Key Points
Platelet-HMGB1 is required for efficient bacterial clearance in intra-abdominal polymicrobial sepsis.
Platelet HMGB1 promotes platelet activation, regulating platelet-neutrophil interactions and ROS production in neutrophils during sepsis.
Abstract
Thrombocytopenia impairs host defense and hemostasis in sepsis. However, the mechanisms of how platelets regulate host defense are not fully understood. High-mobility group box 1 (HMGB1), a danger-associated molecular pattern protein, is released during infection and contributes to the pathogenesis of sepsis. Platelets express HMGB1, which is released on activation and has been shown to play a critical role in thrombosis, monocyte recruitment, and neutrophil extracellular trap (NET) production. However, the contribution of platelet HMGB1 to host defense is unknown. To determine the role of platelet HMGB1 in polymicrobial sepsis, platelet-specific HMGB1 knockout (HMGB1 platelet factor 4 [PF4]) mice were generated and were subjected to cecal ligation and puncture (CLP), a clinically relevant intra-abdominal sepsis model. Compared with HMGB1 Flox mice and wild-type (WT) mice, HMGB1 PF4 mice showed significantly higher bacterial loads in the peritoneum and blood, an exaggerated systemic inflammation response, and significantly greater mortality after CLP. Deletion of HMGB1 in platelets was associated with lower platelet-derived chemokines (PF4 and RANTES) in the peritoneal cavity, and a decrease of platelet-neutrophil interaction in the lung after CLP. In vitro, neutrophils cocultured with activated HMGB1 knockout platelets showed fewer platelet-neutrophil aggregates, reduced reactive oxygen species (ROS) burst as compared with control. Taken together, these data reveal an unrecognized role of platelet HMGB1 in the regulation of neutrophil recruitment and activation via modulation of platelet activation during sepsis.
Introduction
Sepsis remains the most common cause of death in intensive care units.1 Much of the morbidity and mortality in sepsis can be attributed to a dysregulated immune response. Increasing evidence shows that in addition to their roles in hemostasis, platelets are an integral part of the innate immune response to infection.2
Platelets play important roles in pathogen detection as well as immune cell recruitment and activation during infection3 In addition, platelets also have direct antimicrobial functions that are mediated via degranulation and release of antimicrobial peptides, such as PF4 (platelet factor 4) and CCL5.3 It has been shown that platelet depletion impairs host defense and results in increased bacterial loads and viral titers,4 although the molecular mechanisms through which platelets regulate microbial clearance is not known.
High mobility group box 1 (HMGB1) is a nuclear DNA binding protein that can regulate intracellular processes and also be released into extracellular space through active or passive processes. When released into the extracellular milieu as a damage-associated molecular pattern molecule, HMGB1 can contribute to the pathogenesis of sepsis.5,6 The level of circulating HMGB1 is associated with the severity of sepsis in patients.6,7 Neutralization of extracellular HMGB1 after the onset of sepsis is protective in experimental models.6 Furthermore, HMGB1 has been shown to play cell-specific roles in sterile inflammation8-10 and endotoxemia.11
Platelets express and export HMGB1 to the cell surface on activation.12 Activated platelets present HMGB1 to neutrophils, and thus promote the formation of neutrophil extracellular traps in sterile conditions.13 We have shown that HMGB1 in platelets regulates systemic inflammation in a murine model of polytrauma.10 However, the role of platelet HMGB1 in the host response to bacterial infection remains unexplored. In the present study, we generated a strain of mice with HMGB1 specifically deleted from platelets and demonstrated a novel role for platelet HMGB1 in the recruitment of neutrophils and clearance of bacteria via regulation of platelet activation in a murine model of polymicrobial sepsis.
Materials and methods
Animals
Animal protocols were approved by the Institutional Animal Care and Use Committee of the University of Pittsburgh, and the experiments were performed in strict adherence to the NIH Guidelines for the Use of Laboratory Animals. Male C57BL/6 (WT) and PF4-Cre mice were specific pathogen-free, weighed approximately 25 g, and were from Jackson Laboratories. Male HMGB1loxP/loxP (HMGB1 Flox) mice and platelet-specific HMGB1 knockout (HMGB1 PF4) mice were bred and cohoused at our facility and used at the age of 8 to 12 weeks. All mice were developed on a C57BL/6 genetic background. HMGB1 Flox and PF4-HMGB1KO mice were generated and characterized as previously described.10
Cecal ligation and puncture
Sepsis was induced by cecal ligation and puncture (CLP). CLP was performed as previously described.14 Mice used were at the age of 8 to 12 weeks. After disinfecting the skin, laparotomy was performed under isoflurane anesthesia (Piramal Critical Care, Bethlehem, PA). Half the cecum was ligated and punctured twice with a 22-gauge needle, followed by resuscitation with 1 mL saline. Mice were kept warm at the blanket for 1 hour after surgery. Buprenorphine (0.1 mg/kg; Butler Schein, Dublin, OH) for analgesia was injected into mice subcutaneously (s.c.) every 12 hours immediately after surgery. At 18 hours after CLP, mice were euthanized and blood was collected by cardiac puncture. For survival study, 75% of the cecum was ligated and punctured twice with an 18-gauge needle. Mice were monitored twice daily by personnel experienced in recognizing signs of a moribund state. Mice were euthanized with CO2, when they became moribund or at observation endpoint (7 days).
Bacterial culture
Mice were euthanized at 18 hours after CLP. The peritoneal lavage fluid (PLF) was collected by washing the peritoneal cavity with 1 mL sterile phosphate-buffered saline (PBS). The left lung was removed and mechanically homogenized with 1 mL PBS. The PLF, lung homogenates, and blood were subjected to serial 10-fold dilution. Fifty microliters of each sample was applied to 5% sheep blood agar (Teknova). Colony-forming units were counted after overnight incubation at 37°C.
Assessment of cytokines
Plasma, PLF, and cell culture supernatant samples were analyzed using interleukin 6 (IL-6), IL-1β, MCP-1, PF4, and RANTES enzyme-linked immunosorbent assay (ELISA) kits (R&D systems, Inc.), according to manufacturer’s instructions.
Myeloperoxidase assay
Myeloperoxidase (MPO) activity was determined by commercially available MPO assay (Mouse MPO ELISA kit; Hycult Biotech Inc, Netherlands), according to manufacturer’s instructions, using peritoneal lavage fluid.
Isolation of platelets
Mice were anesthetized with isoflurane, and blood was drawn by cardiac puncture. Platelet-rich plasma was obtained by centrifugation at 260g × 5 minutes followed by another centrifugation 640g × 5 minutes to pellet the platelets in the presence of 0.5 mM PGE1 (Sigma-Aldrich). All centrifugations were performed without braking. The pellets were resuspended with HEPES-Tyrodes buffer (pH 7.4; supplemented with CaCl2, 1 mmol/L; MgCl2, 1 mmol/L) containing 0.5 mM PGE1.
Adoptive transfer of platelets
Platelets from HMGB1 flox or HMGB1 PF4 mice were isolated as described earlier, and 2 × 108 platelets were suspended in PBS and injected IV into HMGB1 PF4 mice 30 minutes before CLP.
Assessment of total P-selectin content in platelets
Isolated platelets (1 million) were collected from each strain of mice and lysed with 50 μL lysis buffer (Cell Signaling Technology). The total P-selectin content was measured by commercially available ELISA kit (R&D Systems).
Quantification of neutrophil extracellular traps
To quantify neutrophil extracellular traps (NETs) in peritoneal lavage fluid, a capture ELISA using MPO associated with DNA was performed as described previously.8 For the capture antibody, mouse MPO ELISA kit (Hycult Biotech Inc, Netherlands) was used according to the manufacturer’s directions. A peroxidase-labeled anti-DNA mAb (component No. 2, Cell Death ELISAPLUS, Sigma-Aldrich) was used. Serum nucleosome quantification was performed using Cell Death Kit (Sigma-Aldrich). Free serum DNA levels were quantified with the PicoGreen assay kit (Invitrogen).
Flow cytometry
The peritoneal cavity was washed with 10 mL sterile PBS. After centrifugation at 1800 rpm for 5 minutes at 4°C, the cell pellet was resuspended in 1 mL PBS, and the total cell number was counted under microscopy. The cell concentration was adjusted to 1 × 107/mL, and 1 × 106/mL was added to each tube. Cells were blocked for Fc receptors with anti-mouse CD16/32 (BD Bioscience) for 5 minutes and then were stained with fluorochrome-conjugated Ab for 30 minutes, at 4°C in dark. To detect the neutrophils percentage, peritoneal lavage fluid was labeled with FITC-CD11b (eBioscience), APC-CY7-LY6G (BD Biosciences). Neutrophils were identified as both CD11b- and Ly6G-positive cells. Data were acquired with a BD FACS LSR Fortessa flow cytometer (BD Bioscience) and analyzed with FlowJo analytical software (TreeStar). The neutrophil number in peritoneum lavage fluid was calculated by total cell number multiplied by neutrophil percentage. To assess the expression of P-selectin on the surface of platelets, platelets were stimulated with or without thrombin activated (thrombin 0.2 U/mL; Sigma-Aldrich) and labeled with PE-CD41 (BD Bioscience), APC-CD62P (P-selectin; eBioscience). The mean fluorescence intensity of P-selectin was assessed using flow cytometry.
In vitro neutrophil-platelet aggregation assay
WT neutrophils from bone marrow were isolated using a mouse neutrophil enrichment kit (STEMCELL Technologies, Vancouver, Canada). Twenty million platelets were treated with thrombin (0.2 U/mL) or collagen (0.5 mM) for 2 minutes and incubated with 1 million neutrophils for 5 minutes.13 To assess neutrophil-platelet aggregates, samples were labeled with antibodies against PE-CD41 (BioLegend), APC-Cy7-Ly6G (BD Bioscience). Platelet-neutrophil aggregates were identified as cells both positive to Ly6G and CD41, using flow cytometry.
Chemotactic assay
A chemotactic assay was performed using a Transwell system. Neutrophils (1.5 × 106) were placed in the upper chamber containing a 5-μm polycarbonate membrane (Corning). Inserts were placed in a 24-well plate with or without N-formyl-methionine-leucyl-phenylalanine (20 nM, Sigma-Aldrich) for 1 hour at 37°C. Neutrophil transmigration through the membrane pores into the lower chamber was assessed by flow cytometry. The chemotaxis index was calculated by the ratio of the cell number in the outside compartment with N-formyl-methionine-leucyl-phenylalanine and the cell number in medium-only culture. To assess the effect of platelet-derived HMGB1 in neutrophil recruitment, inserts were placed in a 24-well plate with platelets (2 × 108 cells/mL) plus HMGB1 neutralizing antibody (2G7, 1 μg/mL, from Kevin Tracy, Feinstein Institute for Medical Research, New York) or mouse immunoglobulin G (IgG; 1 μg/mL; R&D Systems). The chemotaxis index was calculated by the ratio of the cell number in the outside compartment with thrombin-treated platelets and the cell number in resting platelets.
Measurement of reactive oxygen species
Neutrophil oxygen radical production was determined by flow cytometry, as previously described.13 After incubation with DCFDA (40 μg/mL, Sigma-Aldrich) for 30 minutes on ice, neutrophils were washed and resuspended in HEPES-Tyrodes buffer. The neutrophils were then incubated with or without thrombin-activated (0.2 U/mL) or collagen-activated (0.5 mM) platelets (platelet:neutrophil ratio, 20:1) for 5 minutes at 37°C. Reactions were stopped by addition of equal volume of fixing solution, and cells were analyzed by flow cytometry to detect the FITC intensity
Immunofluorescence staining
For immunofluorescence staining, the right upper lobe of the lung was inflated, followed by perfusion-fixation with PBS and then 2% paraformaldehyde. Tissue sections (5 mm) were incubated with 5% Normal Goat Serum in PBS for 1 hour, followed by 5 washes with PBS containing 0.5% bovine serum albumin. The samples were then incubated with the following primary antibodies: anti-CD41 polyclonal antibody (2 μg/mL in PBS containing 0.5% bovine serum albumin, hamster IgG; Abcam) for 12 hours at 4°C and anti-Ly6G-APC (2 μg/mL in PBS containing 0.5% bovine serum albumin; BD Pharmigen) for 6 hours at room temperature. The secondary antibodies used in these experiments were as follows: Alexa 488-conjugated goat anti-rat IgG (1:500; for anti-CD41 antibody; Jackson Immuno Research). Nuclear staining was carried out with Bisbenzimide H33258 at a concentration of 20 mM (Sigma-Aldrich). Imaging was performed using a Nikon A1 confocal microscope (Nikon, purchased with 1S10OD019973-01 awarded to Simon C. Watkins). Quantification was performed using NIS Elements (Nikon, Melville, NY).
Cell lysis and immunoblot analysis
Snap-frozen lung (right lung) was homogenized in lysis buffer (Cell Signaling Technology) and centrifuged (13 000 rpm for 15 minutes), and then the supernatant was collected. Protein concentrations were determined using the bicinchoninic acid protein assay kit (Thermo Fisher Scientific). Loading buffer was added to the samples and separated by 10% and 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. The gel was then transferred to a polyvinylidene fluoride membrane at 250 mA for 2 hours. The membrane was blocked in 5% milk for 1 hour, and then incubated in primary antibody with 1% milk overnight. Membranes were washed in TBS-T (Tween) for 10 minutes and then placed in fluorescence conjugated secondary antibody for 1 hour and washed in TBS-T for 10 minutes 4 times. The primary antibodies used were: ICAM (1:1000, R&D Systems) and Rhodamine conjugated actin (1:5000; Bio-Rad). The secondary antibody was IRDye 800W donkey anti-goat (1:20 000, Licor). The signal was acquired and quantified with ChemiDoc MP Imagining System (Bio-Rad).
Statistical analysis
All data were analyzed using GraphPad Prism software (GraphPad Software). Data were analyzed using by Student t test. For measurements of bacterial colony-forming units, groups were compared using nonparametric Mann-Whitney U statistics. Survival data were analyzed using the log-rank test. P < .05 was considered statistically significant for all experiments. All values are presented as the mean ± SD, except for bacterial counts, for which median values are designate.
Results
HMGB1 PF4 mice showed enhanced lethality and impaired bacterial clearance during sepsis
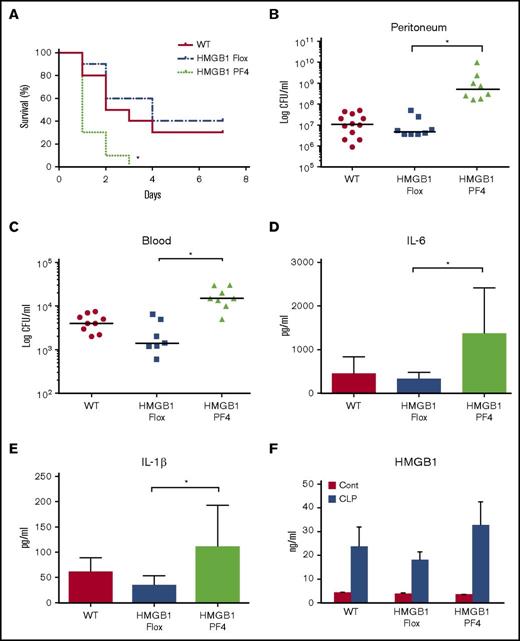
To establish the role of HMGB1 in platelets during polymicrobial sepsis, CLP was performed on HMGB1 flox and platelet-specific HMGB1 knockout (HMGB1 PF4) mice. The mortality rate of HMGB1 PF4 mice reached 100% on day 3 compared with only 40% in control mice (Figure 1A). Blood and peritoneal cavity bacterial counts at 18 hours in HMGB1 PF4 mice were significantly higher than WT or HMGB1 Flox mice (Figure 1B-C). In the CLP model, magnitude of systemic inflammatory response correlates with efficiency of bacterial clearance.14 Consistent with this, the impaired bacterial clearance in HMGB1 PF4 mice resulted in significantly higher circulating IL-6, IL-1β, and HMGB1 levels compared with those in the HMGB1 Flox mice (Figure 1D-F). The baseline levels of IL-6 and IL-1β in all 3 mouse strains were undetectable. These results indicate that platelet HMGB1 is required for efficient bacterial clearance during sepsis.
Platelet-HMGB1 regulates bacterial clearance and thus regulates survival and systemic inflammation after CLP. WT, HMGB1 Flox (cell-specific HMGB1 knockout control), and HMGB1 PF4 mice were subjected to CLP. (A) Seven-day survival after CLP. Data are from 20 mice per group from 2 separate experiments. Statistical difference was tested using the log-rank test. (B-C) Peritoneal lavage fluid and blood were collected at 18 hours after CLP. Bacterial counts in (B) peritoneum and (C) blood. Symbols represent individual mice. Statistical difference was tested using nonparametric Mann-Whitney U statistics. (D-F) Plasma cytokine levels. Blood was collected at 18 hours after CLP. Plasma (D) IL-6, (E) IL-1β, and (F) HMGB1 concentrations were measured by ELISA. Data are means ± SD from 8 mice per group from 2 separate experiments. Statistical difference was tested using Student t test; *P < .05
Platelet-HMGB1 regulates bacterial clearance and thus regulates survival and systemic inflammation after CLP. WT, HMGB1 Flox (cell-specific HMGB1 knockout control), and HMGB1 PF4 mice were subjected to CLP. (A) Seven-day survival after CLP. Data are from 20 mice per group from 2 separate experiments. Statistical difference was tested using the log-rank test. (B-C) Peritoneal lavage fluid and blood were collected at 18 hours after CLP. Bacterial counts in (B) peritoneum and (C) blood. Symbols represent individual mice. Statistical difference was tested using nonparametric Mann-Whitney U statistics. (D-F) Plasma cytokine levels. Blood was collected at 18 hours after CLP. Plasma (D) IL-6, (E) IL-1β, and (F) HMGB1 concentrations were measured by ELISA. Data are means ± SD from 8 mice per group from 2 separate experiments. Statistical difference was tested using Student t test; *P < .05
Platelet-HMGB1 regulates neutrophil recruitment during sepsis
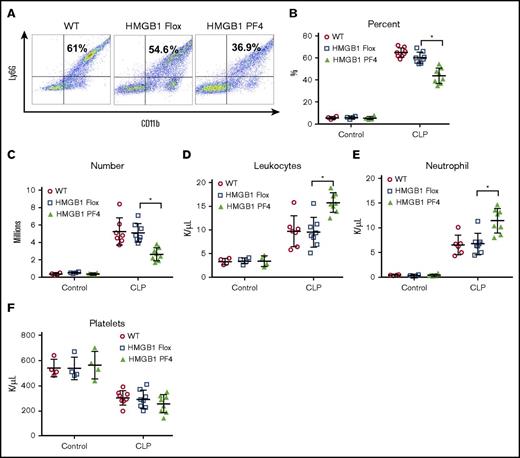
The migration of neutrophils into the peritoneal cavity after CLP is a key marker of an appropriate immune response in abdominal sepsis.15 Neutrophil recruitment to peritoneum was quantified by flow cytometry (Figure 2A). Both the percentage of total peritoneal cells identified as neutrophils and the absolute numbers of neutrophils were significantly lower in the HMGB1 PF4 mice as compared with HMGB1 Flox or WT mice after CLP (Figure 2B-C). This is a remarkable finding, considering that bacterial counts were higher in the peritoneum of the HMGB1 PF4 mice. There was no difference in peritoneal neutrophil numbers at baseline between the mouse strains (Figure 2B-C).
Platelet-HMGB1 regulates neutrophil recruitment during sepsis. WT, HMGB1 Flox, and HMGB1 PF4 mice were subjected to CLP for 18 hours. (A-B) Percentage of neutrophils was assessed by flow cytometry. Numbers indicate the percentage of CD11b and Ly6G double-positive cells. (C) The total number of neutrophils in the peritoneal cavity. (D) Circulating leukocytes, (E) neutrophils, and (F) platelets were measured in WT, HMGB1 Flox, and HMGB1 PF4 mice at 18 hours after CLP. Symbols represent individual mice. Data represent mean ± SD from 3 individual experiments. Statistical difference was tested using Student t test; *P < .05.
Platelet-HMGB1 regulates neutrophil recruitment during sepsis. WT, HMGB1 Flox, and HMGB1 PF4 mice were subjected to CLP for 18 hours. (A-B) Percentage of neutrophils was assessed by flow cytometry. Numbers indicate the percentage of CD11b and Ly6G double-positive cells. (C) The total number of neutrophils in the peritoneal cavity. (D) Circulating leukocytes, (E) neutrophils, and (F) platelets were measured in WT, HMGB1 Flox, and HMGB1 PF4 mice at 18 hours after CLP. Symbols represent individual mice. Data represent mean ± SD from 3 individual experiments. Statistical difference was tested using Student t test; *P < .05.
Despite the lower numbers of neutrophils found in the peritoneum in the PF4 HMGB1 mice, total circulating leukocytes and neutrophils were higher in these mice after CLP (Figure 2D-E). There were no baseline differences in leukocyte counts between the HMGB1 flox and PF4 HMGB1 strains (Figure 2D-E). The circulating platelet numbers dropped to half the baseline levels after CLP (Figure 2F). Importantly, there was no difference in circulating platelet numbers between the 3 strains at baseline or after CLP (Figure 2F), suggesting the observed deficit in peritoneal neutrophil numbers was not a result of difference in platelet numbers between transgenic strains.
We recently demonstrated that neutrophil expression of HMGB1 is identical between HMGB1 Flox mice and HMGB1 PF4 mice.16 Furthermore, there is no difference in the ability of neutrophils to form NETs between both strains.16 To test whether specific deletion of HMGB1 in platelets impairs neutrophil activation, the expression levels of surface CD11b on neutrophils as a marker of neutrophil activation17 were assessed using flow cytometry. The expression of surface CD11b increased significantly after LPS stimulation compared with control (supplemental Figure 1A). In addition, there was no difference in surface CD11b expression on neutrophils between 2 strains (supplemental Figure 1A). To test whether the impairment of neutrophil recruitment in HMGB1 PF4 mice is a result of the defect in neutrophil chemotaxis, a chemotaxis assay was performed using neutrophils isolated from HMGB1 Flox and HMGB1 PF4 mice. There was also no difference in in vitro neutrophil chemotaxis between the 2 strains, suggesting that the observed deficit in peritoneal neutrophil numbers was a result of the lack of HMGB1 on platelets and not an inherent difference in neutrophils between transgenic strains (supplemental Figure 1B).
Adoptive transfer of HMGB1 Flox platelets improves bacterial clearance and neutrophil recruitment in HMGB1 PF4 mice after CLP
To establish whether platelet HMGB1 regulates bacterial clearance and neutrophil influx during CLP-induced sepsis, we carried out adoptive transfer experiments. Thirty minutes before CLP, HMGB1 PF4 mice were injected IV with HMGB1+/+ platelet (Flox-Plt) or HMGB1−/− platelets (PF4-Plt; 2 × 108 cells/ mouse) isolated from HMGB1 Flox mice or HMGB1 PF4 mice, respectively. Mice were euthanized 18 hours after CLP. Bacterial counts in the blood and peritoneal fluid were significantly lower in the HMGB1 PF4 mice that received Flox-Plt than in mice that received HMGB1-deficient platelets (Figure 3A-B). Neutrophil counts in peritoneal fluid were inversely correlated to bacterial load (Figure 3C). As seen in previous experiments, circulating platelet numbers decreased to 50% of baseline level after CLP (Figure 3D). In addition, there was no significant difference in platelet numbers between mice that received platelets derived from either HMGB1 flox or HMGB1 PF4 mice (Figure 3D). Circulating IL-6 and HMGB1 levels correlated to bacterial load (Figure 3E-F). These data provide direct evidence that platelet HMGB1 regulates bacterial clearance and neutrophil recruitment in CLP-induced sepsis.
Adoptive transfer of HMGB1 Flox-platelets improves bacterial clearance and neutrophil recruitment in HMGB1 PF4 mice after CLP. Platelets were isolated from HMGB1 Flox (Flox-plt) or HMGB1 PF4 (PF4-plt) mice. Two hundred million platelets were suspended in PBS and injected IV into HMGB1 PF4 mice 30 minutes before CLP. (A-B) peritoneal lavage fluid and blood were collected at 18 hours after CLP. Bacterial counts in (A) blood and (B) peritoneum. Statistical difference was tested using a nonparametric Mann-Whitney U statistics. (C) The total number of neutrophils in the peritoneal cavity. (D) Circulating platelets numbers at 18 hours after CLP. (E-F) Plasma cytokine levels. Blood was collected at 18 hours after CLP. Plasma (E) IL-6 and (F) HMGB1 concentrations were measured by ELISA. Symbols represent individual mice. Statistical difference was tested using Student t test; *P < .05
Adoptive transfer of HMGB1 Flox-platelets improves bacterial clearance and neutrophil recruitment in HMGB1 PF4 mice after CLP. Platelets were isolated from HMGB1 Flox (Flox-plt) or HMGB1 PF4 (PF4-plt) mice. Two hundred million platelets were suspended in PBS and injected IV into HMGB1 PF4 mice 30 minutes before CLP. (A-B) peritoneal lavage fluid and blood were collected at 18 hours after CLP. Bacterial counts in (A) blood and (B) peritoneum. Statistical difference was tested using a nonparametric Mann-Whitney U statistics. (C) The total number of neutrophils in the peritoneal cavity. (D) Circulating platelets numbers at 18 hours after CLP. (E-F) Plasma cytokine levels. Blood was collected at 18 hours after CLP. Plasma (E) IL-6 and (F) HMGB1 concentrations were measured by ELISA. Symbols represent individual mice. Statistical difference was tested using Student t test; *P < .05
Platelet HMGB1 regulates neutrophil recruitment via modulation of platelet activation
Neutrophil recruitment to the infection site is driven by chemokines.18 Concentrations of MCP-1 and KC in PLF (Figure 4A-B) were significantly higher in HMGB1 PF4 mice than their controls at 18 hours after CLP. The baseline levels of these chemokines in all 3 mouse strains were undetectable. Thus, the levels of these chemokines appeared to correlate with bacterial levels, but not with the number of neutrophils in the peritoneal cavity.
Platelet-HMGB1 regulates neutrophil recruitment via modulation of PF4 release. (A-B) Peritoneal chemokine levels. Peritoneal lavage fluid was collected at 18 hours after CLP. Peritoneal (A) MCP-1 and (B) KC concentrations were measured by ELISA. Data are means ± SD from 8 mice per group from 2 separate experiments. (C) In vitro chemotactic assay. Isolated neutrophils were cocultured with platelets in presence of indicated treatment of 1 hour. Data represent mean ± SD from 2 individual experiments. (D-E) Peritoneal (D) PF4 and (E) RANTES concentrations were measured by ELISA. Data are means ± SD from 8 mice per group from 2 separate experiments. (F-G) Isolated platelets from HMGB1 Flox, and HMGB1 PF4 mice were stimulated with or without thrombin (0.2 U/mL) for 2 hours. The concentrations of (F) PF4 and (G) RANTES in media in resting and stimulated conditions were measured using ELISA. These experiments have been repeated for 2 times. Data represent mean ± SD. Statistical difference was tested using Student t test; *P < .05.
Platelet-HMGB1 regulates neutrophil recruitment via modulation of PF4 release. (A-B) Peritoneal chemokine levels. Peritoneal lavage fluid was collected at 18 hours after CLP. Peritoneal (A) MCP-1 and (B) KC concentrations were measured by ELISA. Data are means ± SD from 8 mice per group from 2 separate experiments. (C) In vitro chemotactic assay. Isolated neutrophils were cocultured with platelets in presence of indicated treatment of 1 hour. Data represent mean ± SD from 2 individual experiments. (D-E) Peritoneal (D) PF4 and (E) RANTES concentrations were measured by ELISA. Data are means ± SD from 8 mice per group from 2 separate experiments. (F-G) Isolated platelets from HMGB1 Flox, and HMGB1 PF4 mice were stimulated with or without thrombin (0.2 U/mL) for 2 hours. The concentrations of (F) PF4 and (G) RANTES in media in resting and stimulated conditions were measured using ELISA. These experiments have been repeated for 2 times. Data represent mean ± SD. Statistical difference was tested using Student t test; *P < .05.
HMGB1 is known to have chemotactic effects on monocytes by forming a complex with CXCL12.19 On activation, platelet can release HMGB1 into the extracellular space.20 To test whether platelet HMGB1 directly regulates neutrophil recruitment, 2G7, an HMGB1 neutralizing antibody, was used to block the activity of platelet-released HMGB1 in the chemotaxis assay. The chemotactic index increased significantly in the presence of thrombin-activated platelets compared with the control. However, there was no significant difference in the chemotactic index between the treatment of HMGB1 neutralizing antibody 2G7 and the IgG control (Figure 4C), suggesting that platelet HMGB1 did not directly regulate neutrophil recruitment.
Activated platelets degranulate α granules21 and release antimicrobial proteins, such as PF4 and chemokine (C-C motif) ligand 5 (RANTES), which are important for neutrophil recruitment.22-24 Interestingly, peritoneal levels of PF4 and RANTES were significantly lower in HMGB1 PF4 mice compared with HMGB1 Flox mice (Figure 4D-E). The baseline levels of these chemokines in the 3 mouse strains were undetectable. Furthermore, isolated HMGB1−/− platelets released significantly less PF4 and RANTES into media after thrombin stimulation compared with HMGB1+/+ platelets (Figure 4F-G). The baseline levels of these chemokines in the 3 mouse strains were undetectable. PF4 and RANTES appeared to be regulated independent of bacterial load and in a platelet HMGB1-dependent manner. These data suggest that platelet HMGB1 plays a critical role in platelet activation, as measured by chemokine release.
Platelets are known to regulate leukocyte recruitment via modulation of endothelial cell activation25 and adhesion molecule expression.26 However, no difference was observed in the circulating ICAM levels and the expression levels of ICAM-1 in the lung in HMGB1 Flox and HMGB1 PF4 mice after CLP (supplemental Figure 2A-B), suggesting the mechanism of platelet HMGB1 in regulation of neutrophil recruitment was not a result of the deficit in endothelial cell activation.
Platelet HMGB1 promotes platelet-neutrophil interactions
Platelets can interact and regulate neutrophil functions through the surface expression of P-selectin.10 We confirmed our previous observation that HMGB1-deficient platelets have less surface P-selectin after activation with thrombin (supplemental Figure 3A-B). Here we also showed that total cellular P-selectin levels are not different between the strains (supplemental Figure 3C), confirming that HMGB1 regulates the surface P-selectin levels after activation, not the total P-selectin expression in the platelets. Platelet-neutrophil interactions play important roles in host defense via regulation of neutrophil recruitment and functions.15,20 Interestingly, we observed that the number of colocalized neutrophils and platelets increased significantly in the lung after CLP (Figure 5A). Importantly, the number of colocalized neutrophils and platelets in the lung of HMGB1 Flox mice was significantly higher than that in HMGB1 PF4 mice (Figure 5A). Thrombin has been shown to induce platelet-neutrophil interactions. In vitro, platelets isolated from either HMGB1 Flox mice or HMGB1 PF4 mice were activated using thrombin or collagen and were then cocultured with neutrophils isolated from WT mice. Platelets lacking HMGB1 formed fewer platelet-neutrophil aggregates compared with HMGB1-containing platelets in response to collagen or thrombin (Figure 5B-C). These data suggest that platelet HMGB1 promotes platelet-neutrophil interaction during sepsis.
Platelet-HMGB1 promotes platelet-neutrophil interactions and neutrophil activations. (A) Lung immunofluorescence in mice at 18 hours after CLP. Green, CD41; Red, Ly6G; blue, nucleus. Scale bar, 50 µm (images in the right column were magnified ×5). Colocalization of CD41 and Ly6G is shown as yellow. Quantification was performed using NIS Elements. Percentage of Ly6G+ nuclei and percentage of CD41+ neutrophils were calculated. (B-C) Platelets were isolated from HMGB1 Flox and HMGB1 PF4 mice. Twenty million platelets were treated with (B) collagen (0.5 mM) or (C) thrombin (0.2 U/mL) for 2 minutes and incubated with 1 million isolated WT neutrophils for 5 minutes. Formation of platelet-neutrophil aggregation was assessed using flow cytometry. These experiments have been repeated 4 times. Data represent mean ± SD. Statistical difference was tested using Student t test; *P < .05.
Platelet-HMGB1 promotes platelet-neutrophil interactions and neutrophil activations. (A) Lung immunofluorescence in mice at 18 hours after CLP. Green, CD41; Red, Ly6G; blue, nucleus. Scale bar, 50 µm (images in the right column were magnified ×5). Colocalization of CD41 and Ly6G is shown as yellow. Quantification was performed using NIS Elements. Percentage of Ly6G+ nuclei and percentage of CD41+ neutrophils were calculated. (B-C) Platelets were isolated from HMGB1 Flox and HMGB1 PF4 mice. Twenty million platelets were treated with (B) collagen (0.5 mM) or (C) thrombin (0.2 U/mL) for 2 minutes and incubated with 1 million isolated WT neutrophils for 5 minutes. Formation of platelet-neutrophil aggregation was assessed using flow cytometry. These experiments have been repeated 4 times. Data represent mean ± SD. Statistical difference was tested using Student t test; *P < .05.
Platelet HMGB1 promotes reactive oxygen species production but not NETs formation by neutrophils
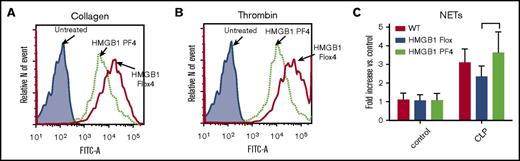
Production of reactive oxygen species (ROS)27 and formation of NETs28 are 2 important mechanisms for effective antimicrobial activity by neutrophils. To ascertain whether platelet HMGB1 had an effect on neutrophil ROS production in vitro, we cocultured neutrophils with platelets and measured ROS production by flow cytometry. ROS production by neutrophils cocultured with activated HMGB1-deficient platelets was significantly lower compared with neutrophils cocultured with platelets that expressed HMGB1 (Figure 6A-B). Thus, platelet HMGB1 also contributes to the capacity of platelets to enhance neutrophil ROS production.
Platelet-HMGB1 promotes ROS production but not NETs formation in neutrophils. (A-B) Platelets were isolated from HMGB1 Flox and HMGB1 PF4 mice. Twenty million platelets were treated with collagen (0.5 mM) or thrombin (0.2 U/mL) for 2 minutes and incubated with 1 million isolated WT neutrophils for 5 minutes. Representative flow cytometry histograms depict the ROS generations in neutrophils in the coculture with (A) collagen- or (B) thrombin-stimulated platelets. The histograms were representative of 4 experiments. (C) MPO-DNA complex levels in the peritoneal cavity. MPO-DNA complex levels in peritoneal lavage fluid were assessed in WT, HMGB1 Flox, and HMGB1 PF4 mice at 18 hours after CLP. Data represent means ± SD from 8 mice per group.
Platelet-HMGB1 promotes ROS production but not NETs formation in neutrophils. (A-B) Platelets were isolated from HMGB1 Flox and HMGB1 PF4 mice. Twenty million platelets were treated with collagen (0.5 mM) or thrombin (0.2 U/mL) for 2 minutes and incubated with 1 million isolated WT neutrophils for 5 minutes. Representative flow cytometry histograms depict the ROS generations in neutrophils in the coculture with (A) collagen- or (B) thrombin-stimulated platelets. The histograms were representative of 4 experiments. (C) MPO-DNA complex levels in the peritoneal cavity. MPO-DNA complex levels in peritoneal lavage fluid were assessed in WT, HMGB1 Flox, and HMGB1 PF4 mice at 18 hours after CLP. Data represent means ± SD from 8 mice per group.
Platelets can induce neutrophils to form NETs during sepsis.28 We recently showed that the capacity of NETs formation by neutrophils from HMGB1 PF4 mice is not altered compared with neutrophils from HMGB1 Flox mice.16 Consistent with this observation, we found that there was no difference in peritoneal MPO-DNA complex level between any of the strains after CLP (Figure 6C). These results indicate that platelet HMGB1 is not required for NET formation during sepsis.
Discussion
HMGB1 has been described as a central mediator of the host response to sepsis.5,6 Although it is expressed by essentially every cell type, many of its cell-specific roles in sepsis are not known. Platelets are known to play an important role in the host response to bacterial infection,2,4,20,28,29 and here we show that platelet HMGB1 is essential for effective bacterial clearance in an intra-abdominal polymicrobial sepsis model. Our studies show that HMGB1 is essential for platelet activation and, through the regulation of platelet activation, also regulates neutrophil chemotaxis and ROS production.
Thrombocytopenia is associated with worse outcomes in septic patients.29,30 Depletion of platelets impairs host defense and results in increased mortality in experimental Klebsiella pneumonia.4 Our study indicates that 1 mechanism for efficient bacterial clearance regulated by platelets is through platelet HMGB1 expression that is essential for platelet activation, and thus promotes neutrophil accumulation and activation at the site of infection. The findings of a critical role for platelet HMGB1 in microbial clearance are in sharp contrast to the deleterious role for platelet HMGB1 that we have recently uncovered in trauma and hemorrhagic shock.10 In this noninfectious model of systemic inflammation, platelet HMGB1 contributes to microthrombi formation in tissues. The observation of unique roles in these different disease states highlights an important difference in platelet response to infectious and noninfectious stimuli, despite similar signaling pathways.
HMGB1 is well characterized as a late instigator of the inflammatory response associated with lethality during sepsis, and is known to have cellular specific roles. Although HMGB1 from other cell types, such macrophages,31 may exert deleterious roles in sepsis, the findings presented here suggest that platelet HMGB1 is essential to survival by promoting bacterial clearance. We postulate that the increase in the systemic inflammatory response seen in the platelet-specific HMGB1 knockout mice after CLP is a result of the failure to efficiently clear bacteria.
HMGB1 is known to promote recruitment of inflammatory cells to damaged tissues by forming a complex with CXCL12.19 Platelet-derived HMGB1 has been demonstrated to be the major source of HMGB1 enhancing the recruitment of monocyte in a mouse deep venous thrombosis model.9 Interestingly, in this polymicrobial sepsis model, systemic HMGB1 levels were higher in the platelet HMGB1-deficient mice after CLP, indicating that platelets may not be an important source of circulating HMGB1 in sepsis. The increased inflammatory response elicited by the higher bacterial counts may lead to HMGB1 release from other sources.
Platelets play important roles in neutrophil recruitment. Activated platelets have been shown to directly interact with neutrophils under inflammatory conditions.32 Platelet-neutrophil interactions are necessary for neutrophil migration and activation.20 Our data indicate that platelet HMGB1 regulates neutrophil recruitment and platelet-neutrophil interaction through release of platelet-derived chemokines, PF4 and RANTES, as well as decreased surface expression of P-selectin. These platelet-derived chemokines are crucial in neutrophil recruitment.22-24 Furthermore, PF4, RANTES, and P-selectin are components in platelet α granules, which are released from platelet on activation.21 Our results suggest that endogenous HMGB1 in platelet regulates α granule degranulation on activation. However, further studies are required to elucidate the mechanism of how HMGB1 regulates this process.
The ROS generation capacity of neutrophils increased significantly when neutrophils interact with activated platelets via P-selectin.13,33 Previously we have shown that extracellular HMGB1 induces NADPH oxidase activation in neutrophils in a TLR4-dependent manner.34 Here we show that HMGB1 in platelets is required for the activation of neutrophils and is associated with ROS production.
In conclusion, we demonstrate that platelet HMGB1 is critical for platelet activation, which in turn regulates neutrophil recruitment and activation at the infection site for effective bacterial clearance during intra-abdominal sepsis. These findings indicate the importance of understanding the diverse and cell-specific roles of HMGB1 in the integrated host response to serious infections.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank Hong Liao for her excellent technical support.
This work was supported by grants from the National Institutes of Health, National Institute of General Medical Sciences (2RO1GM044100 and 2RO1GM050441 [T.R.B]; 1R35GM119526-01 [M.D.N.]; and R01-GM-102146 [M.J.S]).
Authorship
Contribution: M.D., M.D.N., and T.R.B. conceived the project and designed experiments and edited the paper; H.Z., Y.L., C.Y., J.Z., R.H., P.A.L., and M.J.S. performed the experiments and analyzed the data; and H.Z. and M.D. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Timothy R. Billiar, Department of Surgery, UPMC Presbyterian Hospital, Pittsburgh, PA 15213; e-mail:billiartr@upmc.edu; and Matthew D. Neal, UPMC Presbyterian Hospital, Suite F1281, Pittsburgh, 15213; e-mail: nealm2@upmc.edu.
References
Author notes
H.Z. and M.D. are joint first authors.