Key Points
Paradoxically higher NK-cell activity in CTCL patients is associated with increased expression of phosphorylated STAT5.
These highly effective NK cells are associated with poor prognosis in patients with leukemic CTCL.
Introduction
Cutaneous T-cell lymphoma (CTCL) is a type of non-Hodgkin lymphoma characterized by the expansion of malignant CD4+ T cells in the skin. There are two main subtypes of CTCL: an indolent form termed mycosis fungoides (MF), which is largely limited to the skin, and Sézary syndrome (SS), an aggressive leukemic variant of the disease which can manifest systemically.1-3
Previous studies have demonstrated defects in cell-mediated immunity in CTCL patients, including altered cytokine profiles and impaired neutrophil function, which lead to a high incidence of recurrent bacterial and viral infections as a result of decreased Th1-mediated immunity.4-9 It has also been reported that natural killer (NK)–cell function is decreased in CTCL patients,10-14 which could contribute to an overall decrease in the innate immune response to both neoplastic cells and viral or bacterial pathogens. Previous groups have reported that NK cells from SS patients are capable of responding to activation ex vivo, indicating the potential for development of immune-based therapeutics.15
Although MF patients often have a prolonged indolent clinical course of disease that requires localized treatment, there are few effective treatments for the successful management of patients with SS. Because of the lack of success with traditional chemotherapeutic approaches, novel immune-based therapeutics are being developed for use in a multitude of hematologic diseases, including CTCL.4,16-18 Understanding the immune microenvironment in patients with CTCL will be critical to the successful design of targeted therapies for their disease.
Previous studies by our group and by others have shown increased expression of interleukin-15 (IL-15) in malignant CD4+ T cells in CTCL patients.19 IL-15 acts through a trimeric IL-15R complex to enhance NK-cell maturation and function.20-22 Indeed, in a first-in-human phase 1 trial in patients with refractory solid cancer tumors, IL-15 treatment induced profound expansion of circulating NK cells (NCT01885897).23 Considering that IL-15 is produced by malignant cells in CTCL, we sought to study the possible effect of chronically elevated IL-15 on NK-cell function in CTCL patients. In this study, we show that NK-cell activity is significantly enhanced in CTCL, and strikingly, higher NK-cell numbers are associated with increased mortality.
Materials and methods
NK-cell numbers
NK-cell numbers were evaluated by flow cytometric analysis of peripheral blood samples drawn on the same day as the initial diagnostic complete blood cell count with differential and was performed using a 10-color technique with a gating strategy based on CD45 staining and light side scatter characteristics. NK-cell number represents the number of CD56+/CD16+/CD3– NK cells per microliter. Samples were taken from November 2007 through November 2016 from patients at The Ohio State University James Cancer Hospital who were diagnosed with biopsy-proven CTCL (Table 1).
Characteristics of patients diagnosed with biopsy-proven CTCL MF
| Patient ID . | Diagnosis . | Stage at diagnosis . | Histology/subtype . | Age at diagnosis, y . | Sex . | Race . |
|---|---|---|---|---|---|---|
| 9 | CTCL, MF | IA | Woringer-Kolopp variant | 48 | F | White |
| 216 | CTCL, MF | IA | CD30+ LPD: primary CTCL γ-δ T-cells | 68 | M | White |
| 27 | CTCL, MF | IA | Hypopigmented | 60 | F | African American |
| 85 | CTCL, MF | IA | Hypopigmented | 38 | M | African American |
| 65 | CTCL, MF | IA | None | 40 | M | African American |
| 180 | CTCL, MF | IA | None | 56 | M | African American |
| 6 | CTCL, MF | IA | None, lyp, and CD30+ LPD | 61 | M | African American |
| 147 | CTCL, MF | IA | Hypopigmented | 45 | F | Asian |
| 89 | CTCL, MF | IA | Folliculotropic | 48 | F | White |
| 137 | CTCL, MF | IA | Granulomatous | 67 | F | White |
| 28 | CTCL, MF | IA | Hypopigmented | 38 | F | White |
| 38 | CTCL, MF | IA | None | 41 | F | White |
| 42 | CTCL, MF | IA | None | 49 | F | White |
| 52 | CTCL, MF | IA | None | 57 | F | White |
| 55 | CTCL, MF | IA | None | 41 | F | White |
| 61 | CTCL, MF | IA | None | 82 | F | White |
| 73 | CTCL, MF | IA | None | 64 | F | White |
| 82 | CTCL, MF | IA | None | 50 | F | White |
| 107 | CTCL, MF | IA | None | 58 | F | White |
| 111 | CTCL, MF | IA | None | 56 | F | White |
| 144 | CTCL, MF | IA | None | 51 | F | White |
| 145 | CTCL, MF | IA | None | 42 | F | White |
| 150 | CTCL, MF | IA | None | 28 | F | White |
| 152 | CTCL, MF | IA | None | 47 | F | White |
| 183 | CTCL, MF | IA | None | 53 | F | White |
| 200 | CTCL, MF | IA | None | 37 | F | White |
| 215 | CTCL, MF | IA | None | 50 | F | White |
| 226 | CTCL, MF | IA | None | 19 | F | White |
| 166 | CTCL, MF | IA | Poikilodermatous | 31 | F | White |
| 84 | CTCL, MF | IA | Poikilodermatous | 55 | F | White |
| 188 | CTCL, MF | IA | Woringer-Kolopp variant | 31 | F | White |
| 142 | CTCL, MF | IA | Follicular | 50 | F | White |
| 179 | CTCL, MF | IA | Follicular T-helper phenotype | 60 | F | White |
| 50 | CTCL, MF | IA | CD8+ | 58 | M | White |
| 32 | CTCL, MF | IA | Folliculotropic | 37 | M | White |
| 36 | CTCL, MF | IA | Folliculotropic | 61 | M | White |
| 54 | CTCL, MF | IA | Folliculotropic | 81 | M | White |
| 64 | CTCL, MF | IA | Folliculotropic | 25 | M | White |
| 100 | CTCL, MF | IA | Folliculotropic | 62 | M | White |
| 130 | CTCL, MF | IA | Hypopigmented, CD8+ | 18 | M | White |
| 41 | CTCL, MF | IA | None | 84 | M | White |
| 46 | CTCL, MF | IA | None | 40 | M | White |
| 47 | CTCL, MF | IA | None | 78 | M | White |
| 57 | CTCL, MF | IA | None | 63 | M | White |
| 62 | CTCL, MF | IA | None | 57 | M | White |
| 67 | CTCL, MF | IA | None | 77 | M | White |
| 71 | CTCL, MF | IA | None | 45 | M | White |
| 80 | CTCL, MF | IA | None | 26 | M | White |
| 95 | CTCL, MF | IA | None | 60 | M | White |
| 99 | CTCL, MF | IA | None | 74 | M | White |
| 103 | CTCL, MF | IA | None | 82 | M | White |
| 108 | CTCL, MF | IA | None | 42 | M | White |
| 120 | CTCL, MF | IA | None | 62 | M | White |
| 124 | CTCL, MF | IA | None | 57 | M | White |
| 128 | CTCL, MF | IA | None | 54 | M | White |
| 131 | CTCL, MF | IA | None | 37 | M | White |
| 139 | CTCL, MF | IA | None | 26 | M | White |
| 162 | CTCL, MF | IA | None | 51 | M | White |
| 174 | CTCL, MF | IA | None | 76 | M | White |
| 181 | CTCL, MF | IA | None | 53 | M | White |
| 189 | CTCL, MF | IA | None | 44 | M | White |
| 191 | CTCL, MF | IA | None | 34 | M | White |
| 197 | CTCL, MF | IA | None | 70 | M | White |
| 223 | CTCL, MF | IA | None | 72 | M | White |
| 146 | CTCL, MF | IA | None | 48 | M | White |
| 199 | CTCL, MF | IA | None | 63 | M | White |
| 4 | CTCL, MF | IB | None | 52 | M | African American |
| 2 | CTCL, MF | IB | Poikilodermatous | 69 | M | African American |
| 203 | CTCL, MF | IB | None | 46 | F | African American |
| 39 | CTCL, MF | IB | Hypopigmented | 38 | M | African American |
| 56 | CTCL, MF | IB | None | 51 | M | African American |
| 77 | CTCL, MF | IB | CD8+ | 58 | F | White |
| 26 | CTCL, MF | IB | Granulomatous | 66 | F | White |
| 87 | CTCL, MF | IB | None | 51 | F | White |
| 91 | CTCL, MF | IB | None | 66 | F | White |
| 136 | CTCL, MF | IB | None | 72 | F | White |
| 173 | CTCL, MF | IB | None | 57 | F | White |
| 194 | CTCL, MF | IB | None | 73 | F | White |
| 115 | CTCL, MF | IB | CD8+ | 63 | M | White |
| 209 | CTCL, MF | IB | CD8+ | 81 | M | White |
| 88 | CTCL, MF | IB | Folliculotropic | 76 | M | White |
| 97 | CTCL, MF | IB | Folliculotropic | 44 | M | White |
| 114 | CTCL, MF | IB | Folliculotropic | 58 | M | White |
| 134 | CTCL, MF | IB | Folliculotropic | 53 | M | White |
| 37 | CTCL, MF | IB | None | 68 | M | White |
| 58 | CTCL, MF | IB | None | 59 | M | White |
| 75 | CTCL, MF | IB | None | 64 | M | White |
| 78 | CTCL, MF | IB | None | 46 | M | White |
| 104 | CTCL, MF | IB | None | 62 | M | White |
| 112 | CTCL, MF | IB | None | 45 | M | White |
| 121 | CTCL, MF | IB | None | 71 | M | White |
| 122 | CTCL, MF | IB | None | 67 | M | White |
| 123 | CTCL, MF | IB | None | 55 | M | White |
| 220 | CTCL, MF | IB | None | 72 | M | White |
| 227 | CTCL, MF | IB | None | 62 | M | White |
| 217 | CTCL, MF | IB | Follicular T-helper phenotype | 36 | M | White |
| 167 | CTCL, MF | IB | None | 78 | M | White |
| 204 | CTCL, MF | IB | Poikilodermatous | 61 | F | Hispanic/Latino |
| 132 | CTCL, MF | IB | None | 79 | M | Hispanic/Latino |
| 206 | CTCL, MF | IIA | Poikilodermatous | 41 | M | White |
| 33 | CTCL, MF | IIB | Large cell transformation | 50 | F | African American |
| 30 | CTCL, MF | IIB | None | 66 | F | African American |
| 157 | CTCL, MF | IIB | Folliculotropic | 69 | M | African American |
| 168 | CTCL, MF | IIB | Folliculotropic | 53 | M | African American |
| 156 | CTCL, MF | IIB | None | 80 | F | White |
| 90 | CTCL, MF | IIB | Folliculotropic | 62 | M | White |
| 63 | CTCL, MF | IIB | None | 88 | M | White |
| 185 | CTCL, MF | IIB | None | 75 | M | White |
| 153 | CTCL, MF | IIB | Tumor | 64 | M | White |
| 141 | CTCL, MF | IIB | Large cell transformation | 59 | M | White |
| 109 | CTCL, MF | IIIB | Erythrodermic | 55 | F | White |
| 213 | CTCL, MF | IIIB | Erythrodermic | 65 | M | African American |
| Patient ID . | Diagnosis . | Stage at diagnosis . | Histology/subtype . | Age at diagnosis, y . | Sex . | Race . |
|---|---|---|---|---|---|---|
| 9 | CTCL, MF | IA | Woringer-Kolopp variant | 48 | F | White |
| 216 | CTCL, MF | IA | CD30+ LPD: primary CTCL γ-δ T-cells | 68 | M | White |
| 27 | CTCL, MF | IA | Hypopigmented | 60 | F | African American |
| 85 | CTCL, MF | IA | Hypopigmented | 38 | M | African American |
| 65 | CTCL, MF | IA | None | 40 | M | African American |
| 180 | CTCL, MF | IA | None | 56 | M | African American |
| 6 | CTCL, MF | IA | None, lyp, and CD30+ LPD | 61 | M | African American |
| 147 | CTCL, MF | IA | Hypopigmented | 45 | F | Asian |
| 89 | CTCL, MF | IA | Folliculotropic | 48 | F | White |
| 137 | CTCL, MF | IA | Granulomatous | 67 | F | White |
| 28 | CTCL, MF | IA | Hypopigmented | 38 | F | White |
| 38 | CTCL, MF | IA | None | 41 | F | White |
| 42 | CTCL, MF | IA | None | 49 | F | White |
| 52 | CTCL, MF | IA | None | 57 | F | White |
| 55 | CTCL, MF | IA | None | 41 | F | White |
| 61 | CTCL, MF | IA | None | 82 | F | White |
| 73 | CTCL, MF | IA | None | 64 | F | White |
| 82 | CTCL, MF | IA | None | 50 | F | White |
| 107 | CTCL, MF | IA | None | 58 | F | White |
| 111 | CTCL, MF | IA | None | 56 | F | White |
| 144 | CTCL, MF | IA | None | 51 | F | White |
| 145 | CTCL, MF | IA | None | 42 | F | White |
| 150 | CTCL, MF | IA | None | 28 | F | White |
| 152 | CTCL, MF | IA | None | 47 | F | White |
| 183 | CTCL, MF | IA | None | 53 | F | White |
| 200 | CTCL, MF | IA | None | 37 | F | White |
| 215 | CTCL, MF | IA | None | 50 | F | White |
| 226 | CTCL, MF | IA | None | 19 | F | White |
| 166 | CTCL, MF | IA | Poikilodermatous | 31 | F | White |
| 84 | CTCL, MF | IA | Poikilodermatous | 55 | F | White |
| 188 | CTCL, MF | IA | Woringer-Kolopp variant | 31 | F | White |
| 142 | CTCL, MF | IA | Follicular | 50 | F | White |
| 179 | CTCL, MF | IA | Follicular T-helper phenotype | 60 | F | White |
| 50 | CTCL, MF | IA | CD8+ | 58 | M | White |
| 32 | CTCL, MF | IA | Folliculotropic | 37 | M | White |
| 36 | CTCL, MF | IA | Folliculotropic | 61 | M | White |
| 54 | CTCL, MF | IA | Folliculotropic | 81 | M | White |
| 64 | CTCL, MF | IA | Folliculotropic | 25 | M | White |
| 100 | CTCL, MF | IA | Folliculotropic | 62 | M | White |
| 130 | CTCL, MF | IA | Hypopigmented, CD8+ | 18 | M | White |
| 41 | CTCL, MF | IA | None | 84 | M | White |
| 46 | CTCL, MF | IA | None | 40 | M | White |
| 47 | CTCL, MF | IA | None | 78 | M | White |
| 57 | CTCL, MF | IA | None | 63 | M | White |
| 62 | CTCL, MF | IA | None | 57 | M | White |
| 67 | CTCL, MF | IA | None | 77 | M | White |
| 71 | CTCL, MF | IA | None | 45 | M | White |
| 80 | CTCL, MF | IA | None | 26 | M | White |
| 95 | CTCL, MF | IA | None | 60 | M | White |
| 99 | CTCL, MF | IA | None | 74 | M | White |
| 103 | CTCL, MF | IA | None | 82 | M | White |
| 108 | CTCL, MF | IA | None | 42 | M | White |
| 120 | CTCL, MF | IA | None | 62 | M | White |
| 124 | CTCL, MF | IA | None | 57 | M | White |
| 128 | CTCL, MF | IA | None | 54 | M | White |
| 131 | CTCL, MF | IA | None | 37 | M | White |
| 139 | CTCL, MF | IA | None | 26 | M | White |
| 162 | CTCL, MF | IA | None | 51 | M | White |
| 174 | CTCL, MF | IA | None | 76 | M | White |
| 181 | CTCL, MF | IA | None | 53 | M | White |
| 189 | CTCL, MF | IA | None | 44 | M | White |
| 191 | CTCL, MF | IA | None | 34 | M | White |
| 197 | CTCL, MF | IA | None | 70 | M | White |
| 223 | CTCL, MF | IA | None | 72 | M | White |
| 146 | CTCL, MF | IA | None | 48 | M | White |
| 199 | CTCL, MF | IA | None | 63 | M | White |
| 4 | CTCL, MF | IB | None | 52 | M | African American |
| 2 | CTCL, MF | IB | Poikilodermatous | 69 | M | African American |
| 203 | CTCL, MF | IB | None | 46 | F | African American |
| 39 | CTCL, MF | IB | Hypopigmented | 38 | M | African American |
| 56 | CTCL, MF | IB | None | 51 | M | African American |
| 77 | CTCL, MF | IB | CD8+ | 58 | F | White |
| 26 | CTCL, MF | IB | Granulomatous | 66 | F | White |
| 87 | CTCL, MF | IB | None | 51 | F | White |
| 91 | CTCL, MF | IB | None | 66 | F | White |
| 136 | CTCL, MF | IB | None | 72 | F | White |
| 173 | CTCL, MF | IB | None | 57 | F | White |
| 194 | CTCL, MF | IB | None | 73 | F | White |
| 115 | CTCL, MF | IB | CD8+ | 63 | M | White |
| 209 | CTCL, MF | IB | CD8+ | 81 | M | White |
| 88 | CTCL, MF | IB | Folliculotropic | 76 | M | White |
| 97 | CTCL, MF | IB | Folliculotropic | 44 | M | White |
| 114 | CTCL, MF | IB | Folliculotropic | 58 | M | White |
| 134 | CTCL, MF | IB | Folliculotropic | 53 | M | White |
| 37 | CTCL, MF | IB | None | 68 | M | White |
| 58 | CTCL, MF | IB | None | 59 | M | White |
| 75 | CTCL, MF | IB | None | 64 | M | White |
| 78 | CTCL, MF | IB | None | 46 | M | White |
| 104 | CTCL, MF | IB | None | 62 | M | White |
| 112 | CTCL, MF | IB | None | 45 | M | White |
| 121 | CTCL, MF | IB | None | 71 | M | White |
| 122 | CTCL, MF | IB | None | 67 | M | White |
| 123 | CTCL, MF | IB | None | 55 | M | White |
| 220 | CTCL, MF | IB | None | 72 | M | White |
| 227 | CTCL, MF | IB | None | 62 | M | White |
| 217 | CTCL, MF | IB | Follicular T-helper phenotype | 36 | M | White |
| 167 | CTCL, MF | IB | None | 78 | M | White |
| 204 | CTCL, MF | IB | Poikilodermatous | 61 | F | Hispanic/Latino |
| 132 | CTCL, MF | IB | None | 79 | M | Hispanic/Latino |
| 206 | CTCL, MF | IIA | Poikilodermatous | 41 | M | White |
| 33 | CTCL, MF | IIB | Large cell transformation | 50 | F | African American |
| 30 | CTCL, MF | IIB | None | 66 | F | African American |
| 157 | CTCL, MF | IIB | Folliculotropic | 69 | M | African American |
| 168 | CTCL, MF | IIB | Folliculotropic | 53 | M | African American |
| 156 | CTCL, MF | IIB | None | 80 | F | White |
| 90 | CTCL, MF | IIB | Folliculotropic | 62 | M | White |
| 63 | CTCL, MF | IIB | None | 88 | M | White |
| 185 | CTCL, MF | IIB | None | 75 | M | White |
| 153 | CTCL, MF | IIB | Tumor | 64 | M | White |
| 141 | CTCL, MF | IIB | Large cell transformation | 59 | M | White |
| 109 | CTCL, MF | IIIB | Erythrodermic | 55 | F | White |
| 213 | CTCL, MF | IIIB | Erythrodermic | 65 | M | African American |
F, female; LPD, lymphoproliferative disorder; lyp, lymphomatoid papulosis; M, male.
NK-cell isolation and cytotoxicity
NK cells were isolated from fresh peripheral blood samples by negative enrichment (STEMCELL Technologies) followed by sorting on a BD Aria II analyzer. No phenotypic alterations were noted between the presorted and postsorted NK cells. Purified NK cells were co-cultured with chromium-labeled K562 target cells for 4 hours in a standard chromium release cytotoxicity assay.24 K562 cells were obtained from American Type Culture Collection (ATCC) and were kept in culture less than 1 month. Cells were routinely tested for mycoplasma per routine protocol. Tables 2 and 3 provide clinical information on the patients who participated in NK-cell functional studies.
Characteristics of patients with biopsy-proven CTCL SS
| Patient ID . | Diagnosis . | Stage at diagnosis . | Age at diagnosis, y . | Sex . | Race . | Clonality on initial flow cytometric analysis . |
|---|---|---|---|---|---|---|
| 69 | CTCL, SS | IVA1 | 51 | M | White | 84% CD3+/CD26– T-cells; 82% CD4+/CD26–; 75% CD7+/CD26– |
| 76 | CTCL, SS | III | 82 | F | White | 22% CD3+/CD4+/CD7– T-cells |
| 86 | CTCL, SS | IVA1 | 77 | F | White | 92% CD3+/CD4+/CD26–; 22% CD3+/CD4+/CD7– |
| 96 | CTCL, SS | IVA1 | 67 | M | White | 82% CD3+/CD26–/CD2– |
| 135 | CTCL, SS | IVA1 | 59 | F | White | 92% CD3+/CD26– |
| 164 | CTCL, SS | IVA1 | 60 | F | White | 63% CD3+/CD4+/CD7+/CD26–; 34% CD3+/CD4–/CD8–/CD7–/CD26– |
| 187 | CTCL, SS | IVA1 | 64 | M | African American | 60% CD3+/CD26–; 50% CD3 biphasic/CD7– |
| 224 | CTCL, SS | IVA1 | 82 | M | White | 98% CD2+/CD4+/CD5+/CD3–/CD7–/CD8–/CD26– |
| 227 | CTCL, SS | IVA1 | 56 | M | White | 93% CD4+/CD26– |
| Patient ID . | Diagnosis . | Stage at diagnosis . | Age at diagnosis, y . | Sex . | Race . | Clonality on initial flow cytometric analysis . |
|---|---|---|---|---|---|---|
| 69 | CTCL, SS | IVA1 | 51 | M | White | 84% CD3+/CD26– T-cells; 82% CD4+/CD26–; 75% CD7+/CD26– |
| 76 | CTCL, SS | III | 82 | F | White | 22% CD3+/CD4+/CD7– T-cells |
| 86 | CTCL, SS | IVA1 | 77 | F | White | 92% CD3+/CD4+/CD26–; 22% CD3+/CD4+/CD7– |
| 96 | CTCL, SS | IVA1 | 67 | M | White | 82% CD3+/CD26–/CD2– |
| 135 | CTCL, SS | IVA1 | 59 | F | White | 92% CD3+/CD26– |
| 164 | CTCL, SS | IVA1 | 60 | F | White | 63% CD3+/CD4+/CD7+/CD26–; 34% CD3+/CD4–/CD8–/CD7–/CD26– |
| 187 | CTCL, SS | IVA1 | 64 | M | African American | 60% CD3+/CD26–; 50% CD3 biphasic/CD7– |
| 224 | CTCL, SS | IVA1 | 82 | M | White | 98% CD2+/CD4+/CD5+/CD3–/CD7–/CD8–/CD26– |
| 227 | CTCL, SS | IVA1 | 56 | M | White | 93% CD4+/CD26– |
Characteristics of patients with CTCL, treatment, and experimental assay performed
| Patient ID . | Assay . | Sex . | Race . | CTCL type . | Stage at sample procurement . | Treatment at time of collection . |
|---|---|---|---|---|---|---|
| MF1 | Flow cytometry | M | White | MF | IA | Imiquimod |
| MF2 | Flow cytometry | F | White | MF | IA | Desoximetasone |
| MF3 | Flow cytometry | M | White | MF | IA | Clobetasol |
| U16-2023 | Cytotoxicity assay | M | White | MF | IB | PUVA, triamcinolone, imiquimod |
| U16-2101 | Cytotoxicity assay | M | White | MF | IB | Bexarotene |
| U16-2102 | Cytotoxicity assay | M | White | MF | IB | Bexarotene |
| U16-2225 | Cytotoxicity assay | M | White | MF | IIB | Gemcitabine/doxorubicin |
| U16-2228 | Cytotoxicity assay | M | White | MF | IIB | Bexarotene |
| U16-2237 | Cytotoxicity assay | F | African American | MF | IA | Topical steroids |
| U17-0092 | Cytotoxicity assay | M | White | MF | IB | Bexarotene |
| U17-0093 | Cytotoxicity assay | F | White | MF | IB | Topical steroids |
| U17-0094 | Cytotoxicity assay | M | White | MF | IA | nbUVB |
| U17-0095 | Cytotoxicity assay/RNA sequencing | M | White | MF | IA | Bexarotene |
| U17-0253 | Cytotoxicity assay/RNA sequencing | F | White | MF | IA | Topical bexarotene |
| U17-0254 | Cytotoxicity assay/RNA sequencing | F | White | MF | IB | nbUVB |
| U17-0314 | Flow cytometry | F | African American | SS | IVB | Romidepsin |
| U17-0315 | Flow cytometry | F | White | MF | 1A | Topical steroids |
| U17-0316 | Flow cytometry | F | White | MF | IB | Valchlor gel |
| U17-0341 | Flow cytometry | M | White | MF | 1A | Oral or topical bexarotene |
| U17-0342 | Flow cytometry | M | White | MF | 1A | Mechlorethamine gel, clobetasol |
| U17-0343 | Flow cytometry | M | White | MF | 1A | Mechlorethamine gel, TAC |
| U17-0350 | Flow cytometry | F | African American | MF | IA | Topical steroids |
| U17-0352 | Flow cytometry | F | White | MF | IA | Oral bexarotene |
| U17-0353 | Flow cytometry | M | White | MF | 1B | Interferon, nbUVB, TAC, clobetasol |
| U17-0543 | RNA sequencing | M | White | SS | 1VA | Romidepsin, IPH 4102 |
| U17-0593 | RNA sequencing | M | White | MF | 1B | Gemcitabine/ doxorubicin |
| U17-0594 | RNA sequencing | M | White | SS | IVA1 | Topical steroids |
| U17-0595 | RNA sequencing | M | White | MF | IA | TAC |
| U17-1613 | Phospho-specific flow cytometry | F | White | MF | IIB | Brentuximab |
| U17-1614 | Phospho-specific flow cytometry | M | White | SS | 1VA2 | Bendamustine + brentuximab vedotin |
| U17-1615 | Phospho-specific flow cytometry | M | White | MF | IA | nbUVB |
| U17-1616 | Phospho-specific flow cytometry | F | White | MF | 1B | TAC, nbUVB |
| U17-1654 | Phospho-specific flow cytometry | M | White | MF | 1A | Clobetasol |
| U17-1657 | Phospho-specific flow cytometry | M | White | MF | IA | TAC |
| U17-1658 | Phospho-specific flow cytometry | M | White | MF | 1A | TAC, topical imiquimod |
| U17-1649 | Phospho-specific flow cytometry | F | White | MF | 1B | Romidepsin |
| U17-1651 | Phospho-specific flow cytometry | F | White | MF | 1B | nbUVB, TAC, interferon, oral bexarotene |
| U17-1652 | Phospho-specific flow cytometry | F | White | MF | IA | Topical bexarotene |
| U17-1653 | Phospho-specific flow cytometry | M | White | MF | IA | TAC |
| U17-1819 | Phospho-specific flow cytometry | M | White | MF | IA | Romidepsin |
| U17-1936 | Phospho-specific flow cytometry | M | White | SS | IVA | Bexarotene and interferon |
| U17-1937 | Phospho-specific flow cytometry | M | White | SS | IIIA | None |
| U17-1938 | Phospho-specific flow cytometry | F | White | MF | 1A | None |
| Patient ID . | Assay . | Sex . | Race . | CTCL type . | Stage at sample procurement . | Treatment at time of collection . |
|---|---|---|---|---|---|---|
| MF1 | Flow cytometry | M | White | MF | IA | Imiquimod |
| MF2 | Flow cytometry | F | White | MF | IA | Desoximetasone |
| MF3 | Flow cytometry | M | White | MF | IA | Clobetasol |
| U16-2023 | Cytotoxicity assay | M | White | MF | IB | PUVA, triamcinolone, imiquimod |
| U16-2101 | Cytotoxicity assay | M | White | MF | IB | Bexarotene |
| U16-2102 | Cytotoxicity assay | M | White | MF | IB | Bexarotene |
| U16-2225 | Cytotoxicity assay | M | White | MF | IIB | Gemcitabine/doxorubicin |
| U16-2228 | Cytotoxicity assay | M | White | MF | IIB | Bexarotene |
| U16-2237 | Cytotoxicity assay | F | African American | MF | IA | Topical steroids |
| U17-0092 | Cytotoxicity assay | M | White | MF | IB | Bexarotene |
| U17-0093 | Cytotoxicity assay | F | White | MF | IB | Topical steroids |
| U17-0094 | Cytotoxicity assay | M | White | MF | IA | nbUVB |
| U17-0095 | Cytotoxicity assay/RNA sequencing | M | White | MF | IA | Bexarotene |
| U17-0253 | Cytotoxicity assay/RNA sequencing | F | White | MF | IA | Topical bexarotene |
| U17-0254 | Cytotoxicity assay/RNA sequencing | F | White | MF | IB | nbUVB |
| U17-0314 | Flow cytometry | F | African American | SS | IVB | Romidepsin |
| U17-0315 | Flow cytometry | F | White | MF | 1A | Topical steroids |
| U17-0316 | Flow cytometry | F | White | MF | IB | Valchlor gel |
| U17-0341 | Flow cytometry | M | White | MF | 1A | Oral or topical bexarotene |
| U17-0342 | Flow cytometry | M | White | MF | 1A | Mechlorethamine gel, clobetasol |
| U17-0343 | Flow cytometry | M | White | MF | 1A | Mechlorethamine gel, TAC |
| U17-0350 | Flow cytometry | F | African American | MF | IA | Topical steroids |
| U17-0352 | Flow cytometry | F | White | MF | IA | Oral bexarotene |
| U17-0353 | Flow cytometry | M | White | MF | 1B | Interferon, nbUVB, TAC, clobetasol |
| U17-0543 | RNA sequencing | M | White | SS | 1VA | Romidepsin, IPH 4102 |
| U17-0593 | RNA sequencing | M | White | MF | 1B | Gemcitabine/ doxorubicin |
| U17-0594 | RNA sequencing | M | White | SS | IVA1 | Topical steroids |
| U17-0595 | RNA sequencing | M | White | MF | IA | TAC |
| U17-1613 | Phospho-specific flow cytometry | F | White | MF | IIB | Brentuximab |
| U17-1614 | Phospho-specific flow cytometry | M | White | SS | 1VA2 | Bendamustine + brentuximab vedotin |
| U17-1615 | Phospho-specific flow cytometry | M | White | MF | IA | nbUVB |
| U17-1616 | Phospho-specific flow cytometry | F | White | MF | 1B | TAC, nbUVB |
| U17-1654 | Phospho-specific flow cytometry | M | White | MF | 1A | Clobetasol |
| U17-1657 | Phospho-specific flow cytometry | M | White | MF | IA | TAC |
| U17-1658 | Phospho-specific flow cytometry | M | White | MF | 1A | TAC, topical imiquimod |
| U17-1649 | Phospho-specific flow cytometry | F | White | MF | 1B | Romidepsin |
| U17-1651 | Phospho-specific flow cytometry | F | White | MF | 1B | nbUVB, TAC, interferon, oral bexarotene |
| U17-1652 | Phospho-specific flow cytometry | F | White | MF | IA | Topical bexarotene |
| U17-1653 | Phospho-specific flow cytometry | M | White | MF | IA | TAC |
| U17-1819 | Phospho-specific flow cytometry | M | White | MF | IA | Romidepsin |
| U17-1936 | Phospho-specific flow cytometry | M | White | SS | IVA | Bexarotene and interferon |
| U17-1937 | Phospho-specific flow cytometry | M | White | SS | IIIA | None |
| U17-1938 | Phospho-specific flow cytometry | F | White | MF | 1A | None |
nbUVB narrow-band ultraviolet B light therapy; PUVA, psoralen and ultraviolet A (light therapy); Tac, triamcinolone cream.
RNA sequencing analysis
NK cells (CD56+/lineage–) were isolated as described above. Total RNA was isolated from cell samples using standard methods (Active Motif). To generate heat maps of the most differential genes, additional comparative metrics were calculated such as fold-change between averages of fragments per kilobase of transcript per million mapped reads (FPKM) values between groups of interests, and analysis of variance P values comparing 2 or more groups. Genes with no values in all samples or a value in only 1 of the 9 samples were removed.
Phosphorylated STAT staining and analysis
Fresh peripheral blood samples were obtained from CTCL patients and age-matched normal donors. Phosphorylated signal of transducer and activator of transcription 3 (pSTAT3) and STAT5 (pSTAT5) were evaluated by direct whole blood antibody labeling (BD Biosciences). Median fluorescence intensity was calculated for each STAT protein.
Results
The absolute number of NK cells in peripheral blood was evaluated in CTCL patients and compared with that in normal donors (n = 51). There was no statistical difference in absolute number of NK cells when all patients with CTCL were included (Figure 1A); however, SS patients had on average 57.4% fewer NK cells compared with normal donors (supplemental Figure 1). We then evaluated the association between absolute NK-cell counts and overall survival. NK-cell counts were significantly associated with overall survival (P = .041; Figure 1B). To evaluate NK-cell function, NK cells were purified from fresh peripheral blood (Figure 1C) and evaluated for cytotoxic function against K562 target cells.24 CTCL patients had significantly higher levels of NK-cell cytotoxicity compared with normal donors (Figure 1D). Although these findings differ from those in previous reports, earlier work did not use NK cells isolated from fresh peripheral blood,10-12 evaluate frozen samples, or use cytokine stimulation.14
NK-cell number and correlation with CTCL patient survival. (A) Absolute NK-cell numbers were calculated in normal donors (n = 51; mean ± standard error of the mean [SEM], 0.2442 ± 0.02 ) and CTCL patients (n = 121; 0.208 ± 0.01; P = .08). (B) Kaplan-Meier curves for overall survival at possible absolute NK-cell counts in CTCL patients (n = 121). (C) NK cells, CD56+/lineage (CD3/CD14/CD20), were isolated from freshly obtained peripheral blood samples from CTCL patients and normal control donors. (D) Purified NK cells were co-cultured with K562 leukemic targets in a standard chromium release assay at indicated ratios. Data are presented as mean ± SEM. *P ≤ .05; **P ≤ .01; ***P ≤ .001; unpaired 2-tailed Student t test. FSC, forward scatter; ns, not significant; SSC, side scatter.
NK-cell number and correlation with CTCL patient survival. (A) Absolute NK-cell numbers were calculated in normal donors (n = 51; mean ± standard error of the mean [SEM], 0.2442 ± 0.02 ) and CTCL patients (n = 121; 0.208 ± 0.01; P = .08). (B) Kaplan-Meier curves for overall survival at possible absolute NK-cell counts in CTCL patients (n = 121). (C) NK cells, CD56+/lineage (CD3/CD14/CD20), were isolated from freshly obtained peripheral blood samples from CTCL patients and normal control donors. (D) Purified NK cells were co-cultured with K562 leukemic targets in a standard chromium release assay at indicated ratios. Data are presented as mean ± SEM. *P ≤ .05; **P ≤ .01; ***P ≤ .001; unpaired 2-tailed Student t test. FSC, forward scatter; ns, not significant; SSC, side scatter.
A comprehensive surface immunophenotypic analysis of NK cells revealed no significant differences (supplemental Figure 2), so we hypothesized that there could be alterations in pathway activation in CTCL patients. Whole transcriptome analysis was completed to evaluate NK-cell activation. A heat map generated by hierarchical clustering of the differentially expressed genes shows the unique patterns of gene expression in NK cells from CTCL patients and healthy donors (Figure 2A). NK cells from CTCL patients had increased expression of cytolytic mediators, including perforin, granzyme A, granzyme B, Fas, and tumor necrosis factor-alpha–related apoptosis-inducing ligand (TRAIL) (Figure 2B). Furthermore, we observed significantly increased cytokine (interferon-γ [IFN-γ]) production in patient NK cells. Using ingenuity pathway analysis, we determined differential expression of key pathways, including IL-15, IL-15/IL-2 receptors, IFN-γ, and several surface activating molecules such as CD2, CD40, FCER1G, CD80, KLR, SELL, and CD244. Furthermore, NF-kB and ICOS were inhibited in our data set (Figure 2C; supplemental Figures 3 and 4).
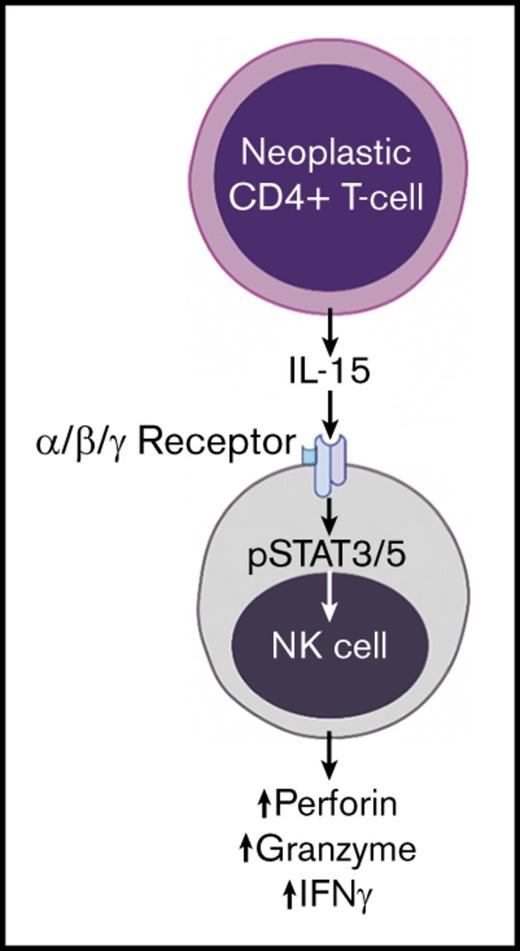
Differential RNA expression analysis in CTCL patients. NK cells were isolated from CTCL patients and normal donors and evaluated by RNA sequencing. (A) Heat map of differentially regulated transcripts among normal donors (n = 3) and CTCL MF (n = 5) and CTCL SS (n = 2) patients. (B) Mediators of cytolytic activity and NK-cell activation were evaluated in all CTCL patients compared with normal controls. Perforin mean ± SEM of transcript in healthy donor vs CTCL patients for NK cells (91.39 ± 9.18 [n = 3] vs 485.7 ± 43.15 [n = 7]; P = .0004), granzyme A (GZMA) (58.15 ± 6.605 [n = 3] vs 218.7 ± 29.78 [n = 7]; P = .0094), granzyme B (GZMB) (143 ± 23.79 [n = 3] vs 431 ± 68.13 [n = 7]; P = .03), Fas (1.413 ± 0.01849 [n = 3] vs 3.967 ± 0.4395 [n = 7]; P = .0063), tumor necrosis factor-alpha–related apoptosis-inducing ligand (TRAIL) (3.756 ± 0.4925 [n = 3] vs 7.634 ± 1.033 [n = 7]; P = .0477), and IFN-G (2.861 ± 0.5007 [n = 3] vs 7.325 ± 1.621 [n = 7]; P = .1218). (C) An ingenuity pathway analysis upstream functional analysis was performed in an enriched data set from the NK cells. Red indicates upregulation in CTCL patients compared with normal donors; green indicates reduced expression. (D) RNA expression of receptor components required for both IL-2 and IL-15 signaling were evaluated in CTCL patients and compared with that in normal donors (IL-15Rα: mean ± SEM of relative RNA in normal vs CTCL, 0.8677 ± 0.1018 [n = 3] vs 1.772 ± 0.2055 [n = 7]; P = .03; IL-15Rγ, 57.02 ± 7.525 [n = 3] vs 154.7 ± 12.14 [n = 7]; P = .001. IL-15Rβ was also elevated (128 ± 23.68 [n = 3] vs 223.2 ± 25.43 [n = 7]; P = .056). (E) Protein expression of pSTAT3 and pSTAT5 on NK cells was determined in freshly obtained whole blood samples from CTCL patients and matched with that of normal donors by flow cytometry. Graph indicates mean fluorescence intensity (MFI) of normal (gray bars [n = 7]) compared with CTCL patients (green bars [n = 14]); pSTAT3: normal, 2417 ± 154.1 (n = 7) vs CTCL, 2734 ± 199; P = .32; pSTAT5: normal, 1253 ± 50.23 (n = 8) vs CTCL, 1405 ± 64.35; P = .028. (F) Schematic of interaction between malignant CD4+ T cells in CTCL patients producing IL-15, which binds to the upregulated IL-15 receptor complex on NK cells and enhances downstream activating pathways in NK cells. Data are presented as mean ± SEM. *P ≤ .05; **P ≤ .01; ***P ≤ .001; unpaired 2-tailed Student t test. FPKM, fragments per kilobase million.
Differential RNA expression analysis in CTCL patients. NK cells were isolated from CTCL patients and normal donors and evaluated by RNA sequencing. (A) Heat map of differentially regulated transcripts among normal donors (n = 3) and CTCL MF (n = 5) and CTCL SS (n = 2) patients. (B) Mediators of cytolytic activity and NK-cell activation were evaluated in all CTCL patients compared with normal controls. Perforin mean ± SEM of transcript in healthy donor vs CTCL patients for NK cells (91.39 ± 9.18 [n = 3] vs 485.7 ± 43.15 [n = 7]; P = .0004), granzyme A (GZMA) (58.15 ± 6.605 [n = 3] vs 218.7 ± 29.78 [n = 7]; P = .0094), granzyme B (GZMB) (143 ± 23.79 [n = 3] vs 431 ± 68.13 [n = 7]; P = .03), Fas (1.413 ± 0.01849 [n = 3] vs 3.967 ± 0.4395 [n = 7]; P = .0063), tumor necrosis factor-alpha–related apoptosis-inducing ligand (TRAIL) (3.756 ± 0.4925 [n = 3] vs 7.634 ± 1.033 [n = 7]; P = .0477), and IFN-G (2.861 ± 0.5007 [n = 3] vs 7.325 ± 1.621 [n = 7]; P = .1218). (C) An ingenuity pathway analysis upstream functional analysis was performed in an enriched data set from the NK cells. Red indicates upregulation in CTCL patients compared with normal donors; green indicates reduced expression. (D) RNA expression of receptor components required for both IL-2 and IL-15 signaling were evaluated in CTCL patients and compared with that in normal donors (IL-15Rα: mean ± SEM of relative RNA in normal vs CTCL, 0.8677 ± 0.1018 [n = 3] vs 1.772 ± 0.2055 [n = 7]; P = .03; IL-15Rγ, 57.02 ± 7.525 [n = 3] vs 154.7 ± 12.14 [n = 7]; P = .001. IL-15Rβ was also elevated (128 ± 23.68 [n = 3] vs 223.2 ± 25.43 [n = 7]; P = .056). (E) Protein expression of pSTAT3 and pSTAT5 on NK cells was determined in freshly obtained whole blood samples from CTCL patients and matched with that of normal donors by flow cytometry. Graph indicates mean fluorescence intensity (MFI) of normal (gray bars [n = 7]) compared with CTCL patients (green bars [n = 14]); pSTAT3: normal, 2417 ± 154.1 (n = 7) vs CTCL, 2734 ± 199; P = .32; pSTAT5: normal, 1253 ± 50.23 (n = 8) vs CTCL, 1405 ± 64.35; P = .028. (F) Schematic of interaction between malignant CD4+ T cells in CTCL patients producing IL-15, which binds to the upregulated IL-15 receptor complex on NK cells and enhances downstream activating pathways in NK cells. Data are presented as mean ± SEM. *P ≤ .05; **P ≤ .01; ***P ≤ .001; unpaired 2-tailed Student t test. FPKM, fragments per kilobase million.
Our group previously demonstrated that CD4+ T cells from CTCL patients exhibit elevated levels of IL-15, a key cytokine that mediates NK-cell activation and homeostasis.19 There was significant upregulation of several IL-15 receptor subunits on NK cells (Figure 2D). To further confirm activation of the IL-15 pathway, we evaluated STAT signaling in fresh peripheral blood NK cells by flow cytometry, because IL-15 signals through both STAT3 and STAT5.25 There was a nonsignificant trend toward increased pSTAT3 expression and a significant upregulation of pSTAT5 in CTCL patients compared with normal donors (Figure 2E). Overall, we propose a mechanism by which malignant CD4+ T cells produce IL-15, which binds to the highly expressed IL-15 receptor complex on NK cells from CTCL patients. NK-cell activation is reflected in enhanced cytotoxicity, STAT5 phosphorylation, and upregulation of downstream effectors (Figure 2F).
Discussion
Although our initial goal with this work was to define the functional capacity of freshly isolated NK cells in CTCL patients, our comprehensive analysis of both NK-cell function and expression profiles uncovered intriguing results. One of the most interesting findings of this study was the significant association between NK-cell number and CTCL patient survival. To the best of our knowledge, this is the first description of higher NK-cell numbers in a malignancy being associated with decreased short-term survivability. The cause of this significant clinical relationship is unknown. Few recent studies have described the tumor-promoting potential of NK cells through their ability to upregulate certain oncogenic pathways such as VEGF-A,25 mediated in part by reduced STAT5 activity. We described increased pSTAT5 and did not identify upregulation of VEGF-A or other immunosuppressive factors. It is possible that there are other yet undiscovered mechanisms of tumor promotion by NK cells, and these may lead to the failure of NK cells to control tumor progression despite elevated cytotoxic activity. It is also possible that there may be additional immunosuppressive mechanisms that are present in the microenvironment of CTCL patients. For example, previous studies by our group and others have demonstrated alterations in NK-cell signaling because of the presence of suppressive myeloid cells such as myeloid-derived suppressor cells and tumor-associated macrophages in cancer patients,26 and both populations have been reported among CTCL patients.27-29
Additional checkpoint inhibitors, such as PD-1, CTLA-4, TIGIT, and TIM-3 may also play a role in decreasing NK-cell function in patients, because previous studies demonstrated that these inhibitors may be increased in the presence of IL-15.30-32 Transcript analysis of PD-1 did show a moderate increase in the NK cells isolated from CTCL patients (supplemental Figure 5), and because K562 cells are known to express low levels of the PD-1 ligand (PD-L1), this inhibitory effect might not be observed in vitro.33 Analysis of inhibitory ligand expression on CTCL cells in circulation in SS patients or in skin biopsies from MF patients suggests that novel potential therapeutic targets could be focused on removing this barrier to immune cell activation.
It is known that γ cytokines such as IL-7 and IL-15 are important for CTCL progression.19,34,35 Indeed, as disease progresses, neoplastic CD4+ T cells express higher levels of IL-15.19 Furthermore, IL-15 overexpression alone can induce CTCL in a murine model of the disease.19 IL-15 is also known to stimulate NK-cell proliferation and cytotoxicity via phosphorylation of STAT5.13,36-38 Thus, we proposed that IL-15 derived from the neoplastic CD4+ T cells in CTCL may contribute to increased NK-cell cytotoxicity observed in patient samples. Indeed, transcriptome analysis reveals upregulation of IL-15–induced signaling pathways, and protein levels of pSTAT5 were significantly elevated in patients with CTCL, further confirming this relationship between increased NK-cell activation and cytotoxicity in CTCL patients. However, recent reports by multiple groups have demonstrated that continuous exposure to IL-15 can lead to a decrease in NK-cell proliferation, cell cycle arrest, and NK-cell exhaustion, suggesting that chronic exposure to IL-15 could also lead to immune-cell exhaustion in CTCL patients.39,40 It is also possible that although NK cells from peripheral blood exhibit higher levels of cytotoxicity, those localized in skin lesions where localized levels of IL-15 may be higher have additional defects that would render them ineffective at lysing the CD4+ malignant T cells.
The reason for this counterintuitive finding is not known; however, we speculate that although NK cells are maintained in a hyperactive state in CTCL patients, malignant cell recognition is impaired. It is also possible that the ability of NK cells to form an effective immune synapse and polarize actin and cytolytic granules is altered in the microenvironment of CTCL patients. In support of this theory, a key mediator of NK-cell polarization, phosphatase and tensin homolog (PTEN), is significantly overexpressed in NK cells from CTCL patients in the RNA sequencing analysis (data not shown). We have previously demonstrated a role for PTEN in the organization of the components of the immunologic synapse and the appropriate convergence of cytolytic granules.41 It is also possible that the process of NK-cell recognition of malignant CD4+ T cells is altered in CTCL patients. Previous studies by Bouaziz et al15 suggest that NK cells are potentially able to be activated to kill autologous CTCL cells, which suggests that malignant CD4+ T cells are susceptible to NK-cell killing, but additional mechanisms of inhibition such as the ones discussed above prevent this in CTCL patients. Although further investigation of multiple facets of NK-cell recognition is warranted, it is clear that the overall immunosuppressive microenvironment in CTCL patients contributes to insufficiency of patient NK cells to effectively control CTCL progression.
The full-text version of this article contains a data supplement.
Acknowledgments
Support for this study was provided by the American Association for Cancer Research (17-20-46-MUND) (B.M.-B.), Spatz Foundation (A.M.), American Skin Association (A.M.), Cutaneous Lymphoma Foundation (A.M.), Pelotonia (A.M.), DeStefano Lymphoma Research funds (A.M.), and the National Institutes of Health, National Cancer Institute (CA016058). The authors gratefully acknowledge The Ohio State University Comprehensive Cancer Center Leukemia Tissue Bank Shared Resource (P30CA016058) for patient samples.
Authorship
Contribution: B.M.-B., L.C., H.C.M., Y.Y., G.L., N.D., and A.M. assisted with experimental design and implementation; E.M. and D.K. assisted with statistical modeling and bioinformatics analysis; P.P., B.W., and S.H. assisted with patient identification and sample acquisition; N.C., D.A.L., and A.M. assisted with ingenuity pathway analysis; B.M.-B., R.K., and A.M. assisted with writing and analyzing experiments; and A.G.F., P.P., and M.A.C. provided critical design and manuscript assistance.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anjali Mishra, Comprehensive Cancer Center, Division of Dermatology, Department of Internal Medicine, The Ohio State University, 886 Biomedical Research Tower, 460 W. 12th Ave, Columbus, OH 43210; e-mail: anjali.mishra@osumc.edu; and Bethany Mundy-Bosse, Comprehensive Cancer Center, Division of Hematology, Department of Internal Medicine, The Ohio State University, 882 Biomedical Research Tower, 460 W. 12th Ave, Columbus, OH 43210; e-mail: bethany.mundy@osumc.edu.

![Figure 1. NK-cell number and correlation with CTCL patient survival. (A) Absolute NK-cell numbers were calculated in normal donors (n = 51; mean ± standard error of the mean [SEM], 0.2442 ± 0.02 ) and CTCL patients (n = 121; 0.208 ± 0.01; P = .08). (B) Kaplan-Meier curves for overall survival at possible absolute NK-cell counts in CTCL patients (n = 121). (C) NK cells, CD56+/lineage (CD3/CD14/CD20), were isolated from freshly obtained peripheral blood samples from CTCL patients and normal control donors. (D) Purified NK cells were co-cultured with K562 leukemic targets in a standard chromium release assay at indicated ratios. Data are presented as mean ± SEM. *P ≤ .05; **P ≤ .01; ***P ≤ .001; unpaired 2-tailed Student t test. FSC, forward scatter; ns, not significant; SSC, side scatter.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/2/15/10.1182_bloodadvances.2018020388/3/m_advances020388f1.jpeg?Expires=1771013919&Signature=vxtXT8ReX3nzd0bn4VRaxMce9FaFcgVczXfOUxwAU0bWN4-g80RhVcBppzueeub4mEeehGNA0LmqjELm~YWt19U-LOQdSOJ9w-x1rnvqN9qk3XLlRGhdpP-a5PO7A-OfHMKzjMUdUIONtD8AQkT7km1QkWNkuyKFXimXVTqMDTCKqApcd6VuR~MVkW8O-~sydjDx8wYJWFppaAv9Ub1-Blrpg9HBlStTl--DOcAI6WlIopbfVRtFEZOn2g9xguWAhDLW2j0fbDGM2~Eav-bLOGWUMMuaEyjyx0TfIxFssMD~HZMIWx0Ip2YUZdbKgm0t47RvfA3MXtoqlNnOmKEdyQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Differential RNA expression analysis in CTCL patients. NK cells were isolated from CTCL patients and normal donors and evaluated by RNA sequencing. (A) Heat map of differentially regulated transcripts among normal donors (n = 3) and CTCL MF (n = 5) and CTCL SS (n = 2) patients. (B) Mediators of cytolytic activity and NK-cell activation were evaluated in all CTCL patients compared with normal controls. Perforin mean ± SEM of transcript in healthy donor vs CTCL patients for NK cells (91.39 ± 9.18 [n = 3] vs 485.7 ± 43.15 [n = 7]; P = .0004), granzyme A (GZMA) (58.15 ± 6.605 [n = 3] vs 218.7 ± 29.78 [n = 7]; P = .0094), granzyme B (GZMB) (143 ± 23.79 [n = 3] vs 431 ± 68.13 [n = 7]; P = .03), Fas (1.413 ± 0.01849 [n = 3] vs 3.967 ± 0.4395 [n = 7]; P = .0063), tumor necrosis factor-alpha–related apoptosis-inducing ligand (TRAIL) (3.756 ± 0.4925 [n = 3] vs 7.634 ± 1.033 [n = 7]; P = .0477), and IFN-G (2.861 ± 0.5007 [n = 3] vs 7.325 ± 1.621 [n = 7]; P = .1218). (C) An ingenuity pathway analysis upstream functional analysis was performed in an enriched data set from the NK cells. Red indicates upregulation in CTCL patients compared with normal donors; green indicates reduced expression. (D) RNA expression of receptor components required for both IL-2 and IL-15 signaling were evaluated in CTCL patients and compared with that in normal donors (IL-15Rα: mean ± SEM of relative RNA in normal vs CTCL, 0.8677 ± 0.1018 [n = 3] vs 1.772 ± 0.2055 [n = 7]; P = .03; IL-15Rγ, 57.02 ± 7.525 [n = 3] vs 154.7 ± 12.14 [n = 7]; P = .001. IL-15Rβ was also elevated (128 ± 23.68 [n = 3] vs 223.2 ± 25.43 [n = 7]; P = .056). (E) Protein expression of pSTAT3 and pSTAT5 on NK cells was determined in freshly obtained whole blood samples from CTCL patients and matched with that of normal donors by flow cytometry. Graph indicates mean fluorescence intensity (MFI) of normal (gray bars [n = 7]) compared with CTCL patients (green bars [n = 14]); pSTAT3: normal, 2417 ± 154.1 (n = 7) vs CTCL, 2734 ± 199; P = .32; pSTAT5: normal, 1253 ± 50.23 (n = 8) vs CTCL, 1405 ± 64.35; P = .028. (F) Schematic of interaction between malignant CD4+ T cells in CTCL patients producing IL-15, which binds to the upregulated IL-15 receptor complex on NK cells and enhances downstream activating pathways in NK cells. Data are presented as mean ± SEM. *P ≤ .05; **P ≤ .01; ***P ≤ .001; unpaired 2-tailed Student t test. FPKM, fragments per kilobase million.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/2/15/10.1182_bloodadvances.2018020388/3/m_advances020388f2.jpeg?Expires=1771013919&Signature=Fm8twinjR9F49CnGILe~GoO6XB3iYtCn~kJsU75~xump834-1JKxAI0H7n5~Il6jNkfr-Yu8wxJewa6hAXfCJf2cnglTAOCaW6WRHx1vTmP94V9zjBsXDqcd8X8tR9FgWrIKODAa2Nldur~5lCSql2d19gZdci0OYwKM8opbaE~lbHxtCeW0pBjsf~ey-H8wQ7eqihIAUuWDjfv5D65IdAlFMZsEW09jg5TTKcS-1xozjMrbwyTT6phs2d1JS88B8-Wu5FDm8Ibyl86KninqwGI-jHMxjJ69HxvG9DlMLo-zLoGQEI8hmMOUuAOii7lW3uCRQWZ50f6squmQlaXeug__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)