr, Ser, and charged amino acids) in CDR3 regions, as has been described for CLL28 and murine models resembling human CLL,29 whatever the genotype of sick mice studied (heterozygous or homozygous Tet2 deletion in Tet2LacZ ANO and CD19-Cre mice). A set of genes known to be important for B-cell differentiation and transformation showed a reduced expression in purified B220low cells in comparison with circulating WT CD19 B cells. The results depicted in Figure 1F clearly show lower expression of Ebf1, Pax5, and Bach2 expression in Tet2-deficient cells. Together, these data establish the malignant status and the cell-autonomous nature of the B-cell accumulation, together with the stability of the abnormal phenotype.
Lack of gross abnormalities in steady state early B-cell development in Tet2-deficient mice
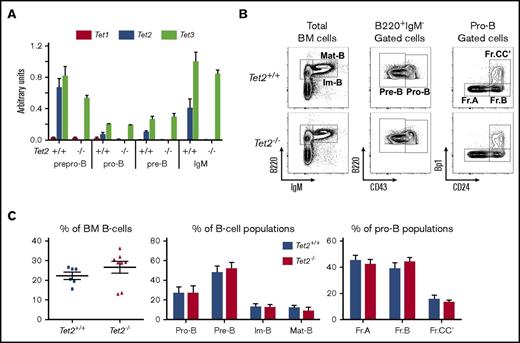
To investigate early B-cell development, we analyzed 2-month-old Tet2−/− mice before the onset of disease. First, to exclude the compensatory effects of Tet2 deficiency by other Tet proteins (ie, Tet1 and Tet3), we sorted Hardy fractions30 : Fr.A (B220+CD43+CD19−IgM−), Fr.BCC’ (B220+CD43+CD19+IgM−), and Fr.D (B220+CD43−CD19+IgM−) cell populations and total IgM+ (B220+IgM+) bone marrow (BM) B cells (Figure 2B); and we evaluated the expression level of all 3 Tet genes. No difference in Tet1 (note that Tet1 is barely detectable) or Tet3 transcription was observed between Tet2+/+ and Tet2−/− cells (Figure 2A).
Normal steady-state early B-cell development in Tet2-deficent mice. (A) qRT-PCR analysis transcription levels of the 3 murine Tet genes normalized by Abl1 expression. Results are the average of 3 independent experiments performed in duplicate. (B) Bone marrow B-cell development analysis. Total BM cells were analyzed for B220 and IgM expression. Immature B cells B220lowIgM+ and mature B cells B220+IgM+ are indicated. Then CD43 expression was evaluated in gated B220+IgM− cells to define pre-B (CD43−) and pro-B (CD43+) populations; finally, the more immature Hardy fractions (Fr.A CD24−Bp1−; Fr.B: CD24+Bp1−; and Fr.CC′: CD24+Bp1+) were analyzed in the pro-B-cell gate. The genotypes of the analyzed mice are indicated on the left side. (C) Average percentage of cell populations. (Left) Total B220+ cells (each dot represents an independent mouse). (Middle) Average percentage of the indicated population in the B-cell compartment for each genotype. (Right) Hardy fraction percentage inside the pro-B-cell compartment. Genotypes are indicated (Tet2+/+ mice, n = 6; Tet2−/− mice, n = 8). Im-B, immature B cells B220lowIgM+; Mat-B, mature B cells B220+IgM+.
Normal steady-state early B-cell development in Tet2-deficent mice. (A) qRT-PCR analysis transcription levels of the 3 murine Tet genes normalized by Abl1 expression. Results are the average of 3 independent experiments performed in duplicate. (B) Bone marrow B-cell development analysis. Total BM cells were analyzed for B220 and IgM expression. Immature B cells B220lowIgM+ and mature B cells B220+IgM+ are indicated. Then CD43 expression was evaluated in gated B220+IgM− cells to define pre-B (CD43−) and pro-B (CD43+) populations; finally, the more immature Hardy fractions (Fr.A CD24−Bp1−; Fr.B: CD24+Bp1−; and Fr.CC′: CD24+Bp1+) were analyzed in the pro-B-cell gate. The genotypes of the analyzed mice are indicated on the left side. (C) Average percentage of cell populations. (Left) Total B220+ cells (each dot represents an independent mouse). (Middle) Average percentage of the indicated population in the B-cell compartment for each genotype. (Right) Hardy fraction percentage inside the pro-B-cell compartment. Genotypes are indicated (Tet2+/+ mice, n = 6; Tet2−/− mice, n = 8). Im-B, immature B cells B220lowIgM+; Mat-B, mature B cells B220+IgM+.
We next analyzed the B-cell development in the bone marrow of constitutively Tet2-inactivated mice. Total bone marrow cells and total B-cell (B220+ cells) numbers were not significantly different between Tet2+/+ and Tet2−/− mice. The percentages of B cells, however, tend to be higher in Tet2-inactivated mice (supplemental Table 1; Figure 2C, left). Analysis of the B-cell stage of differentiation, pro-B (B220+CD43+IgM−), pre-B (B220+CD43−IgM−), immature B (B220lowIgM+), and mature B (B220+IgM+), showed no statistically significant alteration of the percentage as well as of the number of each population (supplemental Table 1; Figure 2C, middle and right). Analysis of the Hardy fractions within the pro-B cells does not reveal any effect of Tet2 deficiency (Figure 2B-C). Similarly, analysis of the more immature compartment of the common lymphoid progenitor (CLP) showed that the percentage and cell number of CLP were not affected by Tet2 inactivation (supplemental Figure 2A-B).
In conclusion, these results indicate that Tet2 deficiency does not markedly affect early B-cell development in marrow before the onset of the disease.
Tet2 deficiency induced alteration of peripheral B-cell populations
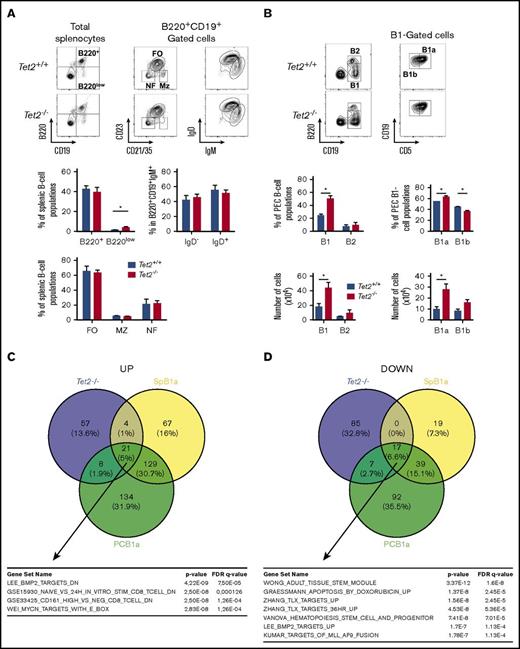
We next analyzed the peripheral B-cell populations in 2-month-old Tet2-deficient mice and controls. In mice, the main subdivision of B cells is between B2 cells, which include marginal zone and follicular B cells, and B1 cells. First, we observed similar spleen weight (Tet2+/+: 0.0899 ± 0.0238 g; and Tet2−/−: 0.1170 ± 0.0626 g) and similar spleen architecture (with identifiable germinal centers) between wild-type and Tet2-deficient mice (supplemental Figure 2C). Splenic B-cell populations are mostly B2 cells (B220+CD19+), but also rare B1 cells (B220lowCD19+) could be detected in wild-type mice. Neither the representation nor the numbers of total B2 B-cell populations was affected in Tet2-deficient mice (Figure 3A). We analyzed the splenic B2-cell subsets by gating in the B220+CD19+ and looking to the expression of CD21/35 and CD23. The 3 populations are defined as follicular (FO: CD21/35+CD23+), marginal zone (MZ: CD21/35+CD23low), and newly formed B cells (NF: CD21/35−CD23−). B cells were easily detectable in Tet2−/− mice and in similar numbers and frequencies as in Tet2+/+ mice (Figure 3A, left panel). No statistically significant alteration was observed in the repartition of the IgM+IgD− and IgM+IgD+ in the B220+CD19+ population (Figure 3A). However, looking carefully at B220lowCD19+ B1 cells, we observed a significant increase of the proportion of those cells expressing low amounts of B220 (B220lowCD19+; Figure 3A).
Tet2 deficiency induces alterations of the proportions of peripheral B-cell populations in old mice. (A) Analysis of splenic B-cell populations in 4-month-old mice. (Left) B220 and CD19 expression analysis on total splenocytes. (Middle) CD23 and CD21 expression in the B220+CD19+ (B220+) gate showing the follicular (FO:CD23+CD21/35medium), marginal zone (MZ: CD23lowCD21/35high), and newly formed (CD23−CD21/35−) B-cell populations. (Right) IgM and IgD staining gated in B220+CD19+ are also shown. Mice genotypes are indicated on the left-hand side. Histograms show the percentages of populations. (Top left) Average percentage ± standard error of the mean (SEM) of B220+ and B220low populations in total splenocytes. (Bottom) Average percentage ± SEM of FO, MZ, and NF. (Top right) IgM+IgD− and IgM+IgD+ populations in the B220+ B-cell compartment (Tet2+/+ mice, n = 5; Tet2−/− mice, n = 8). (B) Analysis of B-cell populations in the peritoneum. Peritoneal cell suspensions were stained with B220 and CD19 to define B2 (B220+CD19+) and B1 (B220lowCD19+) populations. In the B1-cell gate, CD19 and CD5 expressions allow us to define B1a (CD19+CD5+) and B1b (CD19+CD5−) subsets. (Tet2+/+, n = 4; Tet2−/−, n = 5). The histograms show the average percentage ± SEM of the peritoneum populations. (C) Venn diagram illustrating comparison of upregulated genes in Tet2−/− abnormal B cells and B1a cells, both with respect to splenic wild-type B cells. The deregulated pathways, as identified using gene set enrichment analyses, are shown at the bottom. SPB1a: B1a population from spleen. PCB1a: B1a population from peritoneum. (Bottom) List of signatures enriched in genes specifically upregulated in Tet2 samples. (D) Same as in panel C but for downregulated genes. (Bottom) List of signatures enriched in genes specifically downregulated in Tet2 samples. *P < .05.
Tet2 deficiency induces alterations of the proportions of peripheral B-cell populations in old mice. (A) Analysis of splenic B-cell populations in 4-month-old mice. (Left) B220 and CD19 expression analysis on total splenocytes. (Middle) CD23 and CD21 expression in the B220+CD19+ (B220+) gate showing the follicular (FO:CD23+CD21/35medium), marginal zone (MZ: CD23lowCD21/35high), and newly formed (CD23−CD21/35−) B-cell populations. (Right) IgM and IgD staining gated in B220+CD19+ are also shown. Mice genotypes are indicated on the left-hand side. Histograms show the percentages of populations. (Top left) Average percentage ± standard error of the mean (SEM) of B220+ and B220low populations in total splenocytes. (Bottom) Average percentage ± SEM of FO, MZ, and NF. (Top right) IgM+IgD− and IgM+IgD+ populations in the B220+ B-cell compartment (Tet2+/+ mice, n = 5; Tet2−/− mice, n = 8). (B) Analysis of B-cell populations in the peritoneum. Peritoneal cell suspensions were stained with B220 and CD19 to define B2 (B220+CD19+) and B1 (B220lowCD19+) populations. In the B1-cell gate, CD19 and CD5 expressions allow us to define B1a (CD19+CD5+) and B1b (CD19+CD5−) subsets. (Tet2+/+, n = 4; Tet2−/−, n = 5). The histograms show the average percentage ± SEM of the peritoneum populations. (C) Venn diagram illustrating comparison of upregulated genes in Tet2−/− abnormal B cells and B1a cells, both with respect to splenic wild-type B cells. The deregulated pathways, as identified using gene set enrichment analyses, are shown at the bottom. SPB1a: B1a population from spleen. PCB1a: B1a population from peritoneum. (Bottom) List of signatures enriched in genes specifically upregulated in Tet2 samples. (D) Same as in panel C but for downregulated genes. (Bottom) List of signatures enriched in genes specifically downregulated in Tet2 samples. *P < .05.
Although most of the peripheral B cells could be found in the spleen, other B-cell populations differentiate in the peritoneal cavity (PEC), particularly B1 cells (B220lowCD19+), which can be subdivided into CD5+ B1a and CD5− B1b cells. In the PEC of Tet2−/− mice, we showed significantly more B1 cells (Figure 3B), whereas B2 cells were not affected, in comparison with Tet2+/+ mice. Further analysis of the B1-cell subsets showed increased CD5+-expressing cells in Tet2-deficient mice. This significant increase of the B1a population correlates with a significant decrease in the B1b percentage, and both subsets (B1a and B1b) are significantly expanded in numbers in Tet2-deficient mice than in wild-type littermate controls.
Taken together, these results indicate minor effects of the Tet2 deficiency during B-cell development in young animals and suggest the need for additional oncogenic hits to develop full-blown malignancies.
We next compared the gene expression profile of flow-sorted CD19+ B220low IgM+ abnormal B cells to normal CD19+ B220+ IgM+ B cells from both bone marrow and spleen. Microarray analyses identified genes differentially expressed between Tet2-deficient tumors and splenic B cells, which are mostly follicular B cells. Ninety genes were overexpressed, and 109 were underexpressed (supplemental Table 2). To better define the genes that could be specific for the Tet2 population, we compared microarray data from B1a and follicular B-cell populations. This identifies a set of genes that are specifically expressed in the B1a populations, independently of their peritoneal cavity or spleen origin and in the Tet2-deficient tumor cells. It included 2 transcription factors, Mef2c, a well-known lymphoma oncogene, and Satb1 (supplemental Table 2). Analysis of the genes specifically upregulated in the Tet2-deficient tumors showed upregulation of bone morphogenetic protein and MYC-regulated pathways that might be important for cellular transformation. Genes specifically downregulated in the Tet2-deficient cells overlapped with “adult tissue stem module,” perhaps accounting for the indolent nature of those lymphomas. The list includes some hematopoietic tumor suppressor candidates, such as Runx1 and Tle1.
Tet2-deficient tumors harbor AID-mediated somatic mutations
We used exome sequencing to compare the coding sequences of 6 B-cell tumors to matched germline counterparts. Thirty-four acquired mutations were identified. We observed that C and G bases were significantly targeted by mutation events in comparison with A and T bases (P = .0486), with a bias toward C-to-T and G-to-A transition than toward other mutations (Figure 4A). Of note, more than half (8 out of 15) of those highly targeted C-to-T and G-to-A mutations lie in a single-stranded DNA–specific AID/APOBEC (apolipoprotein B messenger RNA editing enzyme, catalytic polypeptide-like) cytidine deaminase–targeted motif: 2 of 15 WRCY/RGYW (W = A or T, R = purine, Y = pyrimidine) (Rbms3 and Rab5b); 5 of 15 WRC (Kmt2a/MLL, Adamts2, Tenm3/Odz3, Zcchc2, and Adam34), and 1 of 15 TCW motifs (Slc2a9).33 These data suggest the involvement of AID activity in the oncogenic transformation of B cells. To confirm this hypothesis, we introduced Tet2 deficiency into an Aicda-deficient background. Comparing the emergence of the B220low B-cell malignancies over time, we observed that nearly 50% of Tet2-deficient mice developed B-cell malignancy, whereas only 1 out of 11 follow-up mice developed similar disease in an Aicda null background, supporting a key role of AID in the leukemic transformation process (Figure 4B).
Tet2-deficient abnormal B cells share features with human B-cell tumors. (A) Coding sequence somatic mutations. Frequencies of the type of mutations observed in the exome analysis of abnormal B-cell populations obtained from 6 independent tumors. The type of point mutations is indicated as well as insertions and deletions (indels). Transversion mutations (green) and transitions (red) are indicated. Note the statistically significant high frequency of C > T and G > A mutations. (B) Survival curves of Aid+/+Tet2+/+ (control [CTR]), Aicda-deficient mice: Aid−/− Tet2+/+ (Aid−/−); Tet2-deficient mice: Aid+/+Tet2−/− (Tet2−/−); and double-deficient mice: Aid−/−Tet2−/− strains. Only mice sacrificed because of B-cell tumors are scored. (C) Venn diagram of exome mutations from the literature of human CLL27,31,34-36 and DLBCL32,37-39 in comparison with exome mutations from the Tet2-deficient sorted abnormal B cells. The name of the mutated genes in the overlaying zones is indicated. *P < .05.
Tet2-deficient abnormal B cells share features with human B-cell tumors. (A) Coding sequence somatic mutations. Frequencies of the type of mutations observed in the exome analysis of abnormal B-cell populations obtained from 6 independent tumors. The type of point mutations is indicated as well as insertions and deletions (indels). Transversion mutations (green) and transitions (red) are indicated. Note the statistically significant high frequency of C > T and G > A mutations. (B) Survival curves of Aid+/+Tet2+/+ (control [CTR]), Aicda-deficient mice: Aid−/− Tet2+/+ (Aid−/−); Tet2-deficient mice: Aid+/+Tet2−/− (Tet2−/−); and double-deficient mice: Aid−/−Tet2−/− strains. Only mice sacrificed because of B-cell tumors are scored. (C) Venn diagram of exome mutations from the literature of human CLL27,31,34-36 and DLBCL32,37-39 in comparison with exome mutations from the Tet2-deficient sorted abnormal B cells. The name of the mutated genes in the overlaying zones is indicated. *P < .05.
Although 5.0 ± 3.7 (range from 2 to 10 mutations) acquired nonsynonymous mutations were found in the tumor B cells in each mouse, no recurrently mutated gene was identified. Comparison with genes mutated in human B-cell malignancies showed that over the 30 orthologous genes, more than half showed overlapping mutations between mouse B220lowCD19+ B cells and CLL27,31,34-36 (Ahi1, Mbtps2, Setd1a, and Vprbp) or DLBCL32,37-39 (Arid2, Atad2, Cnpy2, Mast1, Odz3, Pou2f2, Rbms3, and Slc2a9) or both (Aff3, Asxl3, MLL, Odz1, Odz2, and Pcdh15) human samples (Figure 4C).
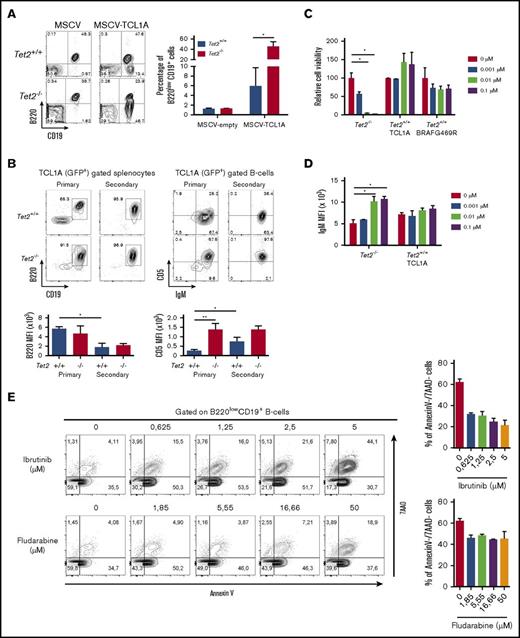
Tet2 deficiency exacerbates the phenotype of primary TCL1A-induced CLL-like B cells
To test whether the loss of Tet2 could promote mature neoplasms’ development/initiation, we investigated the cooperation of Tet2 deficiency with the overexpression of TCL1A, a well-known oncogene, by performing retroviral transduction and bone marrow transplantation to induce lymphoid malignancies. TCL1A was first identified in T-cell leukemia, but it has a more prominent role in the development of B-cell neoplasms.40 TCL1A encodes a coactivator of the AKT family of tyrosine kinases. Indeed, TCL1A overexpression in the B-cell compartment of transgenic mice (Eµ-TCL1A) resulted in the development of mature B-cell neoplasms comparable to human CLL.41 We transduced bone marrow progenitors from Tet2−/− or wild-type littermate mice with TCL1A-expressing MSCV and engrafted the transduced cells into lethally irradiated recipient mice. Expression of TCL1A induced lymphoid transformation in recipient mice within 3 to 8 months. In the wild-type background, 3 mice developed a T-cell leukemia/lymphoma, and 9 developed a CLL-like disease. The 5 engrafted mice in a Tet2-deficient context developed B-cell malignancy. Interestingly, 4 months after engraftment, we observed in the blood of mice engrafted with TCL1A-transduced Tet2−/− cells a higher proportion of B220low population than that in mice receiving TCL1A in Tet2 wild-type background (Figure 5A). Phenotypic analysis of the splenic B-cell population showed a lower expression of the B220 antigen in secondary recipients than in primary engrafted mice, and this tended to be lowest when TCL1A was expressed in a Tet2-deficient context in comparison with wild-type background in primary mice (Figure 5B). In contrast, CD5 expression was increased with serial engraftment for TCL1A-expressing B cells in a Tet2 wild-type background. This increase in CD5 mean fluorescence intensity was not observed in TCL1A- expressing B cells in a Tet2-deficient background because of a higher CD5 expression in primary recipients, indicating a more pronounced phenotype of CLL-like B cells induced by TCL1A overexpression in cells devoid of Tet2 (Figure 5B). These results show that the Tet2-deficient background cooperates with TCL1A overexpression to induce a CLL-like disease.
Tet2-deficient abnormal B-cells are BCR-inhibition sensitive. (A) Abnormal B-cell phenotype and frequencies in the peripheral blood of mice transplanted with TCL1A transduced Tet2+/+ or Tet2−/− hematopoietic progenitors. Dot plots show a representative example of B220 and CD19 expression analysis in the GFP+ total leukocytes 4 months after transplantation. Diagrams depict the average percentages of the B220low population in the blood of n = 3 mice per group 4 months after transplantation. Error bars represent means ± standard deviation (SD). (B) Tet2 deficiency enhances the tumor phenotype of TCL1A (GFP+) overexpressing B cells, as is shown by abnormal cell numbers and CD5 membrane expression level. (Upper left) B220 CD19 expression in gated GFP+ cells expressing TCL1A in primary and secondary recipient mice engrafted with BM cells overexpressing TCL1A in a Tet2+/+ or Tet2−/− background. (Lower left) Average ± SD of the median fluorescence intensity of the B220 antigen at the cell surface of CD19+ B cells in each group. (Upper right) Expression of CD5 and IgM on the gated B cells as in the upper-left contour plots. A representative contour plot is shown for each group. (Lower right) Average ± SD of the median fluorescence intensity of the CD5 antigen at the cell surface of CD19+ B cells in each group. (C) Ibrutinib-mediated BTK inhibition results in growth reduction and increase of surface IgM expression on Tet2-deficient B cells. Dose effects of ibrutinib on the B-cell population of Tet2-deficient or wild-type cell lines are derived from independent primary mice: 2 tumoral B-cell lines, one obtained from a spontaneous B-cell malignancy development in a Tet2−/− mouse, and the other obtained from a mouse from a BMT experiment of wild-type Tet2+/+ BM cells transduced by the TCL1A oncogene; and a T-cell line obtained from a BMT experiment of wild-type Tet2+/+ BM cells transduced by the BRAFG469R oncogene. Diagrams depict variable cell proportion in comparison with the nontreated cells. Cells were counted 3 days after treatment with the indicated inhibitor concentration. (D) Surface expression level (mean fluorescence intensity) of IgM on B-cell population (IgM+B220+) observed after staining of the 2 B-cell lines cultivated as in panel C. (E) Splenocytes from Tet2−/− mice were treated with ibrutinib and fludarabine for 18 hours ex vivo. Cells were stained with CD19-BV510, B220-allophycocyanin (APC)-Cy7, annexin V–APC, and 7-aminoactinomycin D (7-AAD). Gated B220lowCD19+ splenic cells were analyzed for annexin V and 7-AAD staining. Histograms show the mean ± SD of the percentages of viable cells (annexin V and 7-AAD double negative) in gated B220lowCD19+ populations from spleens of Tet2−/− mice (n = 2). *P < .05 (unpaired Student t test). MFI, mean fluorescence intensity.
Tet2-deficient abnormal B-cells are BCR-inhibition sensitive. (A) Abnormal B-cell phenotype and frequencies in the peripheral blood of mice transplanted with TCL1A transduced Tet2+/+ or Tet2−/− hematopoietic progenitors. Dot plots show a representative example of B220 and CD19 expression analysis in the GFP+ total leukocytes 4 months after transplantation. Diagrams depict the average percentages of the B220low population in the blood of n = 3 mice per group 4 months after transplantation. Error bars represent means ± standard deviation (SD). (B) Tet2 deficiency enhances the tumor phenotype of TCL1A (GFP+) overexpressing B cells, as is shown by abnormal cell numbers and CD5 membrane expression level. (Upper left) B220 CD19 expression in gated GFP+ cells expressing TCL1A in primary and secondary recipient mice engrafted with BM cells overexpressing TCL1A in a Tet2+/+ or Tet2−/− background. (Lower left) Average ± SD of the median fluorescence intensity of the B220 antigen at the cell surface of CD19+ B cells in each group. (Upper right) Expression of CD5 and IgM on the gated B cells as in the upper-left contour plots. A representative contour plot is shown for each group. (Lower right) Average ± SD of the median fluorescence intensity of the CD5 antigen at the cell surface of CD19+ B cells in each group. (C) Ibrutinib-mediated BTK inhibition results in growth reduction and increase of surface IgM expression on Tet2-deficient B cells. Dose effects of ibrutinib on the B-cell population of Tet2-deficient or wild-type cell lines are derived from independent primary mice: 2 tumoral B-cell lines, one obtained from a spontaneous B-cell malignancy development in a Tet2−/− mouse, and the other obtained from a mouse from a BMT experiment of wild-type Tet2+/+ BM cells transduced by the TCL1A oncogene; and a T-cell line obtained from a BMT experiment of wild-type Tet2+/+ BM cells transduced by the BRAFG469R oncogene. Diagrams depict variable cell proportion in comparison with the nontreated cells. Cells were counted 3 days after treatment with the indicated inhibitor concentration. (D) Surface expression level (mean fluorescence intensity) of IgM on B-cell population (IgM+B220+) observed after staining of the 2 B-cell lines cultivated as in panel C. (E) Splenocytes from Tet2−/− mice were treated with ibrutinib and fludarabine for 18 hours ex vivo. Cells were stained with CD19-BV510, B220-allophycocyanin (APC)-Cy7, annexin V–APC, and 7-aminoactinomycin D (7-AAD). Gated B220lowCD19+ splenic cells were analyzed for annexin V and 7-AAD staining. Histograms show the mean ± SD of the percentages of viable cells (annexin V and 7-AAD double negative) in gated B220lowCD19+ populations from spleens of Tet2−/− mice (n = 2). *P < .05 (unpaired Student t test). MFI, mean fluorescence intensity.
Abnormal Tet2−/− B-cell survival depends on BCR signaling
Next, we tested a role for BCR signaling in survival of abnormal B cells by monitoring cellular viability in the presence of a Bruton's tyrosine kinase (BTK) inhibitor (ibrutinib). We compared BTK inhibitor susceptibility of 3 cell lines obtained from long-term cultures of cells from mice developing a T-cell lymphoma (BRAFG469R)27 as control and 2 B-cell–type lymphomas, one from the TCL1A overexpression in Tet2+/+ B cells and the other obtained directly from a Tet2−/− mouse. Increasing doses of BTK inhibitor in the culture medium did not affect cell count for the T-cell line, as was expected, nor the TCL1A-transformed Tet2+/+ B cells, in contrast to Tet2−/− B cells that showed decreased survival rate (Figure 5C). It was previously shown that B cells treated with BTK inhibitors show an increase in BCR expression levels at the cell surface.42 Indeed, we detected a specific increase of IgM expression at the cell membrane of Tet2−/− B-cell line in comparison with the TCL1ATet2+/+ B-cell lines (Figure 5D). This result demonstrates a BCR dependency of the Tet2-deficient tumor B cells. On the basis of the cell survival data on the transformed Tet2−/− cell line, we next investigated the effect of ibrutinib on the cell viability of the primary Tet2−/− malignant cells. We found that ibrutinib exhibited a dose-dependent induction of cell death in malignant cells because the percentage of annexin V/7AAD negative cells (viable cells) decreased as doses increased (Figure 5E). Cell death induced by ibrutinib in primary Tet2−/− malignant cells was appreciable in comparison with cell death induced with the purine analog fludarabine (Figure 5E). These data underscore the importance of BCR signaling for the survival of Tet2−/− malignant B cells.
Discussion
Tet2 loss affects the level of 5hmC in murine hematopoietic progenitors, resulting in deregulated cytosine methylation and widespread hematopoietic differentiation defects. Approximately 30% of animals in our published Tet2-deficient models (constitutive Tet2LacZ ANO model and inducible Mx1-Cre model) develop a chronic myelomonocytic leukemia (CMML)–like disease in agreement with the high frequency of Tet2 mutations in human CMML disease. Interestingly, in these models another third of the mice shows accumulation with age of an abnormal B-cell population (B220lowCD19+).8 In the present study, using both constitutive and B-cell–specific (CD19-Cre) Tet2-deficient mice, we showed the malignant and cell-autonomous nature of these B-cell tumors, together with a restricted clonality and stereotypic VH CDR3 IgH rearrangement.
Young Tet2−/− mice show a moderate increase of the B1a compartment in the spleen and PEC. Molecular mechanisms underlying B1a expansion in young Tet2−/− mice need to be further investigated. Similar observations were reported in various mouse models that affect BCR signaling thresholds. When IgH-deficient hematopoiesis is rescued by the expression of the latent membrane protein 2a (LMP2a; which mimics BCR signaling), expressed from transgenic diversity region (DH) or variable region (VH) promoters, the strength of LMP2a signaling is shown to modulate the balance between peripheral B-cell populations: a high level of signaling (VH-LMP2a mice) induces a bias toward B1a development, whereas low signaling levels (DH-LMP2a mice) induce a bias toward follicular B-cell development.43 Further comparative analyses of B1a B cells and their progenitors (in fetal and adult mice) could provide new information on B1-cell differentiation/proliferation and new clues on CLL-like disease development, unraveling new therapeutic targets.
The long latency for developing the B-cell tumors indicates the requirement of cooperating mutations. Our data indicate that Aicda is mainly responsible for those mutations, as is described in CLL and other human B-cell lymphomas, because the frequency of B-cell tumors markedly decrease in Aicda- and Tet2-deficient animals, with respect to Aicda+/+Tet2−/− animals. However, in addition to AID’s function enhancing the ability of antibodies to bind and eliminate pathogens via the mechanism of somatic hypermutation in B cell, several studies in lymphoid and nonlymphoid tissues have demonstrated that AID can also participate in the loss of methylation through its deaminase activity, followed by base excision DNA repair and replacement with unmethylated C.17,33 Therefore we cannot exclude the possibility that AID could also participate, in the absence of Tet2, in B-cell tumor development via its DNA demethylation function. Pan et al18 reported phenotypically similar B-cell tumors in 5% of constitutive Tet2-knockout mice and underscored hypermutagenicity in the hematopoietic stem/progenitor. Our results confirm the development of lymphoid tumors in a Tet2-deficient context and point to AID activity as a major player in disease development. It was shown that Tet1 deficiency also gives rise to follicular lymphoma–like disease development13 in mice. Besides underscoring nonredundant roles for ten-eleven translocation (TET) proteins in normal hematopoiesis and transformation, this reinforces a general function of TET(s) and DNA-methylation control as a tumor suppressor of B-cell malignancies.13,44 A specific role of Tet3 in the B-cell lineage needs to be identified, however, because Tet3 inactivation does not markedly alter lymphopoiesis in mice (Ko et al45 and our unpublished results).
Recent work has shown the presence of somatic TET2 mutations in B-cell malignancies,8 up to 10% of DLBCLs in a specific series.10 Those DLBCLs are associated with the activated B-cell subtype and a specific DNA-methylation signature.10 This lymphoma subtype is highly sensitive to ibrutinib.46 We show that a Tet2-deficient gene signature may separate human FL samples from CLL and DLBCL (supplemental Figure 3C). Tet2-inactivated malignant B cells show transcriptional features of BCR signaling and BTK-inhibitor sensitivity but fail to clearly associate to any of the human lymphoma subtypes. This suggests that Tet2 deficiency paves the way for transformation driven by BCR signaling, which may correspond to DLBCL, CLL, and perhaps additional subtypes of mature B-cell malignancies.
There is growing evidence of hematopoietic progenitor alterations in malignancies of mature cell types,16 and the role of the deregulation of the control of cytosine methylation (through TET2 mutations) has been shown to endow the cells with a growth advantage at the early stage of hematopoietic differentiation. On the basis of observations from both mouse and human studies, it is currently accepted that TET2 deficiency predisposes to, but depends on, additional oncogenic hits to induce the development of full-blown hematological malignancies.47 In summary, our results support a direct effect of Tet2 inactivation on B-cell malignancy development and suggest that DNA-methylation control alterations induce an epigenetic background that predisposes it to the development of myeloid but also lymphoid diseases. Our results also indicate that TET2 loss plays a role at late stages of malignant development through deregulation of BCR signaling.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank the members of the Gustave Roussy Platforms, including Patrick Gonin for excellent mouse care, Yann Lecluse and Philippe Rameau for flow cytometry, and Olivia Bawa for histopathological analysis.
This work was supported by grants from INSERM, the Institut National du Cancer (2013-PLBIO-09, 2016-PLBIO-068, INCa-DGOS-INSERM 6043, and SIRIC-SOCRATE 2.0), the Fondation pour la Recherche Médicale, and the Association Laurette Fugain. This work was also supported by grants from Region Ile de France and Fondation de France (H.G.), Cancéropole Ile de France et Fondation pour la Recherche Médicale (C.K.L.), and the Dutch Cancer Society “KWF Kankerbestrijding” and the Daniel den Hoed Cancer Foundation (I.P.T.).
Authorship
Contribution: E.M., H.G., V.D.-V., L.S., C.Q., D.R.-W., P.P., V.S., and C.K.L. performed experiments and analyzed results; M.K.D. and P.D. analyzed the bioinformatics data; M.F. and I.P.T. analyzed results; E.M., H.G., T.M., S.A., and O.A.B. designed the project and experiments and wrote the manuscript; and all authors reviewed and approved the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Olivier A. Bernard, INSERM U1170, Gustave Roussy, 39 rue Camille Desmoulins, 94805 Villejuif Cedex, France; e-mail: olivier.bernard@inserm.fr.

![Figure 4. Tet2-deficient abnormal B cells share features with human B-cell tumors. (A) Coding sequence somatic mutations. Frequencies of the type of mutations observed in the exome analysis of abnormal B-cell populations obtained from 6 independent tumors. The type of point mutations is indicated as well as insertions and deletions (indels). Transversion mutations (green) and transitions (red) are indicated. Note the statistically significant high frequency of C > T and G > A mutations. (B) Survival curves of Aid+/+Tet2+/+ (control [CTR]), Aicda-deficient mice: Aid−/− Tet2+/+ (Aid−/−); Tet2-deficient mice: Aid+/+Tet2−/− (Tet2−/−); and double-deficient mice: Aid−/−Tet2−/− strains. Only mice sacrificed because of B-cell tumors are scored. (C) Venn diagram of exome mutations from the literature of human CLL27,31,34-36 and DLBCL32,37-39 in comparison with exome mutations from the Tet2-deficient sorted abnormal B cells. The name of the mutated genes in the overlaying zones is indicated. *P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/2/6/10.1182_bloodadvances.2017014118/4/m_advances014118f4.jpeg?Expires=1767776310&Signature=OuHw2rxJRpdkaKzhvfz5LJ5l9gATiezfaPA6BZ-lQ3NSjH6YCgE4JTyy2RDOfS8lK6IKakefrljzKXwrvAMeEx6enMgwBoMFicm0TtUjcneU97tCzrinZGPsIaTbxzO4O5z0UAp1c70GEH6dncF6vDpE6gSm7zPgQB5Z~o1PpYabu-JXaS5XeJgeeNb9NRr-YhhA3yaosPh26Pfyw-Xf2u4IsFvLp7y8h7ukTaNU2yXlujbK~Otit2YdYsQ0esX-dRZYNf27g4b5dWw8azYHEj4XcGtgWfNI7eKh1N5y7nn48MKaDiwLvD4CYfIoFM2hyQ7T~Fb~zvow9YKm-3RgZw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)