Key Points
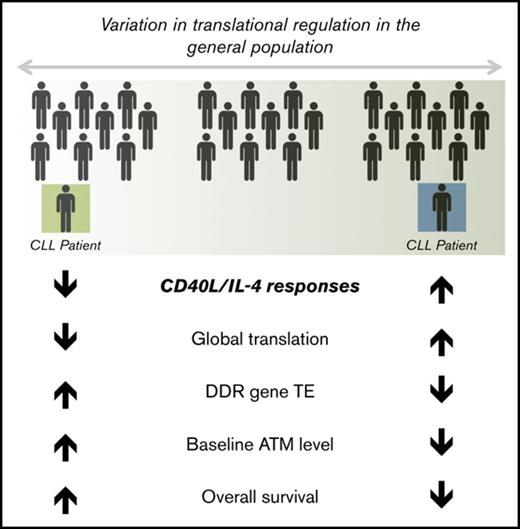
CD40L/IL-4 responses mediate translational regulation of DNA damage repair genes, including ATM, and associate with baseline levels of ATM.
Lower levels of baseline ATM, independent of 11q deletion, associate with reduced overall survival.
Abstract
CD40L/interleukin-4 (IL-4) stimulation occurs in vivo in the tumor microenvironment and induces global translation to varying degrees in individuals with chronic lymphocytic leukemia (CLL) in vitro. However, the implications of CD40L/IL-4 for the translation of specific genes is not known. To determine the most highly translationally regulated genes in response to CD40L/IL-4, we carried out ribosome profiling, a next-generation sequencing method. Significant differences in the translational efficiency of DNA damage response genes, specifically ataxia‐telangiectasia–mutated kinase (ATM) and the MRE11/RAD50/NBN (MRN) complex, were observed between patients, suggesting different patterns of translational regulation. We confirmed associations between CD40L/IL-4 response and baseline ATM levels, induction of ATM, and phosphorylation of the ATM targets, p53 and H2AX. X-irradiation was used to demonstrate that CD40L/IL-4 stimulation tended to improve DNA damage repair. Baseline ATM levels, independent of the presence of 11q deletion, correlated with overall survival (OS). Overall, we suggest that there are individual differences in translation of specific genes, including ATM, in response to CD40L/IL-4 and that these interpatient differences might be clinically important.
Introduction
Increased global messenger RNA (mRNA) translation is a feature of cancer,1 and mouse models show that overexpression of the eukaryotic initiation factor-4E (eIF-4E) is sufficient for the development of lymphoma.2 The translation machinery might also be a target for therapy in hematological malignancy.3,4 It is becoming clear that increased translation in cancer not only has global effects on growth but also alters the expression of specific critical genes to modify the effects of oncogene signaling.5-7
The tumor microenvironment provides signals for the survival and proliferation of chronic lymphocytic leukemia (CLL) cells.8,9 CLL cells cultured with fibroblasts expressing CD40L and IL-4 (CD40L/IL-4)10 demonstrate increased global protein synthesis11 as compared with unstimulated peripheral blood leukemic cells. Similarly, cross-linking of surface immunoglobulin M (IgM) also markedly increases translation.12 These 2 microenvironment model systems, mimicking T-cell engagement and B-cell receptor cross-linking, respectively; therefore, both strongly drive global mRNA translation, possibly through induction of c-MYC.12,13 There is interpatient variation in the magnitude of the downstream effects of CD40L signaling14 or IgM cross-linking15 and these responses are associated with markers of clinical outcome. We hypothesized that translation of specific genes in response to microenvironment stimuli differed between patients and that this variation contributed to variation in clinical outcome, possibly by modifying the effects of recurrent mutations or chromosomal aberrations.
The disease-specific translatome of unstimulated CLL has been determined by polysome profiling coupled to microarray analysis16 to demonstrate reduction in ribosomal proteins compared with control B cells. However, broad application of the polysome-profiling approach is hindered by the technical difficulty of polysome fractionation, which does not cleanly resolve fractions that have more than a few ribosomes per transcript, and by the need to collect and analyze many fractions per sample.
In this report, we use a superior method, ribosome profiling, to dissect the translatome. The principle underlying this technique is that changes to the occupancy of ribosomes by specific mRNAs are directly and quantitatively related to their translation.17,18 Sequencing of ribosome protected fragments (RPFs) of mRNA by next-generation sequencing and simultaneous sequencing of the whole transcriptome, produces a measure called translational efficiency (TE), a ratio of RPF reads to mRNA transcript reads, which gives a measure of ribosome density on specific mRNAs that is used to compare the translation of specific genes.19 In turn, this produces a detailed understanding of how individual genes or pathways are regulated under specific experimental conditions.
We determine, for the first time by ribosome profiling, the CD40L/IL-4–stimulated CLL translatome to demonstrate which genes and signaling pathways are differentially expressed between patients, and show that DNA damage response genes, especially those from the MRE11, RAD50, and NBN (MRN) complex and ataxia‐telangiectasia–mutated kinase (ATM)-signaling pathway, are translationally regulated. This work has implications for understanding individual differences in clinical outcome that may modify the expected effects of recurrent genetic mutations.
Materials and Methods
Patient samples
We used several cohorts of patients in the study: a cohort of 37 patients for analysis of CD40L/IL-4 responses (cohort A), a cohort of 10 patients for ribosome profiling (cohort B, contained within cohort A), a cohort of 11 patients for validation of the ribosome profiling by western blots and functional studies (cohort C), and a cohort of 57 patients for survival analysis (cohort D).
Patients provided written informed consent in accordance with Research Ethics Committee approvals (06/Q2501/122) and the Declaration of Helsinki. Heparinized peripheral blood mononuclear cells (PBMCs) were obtained from patients attending clinics at the Leicester Royal Infirmary (clinical characteristics described in supplemental Tables 1 [cohort A and cohort B], 2 [cohort C], and 3 [cohort D]). Diagnosis of CLL was made according to the International Workshop on Chronic Lymphocytic Leukemia–National Cancer Institute (IWCLL-NCI) 2008 criteria.20 Samples were obtained from patients who had either never been treated (supplemental Tables 1 and 2) or were >6 months from any treatment (supplemental Table 3).
Cell culture and assays
CLL cells (3 × 106/mL) were cultured in RPMI 1640 medium (Thermo Fisher, Waltham, MA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher), nonessential amino acids (Thermo Fisher), penicillin/streptomycin (Thermo Fisher), and N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) buffer (Lonza, Basel, Switzerland) at 37°C in a 5% CO2 incubator. For ribosome profiling, cells were either cultured on tissue-culture plastic or cocultured with 80% to 90% confluent and 35 Gy irradiated nontransfected mouse fibroblast cells (NTL) or human CD40L-expressing mouse fibroblast cells supplemented with recombinant human (rh) IL-4 (10 ng/mL; R&D Systems, Minneapolis, MN) (CD40L/IL-4).21,22 Alternatively, CLL cells were cultured on tissue-culture plastic or activated using 100 ng/mL His-rh soluble CD40L (sCD40L; R&D Systems), 20 ng/mL rh-IL-4 (R&D Systems), and 500 ng/mL anti-His-tag antibody (R&D Systems) for 24 to 48 hours. Adenosine triphosphate (ATP) luminescence was measured by CellTiterGlo (Promega, Madison, WI).
Other methods are found in supplemental Methods.
Results
CD40L/IL-4 responses correlate with global translation and expression of eIF-4E and 4EBP1
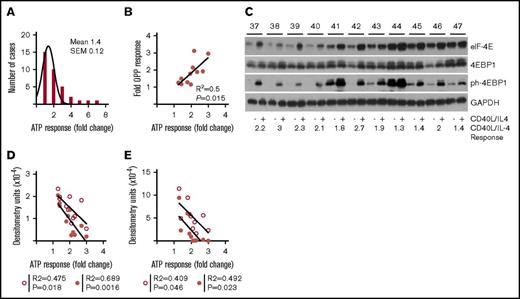
To establish a range of response to CD40L/IL-4, we used ATP luminescence (a combined measure of viability and proliferation) in a group of patients (n = 37) (Figure 1A; supplemental Figure 1A-B; supplemental Table 1). As anticipated, culture on a fibroblast layer (NTL) produced an increment over culture on plastic (paired 2-tailed Student t test, P = .0013), and the addition of CD40L/IL-4 produced a further increment (P < .0001). CD40L/IL-4 produced a 2.3- ± 0.13-fold (mean ± standard error of the mean [SEM]) increase in ATP luminescence compared with tissue-culture plastic (supplemental Figure 1B), but a 1.4- ± 0.12-fold increase in comparison with NTL (Figure 1A). In this paper, we will refer to improvement in viability due to CD40L/IL-4 as ATP response.
Increased global translation is a characteristic of CD40L/IL-4 stimulation. (A) CLL cells were cultured either on tissue culture plastic (PL), untransfected mouse fibroblasts (NTL) or mouse fibroblasts expressing CD40L and with IL-4 in the medium (CD40L/IL-4). Histogram showing fold-increases in ATP luminescence comparing CD40L/IL-4 with NTL. The mean increase is 1.4-fold (SEM = 0.12) (n = 37). (B) Association between fold increase in ATP luminescence (comparing CD40L/IL-4 and PL conditions) and incorporation of OPP (a measure of new protein synthesis; n = 11) (R2 = 0.5, P = .015). (C) Western blots showing the effects of CD40L/IL-4 culture on expression of eIF4E, 4EBP1, and ph-4EBP1 (n = 11). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is the loading control. Patient identification (ID) is given above (labeled 37-47) and fold response to CD40L/IL-4 stimulation below the autorads. (D) Correlation of eIF-4E levels without stimulation (solid circles) or following CD40L/IL-4 stimulation (open circles) with ATP response (without stimulation R2 = 0.689, P = .0016 and following stimulation R2 = 0.475, P = .018). (E) Correlation of phosphorylated-4EBP1 levels without stimulation (solid circles) or following CD40L/IL-4 stimulation (open circles) with ATP response (without stimulation R2 = 0.492, P = .023 and following stimulation R2 = 0.409, P = .046).
Increased global translation is a characteristic of CD40L/IL-4 stimulation. (A) CLL cells were cultured either on tissue culture plastic (PL), untransfected mouse fibroblasts (NTL) or mouse fibroblasts expressing CD40L and with IL-4 in the medium (CD40L/IL-4). Histogram showing fold-increases in ATP luminescence comparing CD40L/IL-4 with NTL. The mean increase is 1.4-fold (SEM = 0.12) (n = 37). (B) Association between fold increase in ATP luminescence (comparing CD40L/IL-4 and PL conditions) and incorporation of OPP (a measure of new protein synthesis; n = 11) (R2 = 0.5, P = .015). (C) Western blots showing the effects of CD40L/IL-4 culture on expression of eIF4E, 4EBP1, and ph-4EBP1 (n = 11). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is the loading control. Patient identification (ID) is given above (labeled 37-47) and fold response to CD40L/IL-4 stimulation below the autorads. (D) Correlation of eIF-4E levels without stimulation (solid circles) or following CD40L/IL-4 stimulation (open circles) with ATP response (without stimulation R2 = 0.689, P = .0016 and following stimulation R2 = 0.475, P = .018). (E) Correlation of phosphorylated-4EBP1 levels without stimulation (solid circles) or following CD40L/IL-4 stimulation (open circles) with ATP response (without stimulation R2 = 0.492, P = .023 and following stimulation R2 = 0.409, P = .046).
To assess the association of ATP response with global translation we determined incorporation of labeled O-propargyl puromycin (OPP), a measure of new peptide synthesis (Figure 1B), and found that a higher ATP response correlated with greater OPP incorporation (R2 = 0.5, P = .015).
Constitutive B-cell expression of eIF-4E is sufficient to drive lymphomagenesis in mice.2 The expression of eIF-4E and an interacting protein in the cap complex, 4EBP1, was determined (n = 11) (supplemental Table 2) to support our hypothesis that there are individual differences in translation between patients (Figure 1C). Interpatient differences in expression of eIF-4E, both at baseline and after CD40L/IL-4 stimulation, were observed (compare patients 37-40 with patients 44-47). There was less variation in levels of 4EBP1, but activated phosphorylated 4EBP1 (ph-4EBP1) again showed interpatient variation in levels, both at baseline and following stimulation. Quantitation by densitometry demonstrated that eIF-4E or phosphorylated-4EBP1 levels at baseline levels or following CD40L/IL-4 stimulation (Figure 1D-E) both associate with a higher ATP responses to CD40L/IL-4 (Mann-Whitney U test, P = .004). There is a significant difference between patients 37 to 40 and patients 44 to 47 in baseline eIF-4E (Mann-Whitney U test, P = .028) but not for induced eIF-4E (P = .057).
The implications of interpatient variation in global translation for the expression of specific genes has not previously been determined in CLL, yet such changes could modify signaling or metabolic pathways. To explore this question, we carried out ribosomal profiling and tandem RNA sequencing (supplemental Figure 2A-B) on primary leukemic cells from 10 patients (supplemental Table 1).
Ribosome profiling of CLL cells: clustering determined by translational efficiency
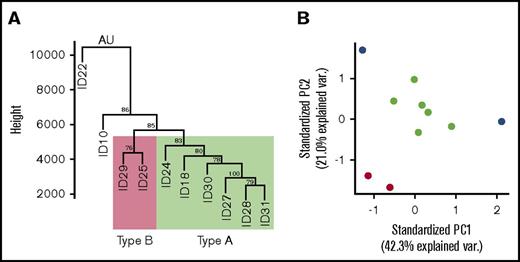
We examined TE following CD40L/IL-4 stimulation to determine interpatient differences in RPFs on specific mRNAs. Cells were stimulated in vitro to drive global translation such that any differences that we observed between the TEs of specific genes are likely to be due to individual differences in translational regulation of specific genes. Hierarchical clustering of TEs showed that patients 18, 24, 27, 28, 30, and 31 (subsequently type A) and patients 25 and 29 (type B) formed clusters with patients 10 and 22 being outliers (Figure 2A). ATP responses were similar between the clusters and this is likely to be because ribosome profiling has a much greater sensitivity to detect interpatient differences than the ATP luminescence read-out for CD40L/IL-4 stimulation. A principal component analysis was carried out to confirm the hierarchical clustering result (Figure 2B). Clusters corresponding to types A and B were again detected, with patients 10 and 22 being outliers.
Clustering analysis of translational efficiencies. (A) The TE values for each detected gene were used to cluster samples using Mcquitty linkage with a Manhattan distance matrix; values at branch points are approximately unbiased (AU) P values, calculated by multiscale bootstrap resampling using the pvclust R package. Patients 18, 24, 27, 28, 30, and 31 (type A) are denoted within the green rectangle; patients 25 and 29 (type B) are denoted within the red rectangle. (B) The TE values for each detected gene were used in a principal components analysis. The outlying patients (10 and 22) are illustrated as blue dots, while type A are green dots and type B are red dots.
Clustering analysis of translational efficiencies. (A) The TE values for each detected gene were used to cluster samples using Mcquitty linkage with a Manhattan distance matrix; values at branch points are approximately unbiased (AU) P values, calculated by multiscale bootstrap resampling using the pvclust R package. Patients 18, 24, 27, 28, 30, and 31 (type A) are denoted within the green rectangle; patients 25 and 29 (type B) are denoted within the red rectangle. (B) The TE values for each detected gene were used in a principal components analysis. The outlying patients (10 and 22) are illustrated as blue dots, while type A are green dots and type B are red dots.
ATM and MRN complex genes differ in their translational efficiencies between the grouped patients
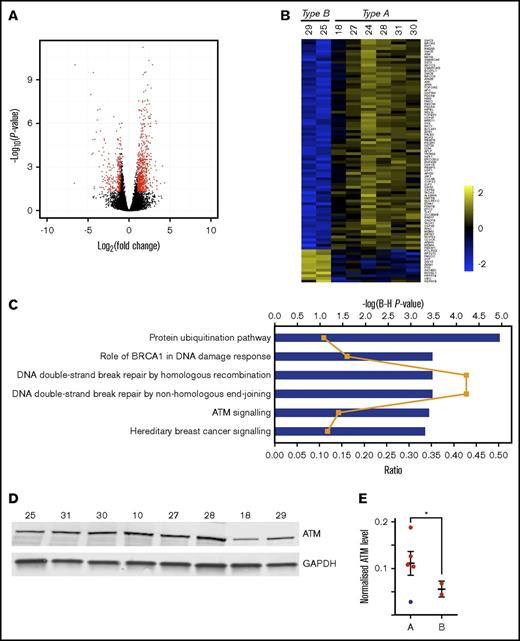
To carry out a functional analysis of genes with TEs that differ between types A and B we used Xtail,23 a tool for quantifying differential translation by comparing changes in mRNA expression with changes in RPF abundance, and finding cases where these changes are dissimilar (supplemental Figure 3). The volcano plot (Figure 3A) demonstrates those genes with a fold change of at least 2, and a P < .05; 176 genes are translated less efficiently, and 572 are translated more efficiently, within type A than type B (Figure 3B; supplemental Table 4).
Significant differences in translational efficiencies of DNA damage repair pathway genes between patient clusters. (A) Volcano plot showing the results of the Xtail analysis. Genes highlighted in red are those with a fold change of at least 2, and an adjusted P < .05. The final list of 748 changing genes discards genes with low raw read counts (supplemental Table 4; “Materials and methods”). (B) Heat map showing the log2 TE values for the 88 genes listed in Figure 4. (C) The 748 genes that are differentially translated between type A and type B were subjected to a function and pathway enrichment analysis using Qiagen’s Ingenuity Pathway Analysis tool (IPA). Plot shows enriched canonical pathways at adjusted P < .001 (−log(Benjamini-Hochberg P value) shown as blue bars), with ratio (the proportion of the genes in the pathway that are also found in our list) shown as orange line. There are some pathways in this plot that do not appear relevant to the functioning of B lymphocytes. Their presence is explained by the reuse of the same signaling proteins in various pathways in different cell lineages. (D) Western blot analysis of ATM protein expression for 8 samples (10, 18, 25, 27, 28, 29, 30, and 31) subjected to ribosome profiling. No material was available for samples 22 and 24. (E) Normalized ATM levels (ATM fluorescence signal to GAPDH fluorescence signal) is shown for type A and type B patients. Error bars show mean ± SEM. Sample 18 (blue dot) in type A patients, was found to bear an ATM mutation (supplemental Table 1) and was excluded from the statistical analysis. Normalized ATM levels are significantly higher in type A than type B (unpaired 2-tailed Student t test, *P = .02).
Significant differences in translational efficiencies of DNA damage repair pathway genes between patient clusters. (A) Volcano plot showing the results of the Xtail analysis. Genes highlighted in red are those with a fold change of at least 2, and an adjusted P < .05. The final list of 748 changing genes discards genes with low raw read counts (supplemental Table 4; “Materials and methods”). (B) Heat map showing the log2 TE values for the 88 genes listed in Figure 4. (C) The 748 genes that are differentially translated between type A and type B were subjected to a function and pathway enrichment analysis using Qiagen’s Ingenuity Pathway Analysis tool (IPA). Plot shows enriched canonical pathways at adjusted P < .001 (−log(Benjamini-Hochberg P value) shown as blue bars), with ratio (the proportion of the genes in the pathway that are also found in our list) shown as orange line. There are some pathways in this plot that do not appear relevant to the functioning of B lymphocytes. Their presence is explained by the reuse of the same signaling proteins in various pathways in different cell lineages. (D) Western blot analysis of ATM protein expression for 8 samples (10, 18, 25, 27, 28, 29, 30, and 31) subjected to ribosome profiling. No material was available for samples 22 and 24. (E) Normalized ATM levels (ATM fluorescence signal to GAPDH fluorescence signal) is shown for type A and type B patients. Error bars show mean ± SEM. Sample 18 (blue dot) in type A patients, was found to bear an ATM mutation (supplemental Table 1) and was excluded from the statistical analysis. Normalized ATM levels are significantly higher in type A than type B (unpaired 2-tailed Student t test, *P = .02).
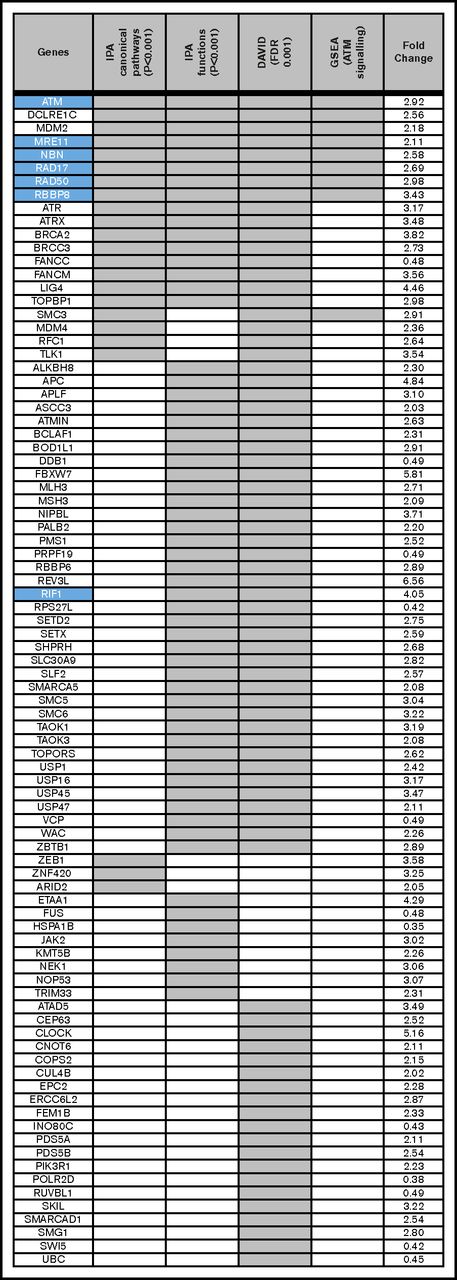
The 748 genes that are differentially translated between types A and B were subjected to a function and pathway enrichment analysis using Qiagen’s Ingenuity Pathway Analysis tool (IPA; qiagenbioinformatics.com/products/ingenuity-pathway-analysis), the DAVID Functional Annotation tool,24 and Gene Set Enrichment Analysis (GSEA).25 Enriched canonical pathways at adjusted P < .001 (Figure 3C) show that the top 2 pathways in terms of ratio (the proportion of the genes in the pathway that are also found in our list) are “DNA double-strand break repair by homologous recombination” and “DNA double-strand break repair by non-homologous end joining.” ATM signaling, which is an important component of the response to DNA damage, is also enriched. The enrichment of genes in the ATM-signaling pathway is confirmed by GSEA (Figure 4). The enrichment of genes involved in DNA damage response is also confirmed by an analysis of Biological Process gene ontology (GO) terms attached to the genes, using the DAVID Functional Annotation tool (enriched functional groups with false discovery rate [FDR] < 0.001)26 (Figures 3B and 4).
Functional analysis of differentially translated genes demonstrates enrichment for DNA repair pathways. The 748 genes that are differentially translated between type A and type B were subjected to a function and pathway enrichment analysis using (1) QIAGEN's Ingenuity Pathway Analysis tool (IPA, https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/), (2) Biological Process GO terms attached to the genes using the DAVID Functional Annotation tool, and (3) GSEA. Eighty-eight genes were extracted from this analysis; of which, 16 are found in IPA's DNA repair-related canonical pathways (adjusted P < .001), are enriched in IPA DNA repair-related functional groupings (adjusted P < .001), and are annotated by DAVID with DNA repair relevant GO terms (FDR < 0.001). Forty-one are found by 2 of these methods and the remainder by 1 method. The Ingenuity canonical pathways are: DNA Double-Strand Break Repair by Homologous Recombination, DNA Double-Strand Break Repair by Non-Homologous End Joining, Role of BRCA1 in DNA Damage Response, and ATM Signaling. The Ingenuity functions are: DNA damage response of cells, double-stranded DNA break repair, double-stranded DNA break repair of cells, and repair of DNA. The DAVID terms are: GO:0006974 (cellular response to DNA damage stimulus) and GO:0006281 (DNA repair). The GSEA results confirm an enrichment of genes involved in the ATM signalling pathway. The right column presents the fold change in translational efficiency of cluster A as compared to cluster B. Twelve genes are downregulated in cluster A, but all other genes are upregulated. Genes in the ATM pathway. either upstream (ie, MRE11, RAD50, and NBN) or interacting (ie, RBBP8, RAD17, and RIF1), are highlighted (blue). Apart from RIF1 all of these genes are present in all 4 of the analyses.
Functional analysis of differentially translated genes demonstrates enrichment for DNA repair pathways. The 748 genes that are differentially translated between type A and type B were subjected to a function and pathway enrichment analysis using (1) QIAGEN's Ingenuity Pathway Analysis tool (IPA, https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/), (2) Biological Process GO terms attached to the genes using the DAVID Functional Annotation tool, and (3) GSEA. Eighty-eight genes were extracted from this analysis; of which, 16 are found in IPA's DNA repair-related canonical pathways (adjusted P < .001), are enriched in IPA DNA repair-related functional groupings (adjusted P < .001), and are annotated by DAVID with DNA repair relevant GO terms (FDR < 0.001). Forty-one are found by 2 of these methods and the remainder by 1 method. The Ingenuity canonical pathways are: DNA Double-Strand Break Repair by Homologous Recombination, DNA Double-Strand Break Repair by Non-Homologous End Joining, Role of BRCA1 in DNA Damage Response, and ATM Signaling. The Ingenuity functions are: DNA damage response of cells, double-stranded DNA break repair, double-stranded DNA break repair of cells, and repair of DNA. The DAVID terms are: GO:0006974 (cellular response to DNA damage stimulus) and GO:0006281 (DNA repair). The GSEA results confirm an enrichment of genes involved in the ATM signalling pathway. The right column presents the fold change in translational efficiency of cluster A as compared to cluster B. Twelve genes are downregulated in cluster A, but all other genes are upregulated. Genes in the ATM pathway. either upstream (ie, MRE11, RAD50, and NBN) or interacting (ie, RBBP8, RAD17, and RIF1), are highlighted (blue). Apart from RIF1 all of these genes are present in all 4 of the analyses.
Given the differential enrichment for genes involved in double-strand break repair pathways and, in particular, ATM signaling, between the types of patient response, western blots were carried out to determine individual differences in the baseline levels of ATM (Figure 3D-E). Type A patients showed higher normalized ATM levels than type B patients (0.131 ± 0.019 vs 0.055 ± 0.011). One patient (ID 18) (represented by the blue dot in Figure 3E) showed an ATM mutation (supplemental Table 1) and had a lower normalized ATM level than the other type A patients. There was a significant difference between type A and type B patients (unpaired 2-tailed Student t test, P = .02) excluding this sample from the analysis. Overall, the ribosome profiling data suggested interpatient differences in translational regulation: 1 type of regulation (type A) appeared to be associated with higher baseline ATM levels.
We also observed differential regulation of ATM, and MRN complex genes (MRE11, RAD50, and NBN), which stabilize and activate ATM by inducing its autophosphorylation on serine 1981, and RIF1, RBBP8, and RAD17, which interact with ATM, such that TEs were higher in type A patients (ATM, 2.92-fold; MRE11, 2.11-fold; RAD50, 2.98-fold; NBN, 2.58-fold; RBBP8, 3.43-fold; RAD17, 2.69-fold; and RIF1, 4.05-fold) (Figures 4 and 5A).
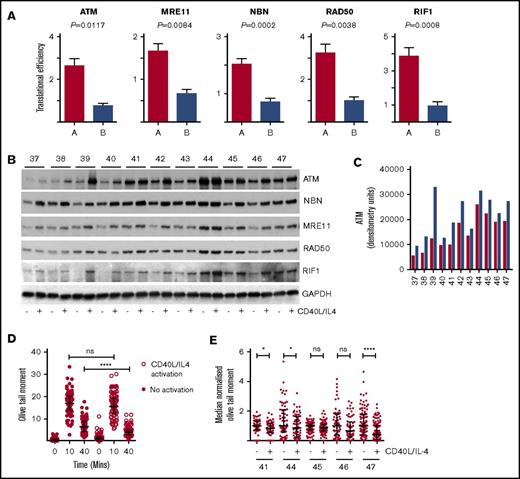
Translational efficiency of ATM, MRN complex, and RIF1. (A) Mean translational efficiency for each gene (ATM, MRE11, RAD50, NBN, and RIF1) in type A or type B is presented as a column and the error bars represent SEM. All 5 genes have significantly different TE (Xtail analysis) between types A and B: ATM, P = .0117; MRE11, P = .0084; NBN, P = .0002; RAD50, P = .0038; and RIF1, P = .0009. (B) Western blots showing the effects of CD40L/IL-4 culture on expression of ATM, MRE11, NBN, RAD50, and RIF1 (n = 11). GAPDH is the loading control. Patient ID is given above (labeled 37-47) the autorads. (C) Densitometry. Western bands were quantitated and the raw data plotted. Red columns are baseline ATM; blue columns are levels of ATM following CD40L/IL-4 stimulation. Patient ID is along the x-axis. There is a significant difference (paired 2-tailed Student t test) between ATM levels at baseline and after stimulation (P = .001). (D) Comet assay. Unstimulated cells (solid red circles) and CD40L/IL-4–activated cells (white circles) were exposed to XR (5 Gy). Olive tail moment was measured at 0, 10, and 40 minutes. Horizontal bars are medians and error bars are interquartile ranges. Olive tail moment is significantly lower in CD40L/IL-4–activated cells (Mann-Whitney U test, ****P < .0001). Patient ID40. (E) Comet assays were carried out in a further 5 patients (IDs 41, 44, 45, 46, and 47) with or without CD40L/IL-4 stimulation. Olive tail moments at 40 minutes have been median normalized to facilitate comparisons. Horizontal bars are medians and error bars are interquartile ranges. There are significant differences in olive tail moment following activation by CD40L/IL-4 for patients ID41, ID44 and ID47, but not for patients ID45 and ID46. (Mann-Whitney U test; patient ID41, *P = .02; ID44, *P = .04; ID45, P = .07; ID46, P = .06; and ID47, ****P = .0001). ns, not significant.
Translational efficiency of ATM, MRN complex, and RIF1. (A) Mean translational efficiency for each gene (ATM, MRE11, RAD50, NBN, and RIF1) in type A or type B is presented as a column and the error bars represent SEM. All 5 genes have significantly different TE (Xtail analysis) between types A and B: ATM, P = .0117; MRE11, P = .0084; NBN, P = .0002; RAD50, P = .0038; and RIF1, P = .0009. (B) Western blots showing the effects of CD40L/IL-4 culture on expression of ATM, MRE11, NBN, RAD50, and RIF1 (n = 11). GAPDH is the loading control. Patient ID is given above (labeled 37-47) the autorads. (C) Densitometry. Western bands were quantitated and the raw data plotted. Red columns are baseline ATM; blue columns are levels of ATM following CD40L/IL-4 stimulation. Patient ID is along the x-axis. There is a significant difference (paired 2-tailed Student t test) between ATM levels at baseline and after stimulation (P = .001). (D) Comet assay. Unstimulated cells (solid red circles) and CD40L/IL-4–activated cells (white circles) were exposed to XR (5 Gy). Olive tail moment was measured at 0, 10, and 40 minutes. Horizontal bars are medians and error bars are interquartile ranges. Olive tail moment is significantly lower in CD40L/IL-4–activated cells (Mann-Whitney U test, ****P < .0001). Patient ID40. (E) Comet assays were carried out in a further 5 patients (IDs 41, 44, 45, 46, and 47) with or without CD40L/IL-4 stimulation. Olive tail moments at 40 minutes have been median normalized to facilitate comparisons. Horizontal bars are medians and error bars are interquartile ranges. There are significant differences in olive tail moment following activation by CD40L/IL-4 for patients ID41, ID44 and ID47, but not for patients ID45 and ID46. (Mann-Whitney U test; patient ID41, *P = .02; ID44, *P = .04; ID45, P = .07; ID46, P = .06; and ID47, ****P = .0001). ns, not significant.
To confirm interpatient variation in ATM and also MRN complex and investigate changes in response to CD40L/IL-4 western blots were carried out in a further group of patients (n = 11) (Figure 5B; supplemental Figure 4; supplemental Table 2). CD40L/IL-4 caused ATM expression to increase in all patients but this varied widely between individuals (Figure 5C). Stimulation also produced significant (paired 2-tailed Student t test) increases in MRE11 (P = .0023) and NBN (P = .0002) protein levels across the cohort of patients suggesting levels of these proteins are translationally regulated, but there was little change to RAD50 levels (P = .0981) and there were variable changes in RIF1 (P = .1071) (supplemental Figure 4). On this evidence translational regulation might contribute directly to expression of ATM, MRE11, and NBN.
CD40L/IL-4 stimulation improves DNA repair
Comet assays were carried out to investigate DNA repair in CD40L/IL-4–stimulated cells following induction of double-strand breaks by X-irradiation (XR). Differences in olive tail moment in unstimulated and CD40L/IL-4–stimulated cells from 1 patient at baseline (before XR) and 10 and 40 minutes after XR demonstrated that XR had the same ability to cause DNA damage both with and without CD40L/IL-4 stimulation (10 minute time point) (Figure 5D). At 40 minutes after XR, both CD40L/IL-4–stimulated and unstimulated cells showed significant reduction in olive tail moment, demonstrating that DNA repair was occurring, but the CD40L/IL-4–stimulated cells showed improved repair compared with unstimulated cells (Mann-Whitney U test, P < .0001), consistent with CD40L/IL-4 promoting DNA repair in this context. In a further group of 5 patients, comet assays (Figure 5E) showed that CD40L/IL-4 stimulation improved DNA repair, but statistical significance was only reached in 3 of these, further suggesting that the DNA damage response to CD40/IL-4 is patient specific.
Interpatient variation in induction of ATM and MRN complex genes following CD40L/IL-4
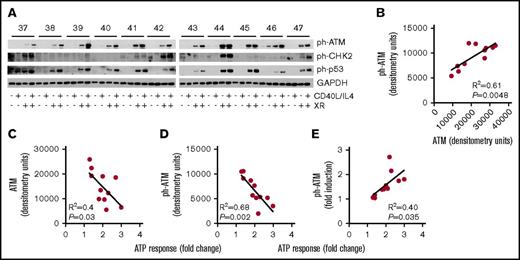
Phosphorylated ATM was detectable after XR (Figure 6A) and increased in line with total ATM (R2 = 0.61, P = .0048) (Figure 6B). We next analyzed the associations of total ATM and ph-ATM with increased viability due to CD40L/IL-4 (the ATP response). Baseline, that is, without CD40L/IL-4 stimulation, levels of both total ATM (R2 = 0.4, P = .03; Figure 6C) and ph-ATM (R2 = 0.68, P = .002; Figure 6D) expression were negatively associated with ATP response. Phosphorylated ATM levels were increased following culture with CD40L/IL-4 (Figure 6A), and the fold increase in ph-ATM (ie, level with CD40L/IL-4: level without CD40L/IL-4) correlated with ATP response due to CD40L/IL-4 (R2 = 0.4, P = .035) (Figure 6E).
Activation of ATM associates with CD40L/IL-4 responses. (A) Western blots showing the effects of CD40L/IL-4 culture and XR (5 Gy) on levels of ph-ATM, ph-p53, and ph-CHK2. GAPDH is the loading control. Patient ID is given above (labeled 37 to 47) the autorads. Those samples that received XR are indicated. (B) Correlation between levels of total and ph-ATM after CD40L/IL-4 stimulation and ATP response (R2 = 0.61, P = .0048). (C) Levels of total ATM after CD40L/IL-4 stimulation correlated with fold-increase in viability due to CD40L/IL-4 (R2 = 0.4, P = .03). (D) Levels of ph-ATM after CD40L/IL-4 stimulation correlated with fold-increase in viability due to CD40L/IL-4 (R2 = 0.68, P = .002). (E) Fold increase in ph-ATM after CD40L/IL-4 stimulation correlated with fold-increase in viability due to CD40L/IL-4 (R2 = 0.4, P = .035).
Activation of ATM associates with CD40L/IL-4 responses. (A) Western blots showing the effects of CD40L/IL-4 culture and XR (5 Gy) on levels of ph-ATM, ph-p53, and ph-CHK2. GAPDH is the loading control. Patient ID is given above (labeled 37 to 47) the autorads. Those samples that received XR are indicated. (B) Correlation between levels of total and ph-ATM after CD40L/IL-4 stimulation and ATP response (R2 = 0.61, P = .0048). (C) Levels of total ATM after CD40L/IL-4 stimulation correlated with fold-increase in viability due to CD40L/IL-4 (R2 = 0.4, P = .03). (D) Levels of ph-ATM after CD40L/IL-4 stimulation correlated with fold-increase in viability due to CD40L/IL-4 (R2 = 0.68, P = .002). (E) Fold increase in ph-ATM after CD40L/IL-4 stimulation correlated with fold-increase in viability due to CD40L/IL-4 (R2 = 0.4, P = .035).
However, with the exception of NBN, ATP responses, both without stimulation and following CD40L/IL-4, were negatively correlated with baseline levels of MRE11, RAD50, and RIF1 (supplemental Figures 5 and 6). Therefore, both total and ph-ATM levels (Figure 6C-D) as well as levels of MRE11, RAD50, and RIF1 correlate negatively with ATP response to CD40L/IL-4.
Expression of ATM targets and CD40L/IL-4 response: CHK2, p53, and H2AX
Functionally important differences in ATM level would be expected to be associated with differences in target protein modifications. The major targets for phosphorylation by ATM are CHK2, p53,27 and H2AX28 and we, therefore, assessed changes in these proteins after XR and CD40L/IL-4 stimulation. Following XR, phosphorylated CHK2 (ph-CHK2) was detectable in only 55% (6 of 11) of patients, with an increase on CD40L/IL-4 stimulation in 36% (4 of 11) (Figure 6A). However, phosphorylated-p53 was detectable, and showed a significant increase in mean protein levels with CD40L/IL-4 stimulation (with XR), (paired 2-tailed Student t test, P < .0001) (Figures 6A and 7A). The fold-increase in phosphorylated-p53 (level of ph-p53 with CD40L/IL-4 stimulation:level of ph-p53 without CD40L/IL-4 stimulation) associated with the ATP response to CD40L/IL-4 (R2 = 0.4, P = .03) (Figure 7B).
Levels of the ATM target proteins ph-p53 and γH2AX increase in association with increased global translation following XR. (A) Densitometry. Western bands (from Figure 6A) were quantitated and the raw data plotted. Red columns are baseline ph-p53 (following XR but without CD40L/IL-4); blue columns are levels of ph-p53 following XR and with CD40L/IL-4 stimulation. Patient ID is along the x-axis. There is a significant difference (paired 2-tailed Student t test) between ph-p53 levels at baseline and after stimulation (P < .0001). (B) Fold increase in ph-p53 after CD40L/IL-4 stimulation of X-irradiated cells correlated with fold-increase in viability due to CD40L/IL-4 (R2 = 0.4, P = .03). (C) Cell viability (ATP luminescence RLU) following different doses of XR (5, 20, 35, and 50 Gy) in samples that were either untreated (orange lines) or CD40L/IL-4–treated (blue lines) (n = 10). Although there was no significant difference (paired 2-tailed Student t tests) in ATP luminescence between CD40L/IL-4–stimulated and unstimulated at 5 Gy (P = .06), there were significant differences at 20 Gy (P < .0001), 35 Gy (P < .0001), and 50 Gy (P = .0002). (D) Relative improvement in cell survival due to CD40L/IL-4 following XR. Cells were stimulated with CD40L/IL-4 and either treated with XR (50 Gy) or not treated. Viability (percentage as compared with no XR) correlated with fold-increase in viability due to CD40L/IL-4 alone (R2 = 0.46, P = .03). The blue dot is a 17p deleted case (patient 46) demonstrating that this had among the highest viability following XR. (E) γH2AX response (percentage as compared with T = 0) in CLL cells at times up to 7 hours following XR (5 Gy) either stimulated with CD40L/IL-4 (blue lines) or unstimulated (orange lines) (n = 11). There were significant differences (paired 2-tailed Student t tests) in ATP luminescence between CD40L/IL-4–stimulated and unstimulated at 40 minutes (P = .0002) and 120 minutes (P = .0002), but not at 420 minutes (P = .06) (F) γH2AX response (percentage as compared with no XR) following doses of XR (2, 5, 10, and 20 Gy) in CLL cells either stimulated with CD40L/IL-4 (blue lines) or unstimulated (orange lines) (n = 5). There were significant differences (paired 2-tailed Student t tests) in γH2AX response between CD40L/IL-4–stimulated and unstimulated at 5 Gy (P = .01) and 10 Gy (P = .01). (G) CD40L/IL-4 response correlates with γH2AX response. The ratio of ATP luminescence in XR-treated and CD40L/IL-4–stimulated cells to that obtained in unstimulated but XR-treated cells is correlated to γH2AX response (determined as ratio of mean fluorescence index from XR-treated and CD40L/IL-4–stimulated cells to that from unstimulated but XR-treated cells) (n = 10) (R2 = 0.59, P = .006). (H) OPP response correlates with γH2AX response (n = 10) (R2 = 0.5, P = .015). The ratio of mean fluorescence index obtained in XR-treated and CD40L/IL-4–stimulated cells to that obtained in unstimulated but XR-treated cells is plotted for both OPP and γH2AX.
Levels of the ATM target proteins ph-p53 and γH2AX increase in association with increased global translation following XR. (A) Densitometry. Western bands (from Figure 6A) were quantitated and the raw data plotted. Red columns are baseline ph-p53 (following XR but without CD40L/IL-4); blue columns are levels of ph-p53 following XR and with CD40L/IL-4 stimulation. Patient ID is along the x-axis. There is a significant difference (paired 2-tailed Student t test) between ph-p53 levels at baseline and after stimulation (P < .0001). (B) Fold increase in ph-p53 after CD40L/IL-4 stimulation of X-irradiated cells correlated with fold-increase in viability due to CD40L/IL-4 (R2 = 0.4, P = .03). (C) Cell viability (ATP luminescence RLU) following different doses of XR (5, 20, 35, and 50 Gy) in samples that were either untreated (orange lines) or CD40L/IL-4–treated (blue lines) (n = 10). Although there was no significant difference (paired 2-tailed Student t tests) in ATP luminescence between CD40L/IL-4–stimulated and unstimulated at 5 Gy (P = .06), there were significant differences at 20 Gy (P < .0001), 35 Gy (P < .0001), and 50 Gy (P = .0002). (D) Relative improvement in cell survival due to CD40L/IL-4 following XR. Cells were stimulated with CD40L/IL-4 and either treated with XR (50 Gy) or not treated. Viability (percentage as compared with no XR) correlated with fold-increase in viability due to CD40L/IL-4 alone (R2 = 0.46, P = .03). The blue dot is a 17p deleted case (patient 46) demonstrating that this had among the highest viability following XR. (E) γH2AX response (percentage as compared with T = 0) in CLL cells at times up to 7 hours following XR (5 Gy) either stimulated with CD40L/IL-4 (blue lines) or unstimulated (orange lines) (n = 11). There were significant differences (paired 2-tailed Student t tests) in ATP luminescence between CD40L/IL-4–stimulated and unstimulated at 40 minutes (P = .0002) and 120 minutes (P = .0002), but not at 420 minutes (P = .06) (F) γH2AX response (percentage as compared with no XR) following doses of XR (2, 5, 10, and 20 Gy) in CLL cells either stimulated with CD40L/IL-4 (blue lines) or unstimulated (orange lines) (n = 5). There were significant differences (paired 2-tailed Student t tests) in γH2AX response between CD40L/IL-4–stimulated and unstimulated at 5 Gy (P = .01) and 10 Gy (P = .01). (G) CD40L/IL-4 response correlates with γH2AX response. The ratio of ATP luminescence in XR-treated and CD40L/IL-4–stimulated cells to that obtained in unstimulated but XR-treated cells is correlated to γH2AX response (determined as ratio of mean fluorescence index from XR-treated and CD40L/IL-4–stimulated cells to that from unstimulated but XR-treated cells) (n = 10) (R2 = 0.59, P = .006). (H) OPP response correlates with γH2AX response (n = 10) (R2 = 0.5, P = .015). The ratio of mean fluorescence index obtained in XR-treated and CD40L/IL-4–stimulated cells to that obtained in unstimulated but XR-treated cells is plotted for both OPP and γH2AX.
ATM is responsible for DNA double-strand break repair after damage due to XR. We determined the effects of CD40L/IL-4 on cell survival following XR. CLL cells showed significantly improved viability after XR following CD40L/IL-4 stimulation (paired 2-tailed Student t test; P < .0001 at 20 Gy, P < .0001 at 35 Gy and P = .0002 at 50 Gy) (n = 10) (Figure 7C), and the proportion of cells that remained viable after XR correlated with the ATP response (R2 = 0.46, P = .03) (Figure 7D).
We next determined γH2AX responses to XR (5 Gy) as a surrogate for ATM activity. The γH2AX response peaked at 45 minutes and then declined such that baseline levels were almost reached by 7 hours (n = 11) (Figure 7E). γH2AX responses (at 45 minutes) were significantly higher in CD40L/IL-4–stimulated cells (median and interquartile range, 458% [447%-525%]) than in nonstimulated cells (361% [330%-409%]) (P = .0002 at 45 minutes and P = .0002 at 120 minutes). For both stimulated and nonstimulated cells γH2AX responses increased with increasing doses of XR (n = 5), but γH2AX responses were significantly higher for CD40L/IL-4–stimulated cells (P = .01 at 5 Gy and P = .01 at 10 Gy) (Figure 7F). Therefore, CD40L/IL-4 stimulation maintains cell viability following XR and this response is associated with increased levels of the ATM targets ph-p53 and γH2AX.
Lower levels of ATM are associated with reduced overall survival
Our data suggested that ATM levels might be distributed continuously in patients who do not have structural rearrangements of chromosome 11q. To test this, we carried out western blots in a cohort of patients (supplemental Table 3; supplemental Figure 7) (n = 57), who were diagnosed between 1984 and 2007 and which included 11 patients showing 11q deletion (Figure 8). The cohort had a median overall survival of 124 months and showed the anticipated relationship to immunoglobulin heavy chain mutational status and Binet stage (Figure 8A). Baseline ATM levels were determined by western blots and densitometry (supplemental Table 3). Across the whole cohort, patients with lower (less than median) ATM levels had a significantly reduced survival as compared with patients with higher ATM levels (log rank, P = .023) (Figure 8B). When patients with 11q deletion were analyzed separately there was no longer a significant difference between patient samples from the less than median and greater than median groups (P = .067), although patients with greater than median levels of ATM continued to have significantly better survival than 11q deleted patients (P = .044) (Figure 8C). However, when only the lower and upper terciles of ATM expression levels were compared there was a significant difference in overall survival (P = .0037) with a hazard ratio for survival of 0.33 (95% confidence interval [CI], 0.128-0.648) (Figure 8D). Therefore, in a cohort of patients without rearrangements of 11q those with lower ATM levels have reduced overall survival.
Lower ATM levels are associated with reduced overall survival in a cohort of patients. (A) Kaplan Meier curves constructed using a cohort of patients (supplemental Table 3) (n = 57). (i) Overall survival of the whole cohort. Median survival 124 months. (ii) Overall survival by immunoglobulin heavy chain gene mutational status (solid red line mutated and dotted blue line unmutated). Median survival of mutated patients 168 months and unmutated 75.5 months. Log-rank test P ≤ .0001. (iii) Overall survival by sex (solid red line, female; dotted blue line, male). Median survival of female patients 168 months and males 117 months. Log-rank test P = .402. (iv) Overall survival by Binet stage (solid red line stage A and dotted blue line stages B and C). Median survival of stage A patients 165.5 months and stages B and C 92.3 months. Log-rank test P = .0037. (B) Overall survival of the entire cohort, including patients with 11q deletion, by ATM level (solid red line patients with ATM levels above the median and dotted blue line patients with ATM levels below the median). Median survival of patients with ATM greater than median (n = 28) 178.5 months and patients less than median (n = 29) 99.5 months. Log-rank test P = .023. (C) Overall survival of 11q deleted patients (n = 11) (dashed green line), patients with ATM levels less than median and intact 11q (n = 21) (dotted blue line) and patients with ATM levels greater than median and intact 11q (n = 25) (solid red line). Median survival of 11q deleted patients 79 months, patients with ATM levels less than median 117 months and patients with ATM levels greater than median 178.5 months. There is a significant difference in survival between patients with ATM levels greater than median and 11q deleted patients (log-rank test P = .044) but not for the other comparisons. (D) Overall survival of patients (excluding 11q deleted cases) with ATM levels in the lower tercile (n = 18) (dotted blue line) and patients with ATM levels in the upper tercile (n = 18) (solid red line). Median survival of lower tercile patients 110.8 months and patients with ATM levels in the upper tercile 290 months. There is a significant difference in survival (log-rank test, P = .0037). (E) Summary. CD40L/IL-4 stimulation promotes global translation and translational regulation of the ATM pathway to different degrees in different individuals. CD40L/IL-4 responses are correlated with survival and DNA repair following XR and CD40L/IL-4 drives ATM steady-state protein levels and levels of ATM target proteins, ph-p53, and γH2AX.
Lower ATM levels are associated with reduced overall survival in a cohort of patients. (A) Kaplan Meier curves constructed using a cohort of patients (supplemental Table 3) (n = 57). (i) Overall survival of the whole cohort. Median survival 124 months. (ii) Overall survival by immunoglobulin heavy chain gene mutational status (solid red line mutated and dotted blue line unmutated). Median survival of mutated patients 168 months and unmutated 75.5 months. Log-rank test P ≤ .0001. (iii) Overall survival by sex (solid red line, female; dotted blue line, male). Median survival of female patients 168 months and males 117 months. Log-rank test P = .402. (iv) Overall survival by Binet stage (solid red line stage A and dotted blue line stages B and C). Median survival of stage A patients 165.5 months and stages B and C 92.3 months. Log-rank test P = .0037. (B) Overall survival of the entire cohort, including patients with 11q deletion, by ATM level (solid red line patients with ATM levels above the median and dotted blue line patients with ATM levels below the median). Median survival of patients with ATM greater than median (n = 28) 178.5 months and patients less than median (n = 29) 99.5 months. Log-rank test P = .023. (C) Overall survival of 11q deleted patients (n = 11) (dashed green line), patients with ATM levels less than median and intact 11q (n = 21) (dotted blue line) and patients with ATM levels greater than median and intact 11q (n = 25) (solid red line). Median survival of 11q deleted patients 79 months, patients with ATM levels less than median 117 months and patients with ATM levels greater than median 178.5 months. There is a significant difference in survival between patients with ATM levels greater than median and 11q deleted patients (log-rank test P = .044) but not for the other comparisons. (D) Overall survival of patients (excluding 11q deleted cases) with ATM levels in the lower tercile (n = 18) (dotted blue line) and patients with ATM levels in the upper tercile (n = 18) (solid red line). Median survival of lower tercile patients 110.8 months and patients with ATM levels in the upper tercile 290 months. There is a significant difference in survival (log-rank test, P = .0037). (E) Summary. CD40L/IL-4 stimulation promotes global translation and translational regulation of the ATM pathway to different degrees in different individuals. CD40L/IL-4 responses are correlated with survival and DNA repair following XR and CD40L/IL-4 drives ATM steady-state protein levels and levels of ATM target proteins, ph-p53, and γH2AX.
A multivariate Cox proportional hazards model was fitted to patients to evaluate the impact of Binet stage at diagnosis, mutational status or ATM level (divided into 2 groups with expression levels above or below the mean) on survival. The model was globally significant (P < .0001, Likelihood ratio test) and all 3 variables were significant at P < .05. A higher risk of death was associated with stage B or C (hazard ratio, 2.244; 95% CI, 1.09-4.621; P = .028), whereas a lower risk of death was associated with mutated immunoglobulin VH genes (hazard ratio, 0.198; 95% CI, 0.098-0.398; P = .000006) and higher levels of ATM (hazard ratio, 0.399; 95% CI, 0.177-0.899; P = .027).
Overall (Figure 8E), we demonstrate associations between the response to CD40L/IL-4 stimulation and levels of global translation (Figure 1B). Ribosome profiling focused our work on ATM as being specifically translationally regulated and we showed that higher levels of baseline ATM (Figure 6C) and ph-ATM (Figure 6D) correlated with lower CD40L/IL-4 responses and better overall patient survival (Figure 8).
Discussion
The observation of profound clinical heterogeneity in CLL was made many years ago.29 We have pursued the hypothesis that individual differences in translational regulation are an alternative, and unexplored, cause of interpatient variation in gene expression that determines clinical course. Our approach has been to use ribosome profiling, a powerful application of next-generation sequencing17 that has been used to dissect the translational landscape of a mouse model of skin cancer30 but has not previously been applied to human cancer. In an initial cohort of patients we demonstrated that the TEs of DNA repair genes differ significantly between patients clustered by their overall pattern of TE per gene. The implication of this result is that these genes or pathways are differentially regulated and that there are interpatient differences in protein expression of DNA damage response genes.
The ribosome profiling results required validation to determine the relationship between CD40L/IL-4 and DNA damage repair. We noted that, as well as ATM, the TEs of several associated genes (MRE11, RAD50, NBN, and RIF1) differed significantly between patient clusters. We focused on analyzing the ATM pathway. There are several lines of evidence to support investigation of this pathway: first, mutations in ATM are clinically significant31,32 ; second, genome-wide association studies have shown that single-nucleotide polymorphisms in ATM are linked to susceptibility to CLL33 ; and third, splicing defects due to SF3B1 mutation, independent of ATM mutation, associate with defects in ATM-associated apoptosis.34 It has been independently demonstrated that there is extensive alternate splicing in the 5′ untranslated region of ATM leading to variant mRNAs with diverse secondary structures and different numbers of AUG codons.35 This illustrates that ATM is potentially a target for translational control in model systems other than CLL and this has been confirmed in work showing that DNA damage caused by UVB shifts ATM mRNA to polysome fractions.36
It has been found that monoallelic deletion of MRE11 can occur in association with 11q deletion, suggesting selection for defects in the proximal DNA damage repair pathways including double-strand break recognition.37 These genetic defects might cooperate with individual differences in the translational regulation of DNA damage responses to enhance differences between individuals and contribute to pronounced differences in clinical outcome. In addition, mouse studies demonstrated that ATM deficiency is sufficient to promote lymphomagenesis,38,39 possibly due to modification of apoptotic signals.39
We have demonstrated associations between the ATP response to CD40L/IL-4 and cap-dependent translation, ATM protein levels, and induction of the ATM targets γH2AX and ph-p53. Although we cannot exclude a contribution from other kinases (ATR and DNA-PK)27 to the phosphorylation of H2AX and p53, a large amount of previous work has shown that ATM is a principal mediator of γH2AX and ph-p53 formation.28,40 Importantly, we also demonstrate that ATP response to CD40L/IL-4 associates with maintenance of viability after genotoxic stress caused by XR and, therefore, there is a potential functional link between CD40L/IL-4 responses and the ATM pathway. The interpatient variation in ATM levels, excluding patients with 11q deletion, which we describe for the first time, is also clinically important. ATM levels associate with overall survival suggesting that the effects of ATM are not binary (absent or present) but are continuous in line with the continuous distribution of protein levels that we observed.
CD40L/IL-4 in vitro promotes proliferation and improved viability. A limitation of our work is that we do not explore the association between viability and changes in DNA damage response genes in response to CD40L/IL-4. Some changes to important survival pathways, that is, PI3K and NF-κB,41 may contribute indirectly to the observed changes in DNA damage response genes. We demonstrate that global translation is, in part, regulated by CD40L/IL-4 and suggest that individual variation in the regulation of translation interacts with CD40L/IL-4 signaling pathways to modify DNA damage response genes. Potential additional factors that may play a role in both the viability response to CD40L/IL-4 and translational regulation are polymorphisms leading to variation in codon usage, biased transfer RNA usage.42,43 The interaction between CD40L/IL-4 signaling, cell viability and DNA repair gene expression has not been fully explored but this report suggests that investigation of the mechanisms responsible will provide insights to pathways playing a role in CLL cell survival in vivo.
Although CD40L/IL-4 stimulation produced elevated levels of ATM it did not produce consistent changes in levels of all MRN complex proteins. Possible reasons for this are: (1) the sensitivities of western blots vs ribosome profiling. Ribosome profiling is highly sensitive and will detect variation that is not detectable by western blot or (2) ribosome profiling does not capture the effects of posttranslational modifications that might be important in determining steady state protein levels.
Why should CD40L/IL-4 stimulation increase protein expression of ATM and associated genes? One possibility might be that ATM is responsible for repairing the subset of “difficult” DNA double-strand breaks in heterochromatin.44 CLL cells, which are in G0/G1 in the peripheral blood, encounter CD40L/IL-4 stimulation in the tissue microenvironment (TME) in which they are driven to express c-MYC, increase translation and proliferate. CD40L/IL-4 stimulation in vitro is responsible for a unique transcriptional program45 likely to be associated with increased heterochromatin. Elevated ATM levels might be an adaptation to ensure genome integrity in this environment.
Using ribosome profiling, we have been able to determine specific genes differing in their translational regulation between patients following CD40L/IL-4 stimulation. We suggest that ATM and certain MRN complex genes are targets for translational regulation and, furthermore, that varying levels of ATM might contribute to heterogeneity in clinical outcome, which is such a prominent clinical feature of CLL, through its effects on DNA damage responses. We propose that investigation of specific translationally regulated genes will be a source of predictive biomarkers or targets for therapy in CLL.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank Sandrine Jayne, manager of the Leicester Haematological Malignancies Biobank, for assistance in obtaining samples for the repository study.
This work was supported in part by a grant from Bloodwise (14038) (A.E.W. and S.D.W.).
Authorship
Contribution: L.L., D.B., C.J., K.M.D., G.D.D.J., and A.B. designed and carried out research; R.V.S. analyzed data and wrote the paper; and L.L., G.P., A.E.W., and S.D.W. designed research, analyzed results, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Simon D. Wagner, University of Leicester, Room 104, Hodgkin Building, Lancaster Rd, Leicester LE1 9HN, United Kingdom; e-mail: sw227@le.ac.uk.
References
Author notes
L.L., R.V.S., and D.B. are joint first authors.