Key Points
MLL-AF9 AML cell–induced BM niche alteration is correlated with leukemia burden.
Ebf2+ MSPCs participate in AML niche formation, and Ebf2+ cell depletion accelerates AML development.
Abstract
Despite increasing evidence for the involvement of bone marrow (BM) hematopoietic stem cell niche in leukemogenesis, how BM mesenchymal stem and progenitor cells (MSPCs) contribute to leukemia niche formation and progression remains unclear. Using an MLL-AF9 acute myeloid leukemia (AML) mouse model, we demonstrate dynamic alterations of BM cellular niche components, including MSPCs and endothelial cells during AML development and its association with AML engraftment. Primary patient AML cells also induced similar niche alterations in xenografted mice. AML cell infiltration in BM causes an expansion of early B-cell factor 2+ (Ebf2+) MSPCs with reduced Cxcl12 expression and enhanced generation of more differentiated mesenchymal progenitor cells. Importantly, in vivo fate-mapping indicates that Ebf2+ MSPCs participated in AML niche formation. Ebf2+ cell deletion accelerated the AML development. These data suggest that native BM MSPCs may suppress AML. However, they can be remodeled by AML cells to form leukemic niche that might contribute to AML progression. AML induced dysregulation of hematopoietic niche factors like Angptl1, Cxcl12, Kitl, Il6, Nov, and Spp1 in AML BM MSPCs, which was associated with AML engraftment and partially appeared before the massive expansion of AML cells, indicating the possible involvement of the niche factors in AML progression. Our study demonstrates distinct dynamic features and roles of BM MSPCs during AML development.
Introduction
Acute myeloid leukemia (AML) is characterized by accumulation of immature myeloid blasts in blood, bone marrow (BM), and other organs.1,2 The efficacy of current treatments for AML targeting leukemic cells has been disappointing, and relapse is common.3,4 Therefore, there is urgent need to identify new therapeutic targets to develop additional treatment strategies for AML. It has been believed that the disease persistence is attributed to residual leukemia-initiating cells or leukemic stem cells (LSCs) that are protected by a specialized BM microenvironment, the so-called hematopoietic stem cell (HSC) niche.5-9 These leukemic cells outcompete normal HSCs for the niche occupancy,10 which ultimately causes disruption of normal hematopoiesis and mortality. Therefore, efforts have been put into untangling the complex interactions between leukemic cells and neighboring stromal cells.11,12 Some of the niche components have been proposed to be critical as the novel candidate target for therapies in AML.13,14
The BM HSC niche is composed of various types of stromal cells, including osteoblasts, adipocytes, perivascular cells, endothelial cells (ECs), mesenchymal stem cells (MSCs), and mesenchymal progenitor cells (MPCs).11,15 MSCs are the precursors of mesenchymal lineages like osteoblasts, adipocytes, and chondrocytes.16 BM MSCs are enriched in CD45−TER119−CD31−CD44− stromal cells17 and can be isolated based on their expression of SCA1 and CD51 or PDGFRA/CD140A.18-20 The MSCs (SCA1+CD51+ or SCA1+PDGRA+) are functionally estimated by their ability to form fibroblast colony-forming units (CFU-Fs) in vitro, and they can generate more differentiated MPCs (SCA1−CD51+) with single or bilineage potential but with little to no CFU-F activity.15,21 The SCA1−CD51+ MPCs, largely overlapping (75%) with Nestin-GFP+ cells, have been shown to be enriched for HSC maintenance genes.22
The HSC niche tightly regulates normal hematopoiesis by controlling HSC fate and contributes to leukemogenesis.11 The niche requirement for AML development was considered to be disease-stage specific in a study showing that the niche was required for leukemogenesis in a preleukemic stage, although it became permissive once leukemia was established.23 Then, malignant hematopoietic cells could actively reprogram the BM niche into a self-reinforcing leukemic niche, thereby propelling leukemia.24,25 A similar finding was reported in acute lymphoid leukemia.26
The niche function and structure in leukemia has been reported to be disease-type specific,27 emphasizing that a thorough BM niche characterization in different types of leukemia is required in order to identify possible disease-specific therapeutic targets. Although BM niche alterations have been reported in the MLL-AF9 AML mouse model,14 the dynamic alterations and the roles of different BM cellular niche components in AML remain largely unexplored.
We here demonstrate dynamic functional and molecular alterations of BM MSPCs in MLL-AF9 AML mouse BM that are associated with AML burden. We also show that early B-cell factor 2 (Ebf2)+ cells, a highly purified primitive MSC population identified in mouse BM,28 are dramatically expanded in AML mouse BM and primed toward differentiation. Ebf2+ cells constitute the major fraction of AML niche. Most importantly, depletion of Ebf2+ cells leads to an accelerated development of the AML. The dysregulations of HSC niche factors in AML MSPCs may aid in identification of novel therapeutic targets for treatment of AML.
Materials and methods
Mice
Wild-type CD45.2 C57BL/6J or CD45.1 B6.SJL-Ptprca Pepcb/BoyJ (The Jackson Laboratory) or Ebf2-Egfp reporter FVB/N mice28 at 8 to 12 weeks were used for transplantation of MLL-AF9 AML cells. Triple-transgenic Ebf2-Egfp × Ebf2-CreER × Rosa26-loxpStoploxp-Tomato mice were generated by crossing Ebf2-Egfp with Ebf2-CreER × Rosa26-loxpStoploxp-Tomato mice and used to trace Ebf2+ cells. Ebf2-CreER × Rosa26-loxpStoploxp-Dta mouse models were used for specific deletion of Ebf2+ cells in vivo. Mice carrying one of the transgenes were used as controls. Mice were injected with tamoxifen (TAM) (Sigma) intraperitoneally at 3 mg/20 g body weight every second day 3 times to induce recombination. All mice were maintained in specific-pathogen–free conditions in the animal facility of Karolinska Institute. Animal procedures were performed with approval from the local ethics committee (ethical number S40-14) at Karolinska Institute (Stockholm, Sweden).
Multicolor fluorescence-activated cell sorting (FACS) of MSCs
Human and mouse MSCs were isolated as described previously.17 See supplemental Materials and methods for the detailed procedure.
Generation of the MLL-AF9–induced AML syngeneic murine model
The AML mouse model was generated by transplanting mouse BM KIT+ cells transduced with MLL-AF9 retrovirus as described previously.29 BM CD45.1+ KIT+ cells for virus transduction were first enriched by magnetic-activated cell sorting using KIT-microbeads (Miltenyi Biotec) and then sorted by FACS from 8- to 10-week-old C57BL/6J or FVB/N mice. Cells were then transduced with MLL-AF9 retrovirus. Transduced cells were clonally propagated and selected by colony assay in methylcellulose M3434 and subsequently by transplantation. MLL-AF9–expressing cells were sorted by FACS from primary recipient mice that developed AML and then expanded in culture in presence of interleukin-3 (10 ng/mL) in RPMI + 10% fetal bovine serum for secondary transplantation to establish the AML mouse model. Finally, 50 000, 250 000, or 1 million MLL-AF9–expressing cells were intravenously transplanted into nonirradiated mice.
Human sample collection and MNC isolation
BM samples were collected from adult or pediatric patients with AML at diagnosis and healthy donors (30-45 years old). The experiments were approved by the local ethical committee at Stockholm (2012/4:10, 2013/3:1 and 2013/1248-31/4), and informed consent was obtained from the patients or guardians and healthy donors. Mononuclear cell isolation from the BM samples was done as previously described.17
Xenograft transplantation of patient AML cells into NSG-SGM3 mice
Primary BM or peripheral blood (PB) mononuclear cells from adult (n = 3) and pediatric patients (n = 3) with AML were transplanted via intrafemoral injection into human cytokine engineered immunodeficient NSG-SGM3 mice (Jackson Laboratory) at doses of 100 000-500 000 cells/mouse. One of 6 patient samples was with MLL-AF9 mutation. The recipient mice were subjected to sublethal irradiation (220 cGy). The BM was collected from the recipients at 6 to 8 weeks after transplantation for analyzing leukemia engraftment and BM stromal cell alterations.
CFU-F assay
CFU-F assay was performed as described previously.17,28
Statistical analysis
Unpaired Student t and Mann-Whitney U tests were used to determine the differences based on data distribution. Pearson or spearman correlation was applied to analyze the correlation analysis. The Kaplan-Meier survival curve of the mice was generated using a log-rank (Mantel-Cox) test in Prism 6.0. All reported P values were obtained using Prism 6.0 software, and P < .05 was considered statistically significant.
See supplemental Materials and methods for additional methods.
Results
AML remodeled the BM cellular niche compartment
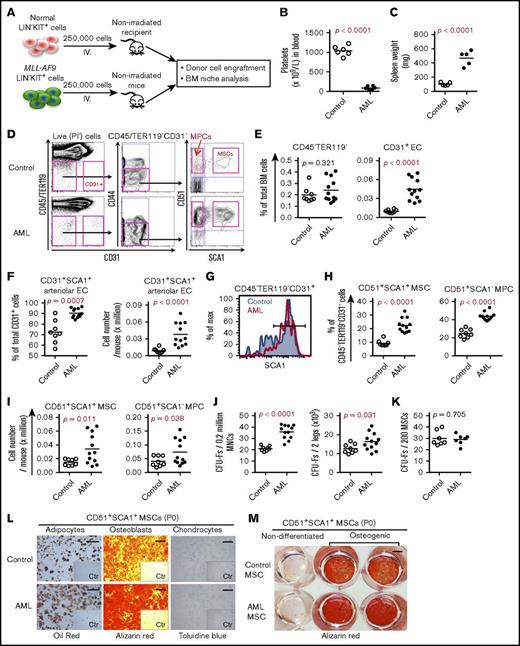
In order to explore leukemia-induced alterations in the BM niche, we used a syngeneic AML mouse model29 by transplanting MLL-AF9–transduced CD45.1+ BM KIT+ cells into nonirradiated CD45.2+ wild-type mice after clonal selection in culture and transplantation. When 250 000 MLL-AF9–expressing cells were transplanted, most C57BL/6 mice developed AML with thrombocytopenia and splenomegaly within 27 to 30 days after transplantation (Figure 1A-C). At the late stage of AML, the number and frequencies of total stromal cells (CD45−TER119−) in the BM remained unchanged in AML mice compared with control mice transplanted with normal KIT+ hematopoietic stem and progenitor cells (HSPCs) (Figure 1D-E; supplemental Figure 1A). However, consistent with the previous findings,14 total CD45−TER119−CD31+ ECs and SCA1+CD31+ arteriolar ECs, but not sinusoidal ECs, were dramatically increased in AML mice (Figure 1E-G; supplemental Figure 1B). The proportions of CD45−TER119− CD31−CD44−CD51+SCA1+ MSCs and CD45−TER119−CD31−CD44−CD51+SCA1− MPCs were significantly increased in AML mice compared with control mice (Figure 1D,H-I). This finding was further supported by the increased frequencies and absolute counts of CFU-Fs in AML mouse BM. The expansion of AML MSPCs might be due to reduced apoptosis, since we detected increased expression of the antiapoptotic gene Bcl2 and reduced apoptotic cells in AML MSPCs (supplemental Figure 2A-C). There was no significant change in either cell-cycle status or cell-cycle regulator expression in AML MSCs (supplemental Figure 2D-F). Nevertheless, we observed an increase of AML MPCs residing in S/G2/M stage, even though Cdk6 expression declined in AML MPCs (supplemental Figure 2E-F).
BM cellular niche alterations in MLL-AF9 AML mice. (A) Strategy for establishing the MLL-AF9 mouse–induced AML mouse model. Normal and MLL-AF9–expressing BM KIT+ cells were transplanted to nonirradiated wild-type C57BL/6 mice. BM stromal cells from control and AML mice were collected and sorted by FACS for subsequent analysis when the mice receiving MLL-AF9–expressing cells developed AML. (B) Platelet numbers in the blood of the AML mice. (C) Increased spleen weight of AML mice. (D) Representative FACS profile showing the gating strategy used for analysis and sorting of BM MSCs and MPCs from control and AML mice. SCA1+CD51+ MSCs and SCA1−CD51+ MPCs were first gated within CD45−TER119−CD31−CD44− cells. CD31+ ECs were gated within CD45−TER119− cells. PI, propidium iodide. (E) Frequencies of CD45−TER119− cells and CD31+ ECs in AML mouse BM. (F) Frequency and absolute number of SCA1+CD31+ arteriolar ECs and SCA1−CD31+ sinusoidal ECs in AML mice. (G) FACS histogram of SCA1+ arteriolar ECs in AML mice. Cells were gated from CD45−TER119−CD31+ cells. (H) Frequencies of BM MSCs and MPCs in AML mice. (I) Absolute numbers of BM MSCs and MPCs in AML mice. (J) Increased frequency and absolute number of CFU-Fs in AML mice. (K) CFU-F activities in freshly sorted MSCs from control mice and AML mice. (L) Enhanced adipocyte and osteoblast differentiation potential of AML MSCs in vitro. The adipogenic, osteogenic, and chondrogenic differentiations were stained with oil red, alizarin red, and toluidine blue, respectively. Scale bars represent 250 μm (left), 500 μm (middle), and 100 μm (right). The inserts are the control (Ctr) images of the nondifferentiated culture. P0, postnatal day 0. (M) An overview image of alizarin red staining of the induced osteoblast differentiation of control and AML MSCs. Data were generated from the same experiment as shown in panel L. Scale bar, 500 μm. Statistical analysis was performed using an unpaired 1-tailed (B-C) or 2-tailed (E-K) Student t test or Mann-Whitney U test. Each dot represents data from an individual mouse. The horizontal bars show mean values. Data are from 2 independent experiments.
BM cellular niche alterations in MLL-AF9 AML mice. (A) Strategy for establishing the MLL-AF9 mouse–induced AML mouse model. Normal and MLL-AF9–expressing BM KIT+ cells were transplanted to nonirradiated wild-type C57BL/6 mice. BM stromal cells from control and AML mice were collected and sorted by FACS for subsequent analysis when the mice receiving MLL-AF9–expressing cells developed AML. (B) Platelet numbers in the blood of the AML mice. (C) Increased spleen weight of AML mice. (D) Representative FACS profile showing the gating strategy used for analysis and sorting of BM MSCs and MPCs from control and AML mice. SCA1+CD51+ MSCs and SCA1−CD51+ MPCs were first gated within CD45−TER119−CD31−CD44− cells. CD31+ ECs were gated within CD45−TER119− cells. PI, propidium iodide. (E) Frequencies of CD45−TER119− cells and CD31+ ECs in AML mouse BM. (F) Frequency and absolute number of SCA1+CD31+ arteriolar ECs and SCA1−CD31+ sinusoidal ECs in AML mice. (G) FACS histogram of SCA1+ arteriolar ECs in AML mice. Cells were gated from CD45−TER119−CD31+ cells. (H) Frequencies of BM MSCs and MPCs in AML mice. (I) Absolute numbers of BM MSCs and MPCs in AML mice. (J) Increased frequency and absolute number of CFU-Fs in AML mice. (K) CFU-F activities in freshly sorted MSCs from control mice and AML mice. (L) Enhanced adipocyte and osteoblast differentiation potential of AML MSCs in vitro. The adipogenic, osteogenic, and chondrogenic differentiations were stained with oil red, alizarin red, and toluidine blue, respectively. Scale bars represent 250 μm (left), 500 μm (middle), and 100 μm (right). The inserts are the control (Ctr) images of the nondifferentiated culture. P0, postnatal day 0. (M) An overview image of alizarin red staining of the induced osteoblast differentiation of control and AML MSCs. Data were generated from the same experiment as shown in panel L. Scale bar, 500 μm. Statistical analysis was performed using an unpaired 1-tailed (B-C) or 2-tailed (E-K) Student t test or Mann-Whitney U test. Each dot represents data from an individual mouse. The horizontal bars show mean values. Data are from 2 independent experiments.
To assess the functionality of the AML MSCs, we performed a CFU-F and multilineage differentiation assay on the sorted MSCs. AML MSCs contained a frequency of CFU-Fs similar to that of control mice (Figure 1J-K), and the colony size derived from AML MSCs was comparable to control MSCs (supplemental Figure 3A). However, AML MPCs appeared to form more CFU-Fs but fewer osteoblast colonies (supplemental Figure 3B-C). We did not observe any difference in population doubling time between AML MSCs and control MSCs (supplemental Figure 3D), suggesting normal proliferation capacity of the AML MSCs.
However, AML MSCs exhibited enhanced osteogenic and adipogenic differentiation potential (Figure 1L-M; supplemental Figure 3E). Similar to a previous report,14 micro–computed tomography analysis indicated a reduced femoral bone density in AML mice (supplemental Figure 3F). However, we did not detect significant difference in osteocalcin expression between control and AML bones (n = 3; supplemental Figure 3G-H).
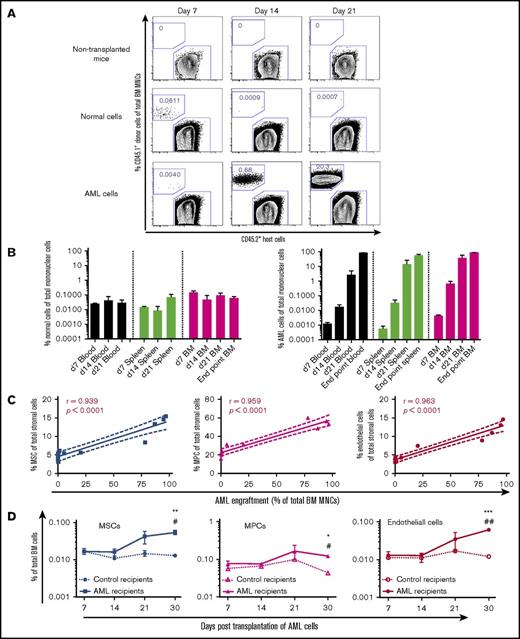
To determine the dynamic changes of the BM niche in relation to AML engraftment, we analyzed the kinetics of BM cellular niche composition and AML engraftment. As expected, normal HSPC engraftment in PB, spleen, and BM remained <0.1% in the nonirradiated recipients after transplantation, whereas AML engraftment was <0.01% of total CD45+ cells within 7 days after transplantation but progressively increased with time thereafter (Figure 2A-B). BM cellular niche alterations closely correlated with AML burden in recipient BM (Figure 2C) and appeared at 21 days after transplantation (Figure 2D; supplemental Figure S4). However, BM cellular niche components remained relatively constant after transplantation of normal HSPCs (supplemental Figure 4A), indicating the niche alterations are AML specific.
Kinetics of BM engraftment of MLL-AF9+AML cells and its impact on BM stromal cells after transplantation. (A) Representative FACS profile showing the engraftment of normal BM KIT+ hematopoietic progenitors and MLL-AF9+ AML cells at different time points posttransplantation. (B) Engraftment kinetics of normal BM KIT+ hematopoietic progenitors and MLL-AF9+ AML cells posttransplantation. Data represent mean ± standard error of the mean from 3 to 4 mice per group per time point. (C) Correlation between BM stromal cell frequencies and AML cell engraftment posttransplantation of MLL-AF9 AML cells. (D) Kinetics of BM cellular niche alterations during development of AML. MLL-AF9 AML cells and BM stromal cell subsets were analyzed in parallel at days 7, 14, 21, and 30 after transplantation of AML cells. Statistical analysis was performed by 2-way analysis of variance analysis with multiple comparisons. *P < .05, **P < .01, and ***P < .001 compared with the control mice. #P < .05 and ##P < .01 to determine the time-related difference in stromal cell frequency. Data represent mean ± standard error of the mean from 3 to 5 mice per group per time point from 3 independent experiments.
Kinetics of BM engraftment of MLL-AF9+AML cells and its impact on BM stromal cells after transplantation. (A) Representative FACS profile showing the engraftment of normal BM KIT+ hematopoietic progenitors and MLL-AF9+ AML cells at different time points posttransplantation. (B) Engraftment kinetics of normal BM KIT+ hematopoietic progenitors and MLL-AF9+ AML cells posttransplantation. Data represent mean ± standard error of the mean from 3 to 4 mice per group per time point. (C) Correlation between BM stromal cell frequencies and AML cell engraftment posttransplantation of MLL-AF9 AML cells. (D) Kinetics of BM cellular niche alterations during development of AML. MLL-AF9 AML cells and BM stromal cell subsets were analyzed in parallel at days 7, 14, 21, and 30 after transplantation of AML cells. Statistical analysis was performed by 2-way analysis of variance analysis with multiple comparisons. *P < .05, **P < .01, and ***P < .001 compared with the control mice. #P < .05 and ##P < .01 to determine the time-related difference in stromal cell frequency. Data represent mean ± standard error of the mean from 3 to 5 mice per group per time point from 3 independent experiments.
Taken together, our data suggest that AML cells alter BM cellular niche composition in a leukemia burden–dependent manner. Moreover, the AML skewed MSC differentiation potential toward adipocytes and osteoblasts.
Primary patient AML cells induced BM niche reconstruction in NSG mice
To test whether primary BM AML cells from patients and mouse AML cells exert a similar effect on the BM niche, we transplanted primary AML mononuclear cells from adult or pediatric patients directly into femurs of sublethally irradiated NSG-SGM3 mice.30 Control mice were injected with phosphate-buffered saline (n = 12) or healthy donor CD34+ cells (n = 2). Human AML cell engraftment in the injected femur varied from 3.8% to 94.6% at 6 to 20 weeks after transplantation (Figure 3A-B). The engraftment of patient AML cells did not lead to any changes in total BM cellularity of the mice (Figure 3D). Similar to the finding in the MLL-AF9 mouse model, both the frequencies and numbers of the ECs, MSCs, and MPCs were significantly increased in recipient BM engrafted with patient AML cells compared with control mice (Figure 3C,E). These data suggest that primary AML cells from patients could remodel BM stroma.
Primary human AML cells resulted in BM stromal cell alterations in NSG-SGM3 mice. Primary patient AML cells with or without MLL-AF9 mutations were transplanted by intrafemoral injection into sublethally irradiated NSG-SGM3 mice. Femurs and tibias were collected to analyze AML cell engraftment and BM stromal cell phenotype 6 to 20 weeks after transplantation. Engraftment levels (3.8% to 94.6% of total BM cells) in NSG-SGM3 mice were included in the analysis. (A) Representative FACS profile showing the gating strategy used for phenotypic analysis of BM stromal cells, including ECs, MSCs, and MPCs, and human CD45 (hCD45)+ cells in NSG-SGM3 mice transplanted with phosphate-buffered saline (control) or normal BM CD34+ cells or patient AML mononuclear cells (AML). CD44− cells were first gated within CD45−TER119−CD31− cells and then subdivided into SCA1+CD51+ MSCs and SCA1−CD51+ MPCs, as indicated. CD31+ ECs were gated within CD45−TER119− cells. Human CD45+ cells were gated within live (propidium iodide negative [PI−]) mononuclear cells. (B) Engraftment of primary AML cells from patient samples and CD34+ cells from healthy donors in NSG-SGM3 mice. (C) Increased frequencies of BM SCA1+CD51+ MSCs, SCA1−CD51+ MPCs, CD45−TER119−CD31+ ECs, and CD45−TER119−CD31+SCA1+ arteriolar ECs in NSG-SGM3 mice after transplantation. (D) Total BM cell counts in xenografted NSG-SGM3 recipient mice. (E) Absolute counts of ECs (CD45−TER119−CD31+), SCA1+CD51+ MSCs, and SCA1−CD51+ MPCs in NSG-SGM3 recipient mice. The green dots in panels B-E represent the cell counts in recipient mice 8 weeks after transplantation of healthy CD34+ BM cells. The blue dot represents cell count in the NSG mouse with 94.6% AML engraftment. Black dots indicate data from adult AML cells transplantation; red dots, pediatric AML donor; orange dots, pediatric AML donor with MLL-AF9 mutation. The horizontal bars in panels B-C,E show median values. Data are from 4 independent experiments. Differences were determined using an unpaired Student t test or Mann-Whitney U test.
Primary human AML cells resulted in BM stromal cell alterations in NSG-SGM3 mice. Primary patient AML cells with or without MLL-AF9 mutations were transplanted by intrafemoral injection into sublethally irradiated NSG-SGM3 mice. Femurs and tibias were collected to analyze AML cell engraftment and BM stromal cell phenotype 6 to 20 weeks after transplantation. Engraftment levels (3.8% to 94.6% of total BM cells) in NSG-SGM3 mice were included in the analysis. (A) Representative FACS profile showing the gating strategy used for phenotypic analysis of BM stromal cells, including ECs, MSCs, and MPCs, and human CD45 (hCD45)+ cells in NSG-SGM3 mice transplanted with phosphate-buffered saline (control) or normal BM CD34+ cells or patient AML mononuclear cells (AML). CD44− cells were first gated within CD45−TER119−CD31− cells and then subdivided into SCA1+CD51+ MSCs and SCA1−CD51+ MPCs, as indicated. CD31+ ECs were gated within CD45−TER119− cells. Human CD45+ cells were gated within live (propidium iodide negative [PI−]) mononuclear cells. (B) Engraftment of primary AML cells from patient samples and CD34+ cells from healthy donors in NSG-SGM3 mice. (C) Increased frequencies of BM SCA1+CD51+ MSCs, SCA1−CD51+ MPCs, CD45−TER119−CD31+ ECs, and CD45−TER119−CD31+SCA1+ arteriolar ECs in NSG-SGM3 mice after transplantation. (D) Total BM cell counts in xenografted NSG-SGM3 recipient mice. (E) Absolute counts of ECs (CD45−TER119−CD31+), SCA1+CD51+ MSCs, and SCA1−CD51+ MPCs in NSG-SGM3 recipient mice. The green dots in panels B-E represent the cell counts in recipient mice 8 weeks after transplantation of healthy CD34+ BM cells. The blue dot represents cell count in the NSG mouse with 94.6% AML engraftment. Black dots indicate data from adult AML cells transplantation; red dots, pediatric AML donor; orange dots, pediatric AML donor with MLL-AF9 mutation. The horizontal bars in panels B-C,E show median values. Data are from 4 independent experiments. Differences were determined using an unpaired Student t test or Mann-Whitney U test.
Dynamic molecular alteration of BM cellular niches in AML mice
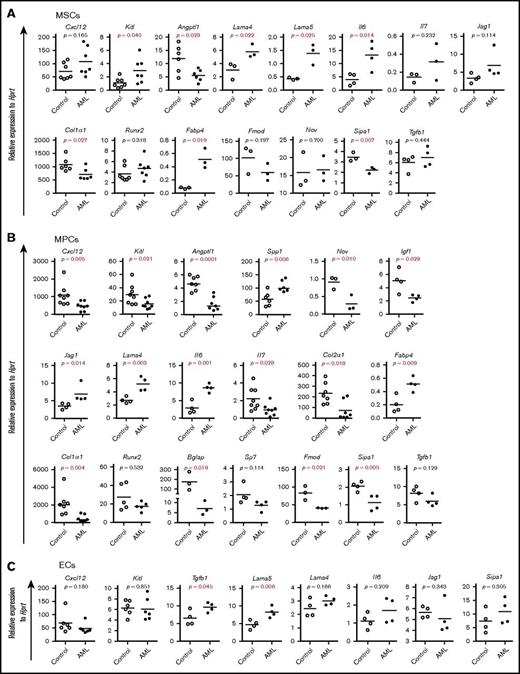
To investigate the molecular mechanisms underlying the massive proliferation of AML cells and impaired normal hematopoiesis, we next examined the expression of HSC niche genes and mesenchymal lineage–associated genes by quantitative polymerase chain reaction (qPCR) (Figure 4A-C). Angiopoietin-like 1(Angptl1) and collagen type I (Col1a1) were significantly reduced, while Kit ligand (Kitl) was higher in AML MSCs than control MSCs (Figure 4A). These genes as well as C-X-C motif chemokine ligand 12 (Cxcl12), interleukin-7 (Il7), and Nov were downregulated in AML MPCs, while osteopontin (secreted phosphoprotein 1 [Spp1]), Il6, Lama4, and Jag1 were upregulated in these cells (Figure 4B). Tgfb1 and Lama5 were significantly increased in AML ECs (Figure 4C). Consistent with the increased in vitro adipogenic and osteogenic differentiation potential of AML MSCs, Fabp4 was increased in AML MSPCs. However, the early osteoblast differentiation gene Sp7 (osterix) and Runx2 remained unaltered, while the late-stage osteoblast marker Col1a1 and Bglap (osteocalcin) decreased in AML MSPCs, indicating a possible osteoblast maturation block in AML. Interestingly, Sipa1, whose expression in the BM niche is critical for maintaining normal hematopoiesis,31 was significantly decreased in AML MSCs and MPCs (Figure 4A-B).
Altered gene expression in AML BM stromal cells. Gene expression analysis was done by qPCR on freshly FACS-sorted BM SCA1+CD51+ MSCs, SCA1−CD51+ MPCs, and CD31+ ECs after AML development. Data are presented as mean values of duplicate or triplicate qPCR measurements from 3 to 5 independent sorting experiments. (A) Relative messenger RNA (mRNA) expression of HSC niche genes and mesenchymal lineage-associated genes in MSCs. (B) Relative expression of HSC niche genes and mesenchymal lineage–associated genes in MPCs. (C) Relative expression of HSC niche genes in CD31+ ECs. Differences were determined using an unpaired parametric Student t test or Mann-Whitney U test. Horizontal bars represent mean values.
Altered gene expression in AML BM stromal cells. Gene expression analysis was done by qPCR on freshly FACS-sorted BM SCA1+CD51+ MSCs, SCA1−CD51+ MPCs, and CD31+ ECs after AML development. Data are presented as mean values of duplicate or triplicate qPCR measurements from 3 to 5 independent sorting experiments. (A) Relative messenger RNA (mRNA) expression of HSC niche genes and mesenchymal lineage-associated genes in MSCs. (B) Relative expression of HSC niche genes and mesenchymal lineage–associated genes in MPCs. (C) Relative expression of HSC niche genes in CD31+ ECs. Differences were determined using an unpaired parametric Student t test or Mann-Whitney U test. Horizontal bars represent mean values.
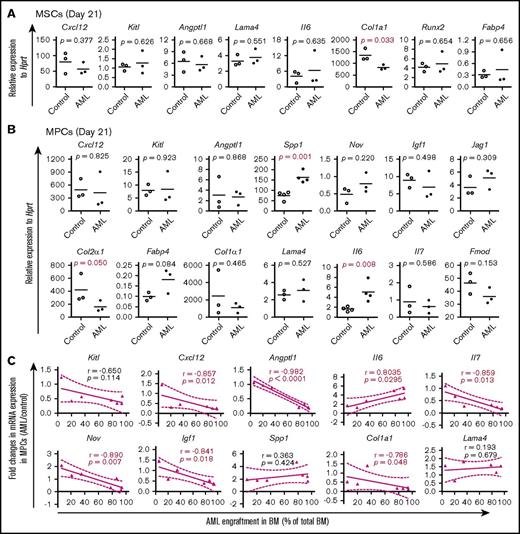
In line with the dynamic changes in BM cellular niche composition in AML mice, the major BM niche population MPCs showed molecular alterations 14 days after transplantation (supplemental Figure S5; Figure 5) and prior to the massive expansion and egress of AML cells (Figure 2). Alterations in most of the HSC niche factors in AML MSPCs seemed to be closely associated with the level of AML engraftment (supplemental Figure 5C; Figure 5C). These data provide molecular evidence for dynamic changes of HSC niche factors in AML MSPCs.
Dynamic alteration of hematopoiesis-supportive gene expression in BM stromal cells during AML development. Gene expression was determined by qPCR on freshly FACS-sorted BM SCA1+CD51+ MSCs, SCA1−CD51+ MPCs, and CD31+ ECs from recipient mice at different time points posttransplantation of MLL-AF9+ AML cells or normal KIT+ HSPCs. Data are presented as mean values of duplicate or triplicate qPCR measurements from 3 to 4 independent sorting experiments. Statistical differences were determined using an unpaired parametric 1-sided Student t test and Pearson or Spearman correlation. (A) Relative mRNA expression of hematopoiesis-supportive genes and mesenchymal lineage–associated genes in BM MSCs 21 days after transplantation of AML cells. (B) Relative expression of hematopoiesis-supportive genes and mesenchymal lineage–associated genes in BM MPCs 21 days after transplantation of AML cells. (C) Correlation between AML cell engraftment and relative expression of the hematopoiesis-supportive genes in recipient BM MPCs after transplantation of the AML cells.
Dynamic alteration of hematopoiesis-supportive gene expression in BM stromal cells during AML development. Gene expression was determined by qPCR on freshly FACS-sorted BM SCA1+CD51+ MSCs, SCA1−CD51+ MPCs, and CD31+ ECs from recipient mice at different time points posttransplantation of MLL-AF9+ AML cells or normal KIT+ HSPCs. Data are presented as mean values of duplicate or triplicate qPCR measurements from 3 to 4 independent sorting experiments. Statistical differences were determined using an unpaired parametric 1-sided Student t test and Pearson or Spearman correlation. (A) Relative mRNA expression of hematopoiesis-supportive genes and mesenchymal lineage–associated genes in BM MSCs 21 days after transplantation of AML cells. (B) Relative expression of hematopoiesis-supportive genes and mesenchymal lineage–associated genes in BM MPCs 21 days after transplantation of AML cells. (C) Correlation between AML cell engraftment and relative expression of the hematopoiesis-supportive genes in recipient BM MPCs after transplantation of the AML cells.
Abnormal expansion of BM Ebf2+ MSPCs in AML mice
We recently identified a stromal cell population highly enriched with MSCs marked by Ebf2 in mouse BM.28 Ebf2+ cells account for ∼6% of total CD51+SCA1+ MSCs and partly overlap with nestin+ cells in mouse BM.28 To evaluate the involvement of Ebf2+ cells in AML niche reprogramming, we next analyzed Ebf2+ MSPCs after AML establishment using Ebf2-reporter mice (Ebf2-Egfp) as recipients for the transplantation (Figure 6A). We found an expansion of Ebf2+ cells in AML mouse BM compared with that in control mice (Figure 6B-C). However, further analysis revealed a reduced MSC fraction (SCA1+CD51+) but an increased fraction of the more differentiated MPC population (SCA1−CD51+) within Ebf2+ cells (Figure 6D). CFU-Fs in AML Ebf2+ cells seemed to be smaller than those in control Ebf2+ cells (supplemental Figure 6A), even though CFU-F frequencies in Ebf2+ cells were similar between AML and normal mice (Figure 6E). Notably, more CFU-Fs from AML Ebf2+ cells contain adipocytes (8 out of 9 in AML and 1 out of 3 in control) (supplemental Figure 6B), supporting the notion of functional alterations of AML Ebf2+ cells with a biased differentiated capacity toward adipocytes. Furthermore, Cxcl12 mRNA was significantly reduced in AML Ebf2+ cells (Figure 6F). These data suggest that AML cells induce a shift of Ebf2+ MSCs toward differentiation into a less primitive MPC population. Similar to a previous report,14 more MSPCs, including Ebf2+ cells, appeared in the central marrow of AML mice than in controls (supplemental Figure 6C-D).
Participation of Ebf2+cells and their progeny in leukemic niche formation during AML progression. (A) Experimental setup for analyzing Ebf2+ cells in the AML BM. Normal and MLL-AF9–expressing lineage (LIN) −KIT+ cells were generated from FVB/N mouse BM LIN−KIT+ cells and transplanted to nonirradiated Ebf2-Egfp reporter FVB/N mice. Ebf2+ cells were detected by their GFP expression. BM stromal cells in control and AML mice were collected when the mice developed AML. BM stromal cell subsets were analyzed and sorted by FACS for subsequent functional assays and gene expression analysis. (B) Representative FACS profiles showing abnormal expansion of Ebf2+ MSPCs in AML mice. The gating strategy used for phenotypic analysis and sorting of Ebf2+ cells and MSCs and MPCs within the Ebf2+ cell fraction in the BM of control and AML mice. (C) Increased frequencies and numbers of Ebf2+ MSPCs within total CD45−TER119−CD31− stromal cells in AML mice. Data are from 2 independent experiments. (D) Reduced proportion of MSCs and increased proportion of MPCs within Ebf2+ cells in AML mice. Data are from 2 independent experiments. (E) Frequency of CFU-Fs in sorted Ebf2+ cells from control and AML BM. Each dot in the left panel represents mean values of duplicates or triplicate measurement. Horizontal bars in panels C-E show median values. Data are from 3 independent sorting experiments. (F) qPCR analysis of Cxcl12, Kitl, and Il6 expression in Ebf2+ cells from AML and control mice. Data are from 2 to 4 independent experiments. (G) Experimental setup. Eight- to 10-week old Ebf2-Egfp × Ebf2-CreER × Rosa26-loxpStoploxp-Tomato mice were first injected with TAM to activate expression of tomato in Ebf2+ cells and their progeny. One month later, mice were transplanted either with normal or MLL-AF9–expressing LIN−KIT+ cells without irradiation. The BM stromal cells in the control and AML mice were collected and analyzed by FACS at the onset of AML. (H) FACS profile showing gating of tomato-expressing stromal cells. Tomato+ cells were first gated within CD45−TER119−CD31−7AAD− live cells. Cells were analyzed for CD44− and CD44+ cell fraction, and CD44−Tomato+ cells were then further subdivided into SCA1+PDGFRA/CD140A+ MSCs (PaS), SCA1−CD140A+ MPCs, and SCA1−CD140A/PDGFRA− mature stromal cells. (I) Frequencies of the total tomato-expressing cells and tomato+ MSPCs within the BM stromal cell compartment (CD45−TER119−CD31−) in control and AML mice. (J) Frequencies of the Tomato− MSCs and MPCs within the BM stromal cell compartment (CD45−TER119−CD31−) in control and AML mice. (K) FACS plots showing the distribution of MSCs and MPCs within Tomato− and Tomato+ cells in AML mice. SCA1+PDGFRA/CD140A+ MSCs (PaS) and SCA1−CD140A+ MPCs were first gated within stromal cells (CD45−TER119−CD31−). Cells were further analyzed for their expression of Tomato and GFP. Gates were made based nontransgenic mice and fluorescence-minus-one control. (L) Dot plots represent the gating strategy used for the expression of Tomato and GFP. Differences were determined using an unpaired parametric Student t test (C [right],D-F,I [left],L), unpaired parametric Student t test with Welch’s correction (F), or Mann-Whitney U test (C [left],I). Horizontal bars represent mean values in panels C-F,I-K. Data are from 2 independent experiments.
Participation of Ebf2+cells and their progeny in leukemic niche formation during AML progression. (A) Experimental setup for analyzing Ebf2+ cells in the AML BM. Normal and MLL-AF9–expressing lineage (LIN) −KIT+ cells were generated from FVB/N mouse BM LIN−KIT+ cells and transplanted to nonirradiated Ebf2-Egfp reporter FVB/N mice. Ebf2+ cells were detected by their GFP expression. BM stromal cells in control and AML mice were collected when the mice developed AML. BM stromal cell subsets were analyzed and sorted by FACS for subsequent functional assays and gene expression analysis. (B) Representative FACS profiles showing abnormal expansion of Ebf2+ MSPCs in AML mice. The gating strategy used for phenotypic analysis and sorting of Ebf2+ cells and MSCs and MPCs within the Ebf2+ cell fraction in the BM of control and AML mice. (C) Increased frequencies and numbers of Ebf2+ MSPCs within total CD45−TER119−CD31− stromal cells in AML mice. Data are from 2 independent experiments. (D) Reduced proportion of MSCs and increased proportion of MPCs within Ebf2+ cells in AML mice. Data are from 2 independent experiments. (E) Frequency of CFU-Fs in sorted Ebf2+ cells from control and AML BM. Each dot in the left panel represents mean values of duplicates or triplicate measurement. Horizontal bars in panels C-E show median values. Data are from 3 independent sorting experiments. (F) qPCR analysis of Cxcl12, Kitl, and Il6 expression in Ebf2+ cells from AML and control mice. Data are from 2 to 4 independent experiments. (G) Experimental setup. Eight- to 10-week old Ebf2-Egfp × Ebf2-CreER × Rosa26-loxpStoploxp-Tomato mice were first injected with TAM to activate expression of tomato in Ebf2+ cells and their progeny. One month later, mice were transplanted either with normal or MLL-AF9–expressing LIN−KIT+ cells without irradiation. The BM stromal cells in the control and AML mice were collected and analyzed by FACS at the onset of AML. (H) FACS profile showing gating of tomato-expressing stromal cells. Tomato+ cells were first gated within CD45−TER119−CD31−7AAD− live cells. Cells were analyzed for CD44− and CD44+ cell fraction, and CD44−Tomato+ cells were then further subdivided into SCA1+PDGFRA/CD140A+ MSCs (PaS), SCA1−CD140A+ MPCs, and SCA1−CD140A/PDGFRA− mature stromal cells. (I) Frequencies of the total tomato-expressing cells and tomato+ MSPCs within the BM stromal cell compartment (CD45−TER119−CD31−) in control and AML mice. (J) Frequencies of the Tomato− MSCs and MPCs within the BM stromal cell compartment (CD45−TER119−CD31−) in control and AML mice. (K) FACS plots showing the distribution of MSCs and MPCs within Tomato− and Tomato+ cells in AML mice. SCA1+PDGFRA/CD140A+ MSCs (PaS) and SCA1−CD140A+ MPCs were first gated within stromal cells (CD45−TER119−CD31−). Cells were further analyzed for their expression of Tomato and GFP. Gates were made based nontransgenic mice and fluorescence-minus-one control. (L) Dot plots represent the gating strategy used for the expression of Tomato and GFP. Differences were determined using an unpaired parametric Student t test (C [right],D-F,I [left],L), unpaired parametric Student t test with Welch’s correction (F), or Mann-Whitney U test (C [left],I). Horizontal bars represent mean values in panels C-F,I-K. Data are from 2 independent experiments.
In vivo contribution of Ebf2+ cells to niche formation of AML cells
To further determine the contribution of Ebf2+ cells during AML progression, we took advantage of triple-transgenic Ebf2-Egfp × Ebf2-CreER × Rosa26-loxpStoploxp-Tomato mice, where Ebf2+ cells and their progeny are reported by enhanced GFP and tomato protein, respectively. AML cells were injected 1 month after activation of tomato expression by TAM treatment. The progeny (tomato+) derived from Ebf2+ cells were detected by FACS (Figure 6G). Interestingly, Ebf2+ MSPCs could give rise to all fractions of BM stromal cells, including the CD44− stromal cells enriched with MSPCs and CD44+ mature stromal cells (Figure 6H). In line with the enhanced proliferation of Ebf2+ MSPCs in AML BM, the frequency of the Ebf2−tomato+ stromal cell fraction representing cells generated from Ebf2+ MSPCs was significantly increased in AML mouse BM (Figure 6I). Both BM MSCs and MPCs within tomato+ cells were significantly increased in AML mice (Figure 6I). Tomato− MSPCs, possibly derived from non-Ebf2+ cells or Ebf2+ cells that escaped TAM treatment, were also increased in the AML BM (Figure 6J). However, tomato+ MSCs and MPCs accounted for approximately two-thirds of the total MSPC population (Figure 6K), indicating significant contribution of Ebf2+ cells to AML niche formation. This result suggests that Ebf2+ cells might be a key niche element contributing to the formation of the AML BM niche.
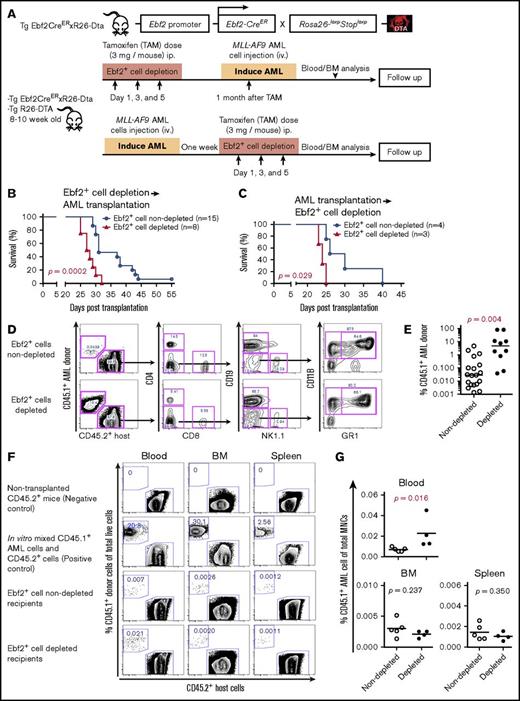
In vivo deletion of Ebf2+ cells accelerated AML establishment
The alteration of Ebf2+ cells and their participation in AML BM niche formation suggest that these cells have a potentially important role in symptomatic AML onset and progression. To functionally determine the role of Ebf2+ cells in AML development, we used transgenic Ebf2-CreER × Rosa26-loxpStoploxp-Dta mice as recipients for establishing an AML mouse model. At steady state, in vivo depletion of Ebf2+ cells led to enhanced myelopoiesis 2 months after TAM injection (supplemental Figure 7), suggesting an important role for these cells in maintaining normal hematopoiesis. We intravenously transplanted CD45.1+MLL-AF9 AML cells into nonirradiated CD45.2+ double-transgenic Ebf2-CreER × Rosa26-loxpStoploxp-Dta or single-transgenic control mice either 1 month after Ebf2+ cell depletion or 1 week prior to depletion (Figure 7A). Surprisingly, we observed a shorter latency of symptomatic AML and an overall reduced survival time in Ebf2+ cell–depleted recipient mice compared with control mice, regardless of whether Ebf2+ cell depletion was induced before or after AML cell transplantation (Figure 7B-C). The onset of symptomatic AML was observed at ∼29 to 55 days after transplantation in control mice and 22 to 32 days in Ebf2+ cell–depleted mice. FACS analysis indicated much higher total engraftment of MLL-AF9 AML cells (CD45.1+) in the PB of Ebf2+ cell–depleted recipients than in nondepleted recipient mice 22 days after transplantation (Figure 7D-E). Disease acceleration following Ebf2+ cell depletion was manifested by faster development of splenomegaly, increased circulating leukemic blasts in the PB, and more profound AML cell infiltration in the spleen and liver of Ebf2+ cell–depleted recipient mice compared with control mice (supplemental Figure 8A-B; Figure 7E). Normal HSPCs were lost in Ebf2+ cell–depleted recipient BM (supplemental Figure 8D-E). While HSCs, granulocyte/macrophage progenitors, and pre–erythrocyte colony-forming units remained in the Ebf2+ cell–depleted recipient spleen, pre–megakaryocyte-erythroids were diminished in recipient spleen (supplemental Figure 8C-D). In line with this, total AML cell engraftment and the frequency of AML KIT+ cells enriched with LSCs29 were dramatically higher in the Ebf2+ cell–depleted recipient BM and spleen than in nondepleted mice (supplemental Figure 8E). Notably, the increased AML cell engraftment and AML acceleration were not related to the initial homing (3 hours after transplantation) of AML cells, since there was no obvious homing advantage of AML cells into Ebf2+ cell–depleted recipient BM and spleen (Figure 7F-G). Taken together, our data suggest that deletion of Ebf2+ cells accelerated AML development. Thus, Ebf2+ cells are important for maintaining normal hematopoiesis and may act as a suppressor for AML development.
Ebf2+cell depletion accelerates AML development. (A) Experimental design. Eight- to 10-week-old Ebf2-CreER x Rosa26-loxpStoploxp-Dta and control single-transgenic mice were injected with TAM to induce specific deletion of Ebf2+ cells either 1 month prior to or 1 week after the transplantation of CD45.1+MLL-AF9 AML cells without irradiation. Survival of the mice was monitored after transplantation. BM stromal cells from the control and AML mice were collected and analyzed by FACS at the onset of AML. (B) Predepletion of Ebf2+ cells led to reduced survival of depleted mice compared with that of nondepleted recipient mice after transplantation of AML cells. Because the TAM injection was given just 1 week after AML cell transplantation, all recipients were male mice to avoid TAM-induced effects. Survival differences were tested using a log-rank (Mantel-Cox) test in Prism 6.0. (C) Deletion of Ebf2+ cells after AML cell transplantation caused shorter latency and reduced survival of Ebf2+ cell–depleted recipients compared with nondepleted recipient mice. The difference in survival rate was tested using a log-rank (Mantel-Cox) test in Prism 6.0. (D) Representative FACS profiles showing AML engraftment and host-derived total cells and blood lineages, including T (CD4+/CD8+), B (CD19+), natural killer (NK1.1+), and myeloid (CD11B+GR1+) cells 22 days after transplantation. (E) Increased total AML cell engraftment in the PB of the Ebf2+ cell–depleted recipient. (F-G) Homing of AML cells into Ebf2+ cell–depleted recipient mice after transplantation. CD45.1+ AML cells (10 million per mouse) were transplanted into Ebf2+ cell–nondepleted (n = 5) and Ebf2+ cell–depleted (n = 4) mice via tail vein injection 1 month after TAM treatment. Blood, femur, and spleen of recipients were harvested 3 hours after transplantation, and homing of donor CD45.1+ AML cells were examined by FACS based on CD45.1 expression. (F) Representative FACS profiles showing AML cells in PB, BM, and spleen after transplantation. The numbers in the panels are the frequencies of donor AML cells among the total number of nucleated cells in each organ. (G) Frequencies of donor cells among the total number of nucleated cells in recipient PB, BM, and spleen. Differences were determined using an unpaired parametric Student t test or Mann-Whitney U test. Horizontal bars represent mean values in panels E,G. Data are from 2 independent experiments.
Ebf2+cell depletion accelerates AML development. (A) Experimental design. Eight- to 10-week-old Ebf2-CreER x Rosa26-loxpStoploxp-Dta and control single-transgenic mice were injected with TAM to induce specific deletion of Ebf2+ cells either 1 month prior to or 1 week after the transplantation of CD45.1+MLL-AF9 AML cells without irradiation. Survival of the mice was monitored after transplantation. BM stromal cells from the control and AML mice were collected and analyzed by FACS at the onset of AML. (B) Predepletion of Ebf2+ cells led to reduced survival of depleted mice compared with that of nondepleted recipient mice after transplantation of AML cells. Because the TAM injection was given just 1 week after AML cell transplantation, all recipients were male mice to avoid TAM-induced effects. Survival differences were tested using a log-rank (Mantel-Cox) test in Prism 6.0. (C) Deletion of Ebf2+ cells after AML cell transplantation caused shorter latency and reduced survival of Ebf2+ cell–depleted recipients compared with nondepleted recipient mice. The difference in survival rate was tested using a log-rank (Mantel-Cox) test in Prism 6.0. (D) Representative FACS profiles showing AML engraftment and host-derived total cells and blood lineages, including T (CD4+/CD8+), B (CD19+), natural killer (NK1.1+), and myeloid (CD11B+GR1+) cells 22 days after transplantation. (E) Increased total AML cell engraftment in the PB of the Ebf2+ cell–depleted recipient. (F-G) Homing of AML cells into Ebf2+ cell–depleted recipient mice after transplantation. CD45.1+ AML cells (10 million per mouse) were transplanted into Ebf2+ cell–nondepleted (n = 5) and Ebf2+ cell–depleted (n = 4) mice via tail vein injection 1 month after TAM treatment. Blood, femur, and spleen of recipients were harvested 3 hours after transplantation, and homing of donor CD45.1+ AML cells were examined by FACS based on CD45.1 expression. (F) Representative FACS profiles showing AML cells in PB, BM, and spleen after transplantation. The numbers in the panels are the frequencies of donor AML cells among the total number of nucleated cells in each organ. (G) Frequencies of donor cells among the total number of nucleated cells in recipient PB, BM, and spleen. Differences were determined using an unpaired parametric Student t test or Mann-Whitney U test. Horizontal bars represent mean values in panels E,G. Data are from 2 independent experiments.
Discussion
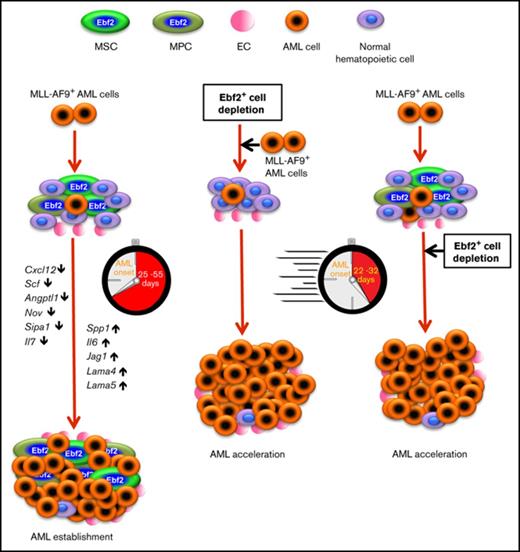
BM HSC niche contribution to leukemia initiation and progression has been increasingly recognized.11,12 However, the contributions of BM MSPCs to leukemia niche formation and progression remain poorly defined. Thus, there is great need to understand the role of BM MSPCs in leukemia in order to identify disease-specific therapeutic targets in the niche. We here demonstrate that MLL-AF9+ AML cells reconstructed the BM HSC niche by inducing abnormal expansion and differentiation of BM MSPCs. In vivo fate-mapping experiments suggested an involvement of Ebf2+ MSPCs in AML niche formation. Strikingly, the depletion of Ebf2+ MSPCs resulted in AML acceleration, emphasizing a critical role for Ebf2+ cells in AML onset and progression and providing new evidence for the contribution of MSPCs to AML development.
Our dynamic characterization reveals a correlation between BM cellular niche alterations and AML cell engraftment. The molecular alterations partially appeared before the massive expansion of AML cells. The progressive deregulation of HSC maintenance genes and inflammatory cytokines in AML BM MSPCs was closely associated with AML burden. This may represent molecular mechanisms by which normal hematopoiesis is impaired while AML cells massively proliferate. Altogether, our findings provide cellular and molecular evidence for dynamic alterations of the BM HSC niche in AML. AML cells may actively reconstruct the HSC cellular niche into a potentially self-reinforcing leukemic niche. The “educated” BM niche could, in turn, contribute to the massive proliferation of leukemic cells and failure of normal hematopoiesis. Strategies to restore BM niche homeostasis may provide a means to eliminate dormant LSCs after treatment and thereby impede AML progression.
Previous studies showed that MLL-AF9 AML14 and BCR-ABL CML cells25 led to increased bone remodeling with an accumulation of osteoblast-primed MSPCs accompanied by reduced numbers of mature osteoblasts. In line with these findings, our data suggest an enhanced capacity of AML MSCs to generate more differentiated MPCs than MSCs, which is reflected in the expansion of MPCs but the reduction of the MSC fraction within the defined Ebf2+ MSPCs. The differentiation bias of MSCs is consistent with their stronger potential to generate osteoblasts and adipocytes in vitro. However, the reduction of Col1a1 and Bglap/osteocalcin transcripts in native AML MPCs suggests that the differentiation of MPCs to mature osteoblasts is blocked at a late development stage in vivo, as supported by micro–computed tomography and a previous study,14 and this block could also contribute to the accumulation of MPCs in AML.
Importantly, this study shows the critical role of Ebf2+ cells in AML BM niche remodeling, as illustrated by their contribution to BM stromal cell turnover. Consistent with our previous finding,28 lineage-tracing experiments demonstrated that Ebf2+ cells could generate all BM stromal cell subsets, including MSPCs. These findings suggest a high hierarchical position of Ebf2+ MSCs in BM mesenchymal cell development and a potential role of these cells in maintaining BM cellular niche homeostasis and in AML niche formation. During AML establishment, Ebf2+ cells preferentially generated more differentiated MPCs than the MSCs. The depletion of Ebf2+ MSPCs may further disrupt the balance between primitive MSCs and more differentiated MPCs, thereby accelerating AML progression.
During AML progression, leukemic cells outcompete normal hematopoietic cells, which leads to pancytopenia and infiltration of leukocytes in the BM, blood, and other organs.32 However, the underlying molecular mechanisms remain to be defined. Cxcl12, Kitl, Nov, Igf1, and Angptl1 are critical for maintaining normal hematopoiesis through keeping HSC in quiescence21,33-36 and lodgment in their niche.37 Decreased expression of these genes in AML MSCs and MPCs could affect HSC retention and quiescence and eventually exhaust the normal HSC pool while promoting leukemic cell proliferation, leading to the progression of leukemia, as reported previously.13,14,38
Accumulated evidence indicates that inflammatory cytokines such as Tgfb1 and Il6 are involved in the development of myeloid malignancies11,12 and are often increased in patients with myeloproliferative neoplasms.39,40 We here report that Il6 and Tgfb1 transcripts are upregulated in AML BM MSPCs and ECs, respectively. In addition, it has been shown that SPP1 (OPN) is a critical negative regulator of normal HSC self-renewal,41 and the interactions between SPP1 and lymphoblastic leukemic cells can induce dormancy of leukemic cells.42 In this study, the upregulation of Spp1 and Il6 in the BM MSPCs has already occurred before the expansion of AML. Thus, the increased Spp1 in the AML BM niche might contribute to the exhaustion of normal HSCs and thereby to AML progression, although more work is required to test this hypothesis. Spp1 upregulation is also consistent with the biased osteoblastic differentiation of AML MSCs, which was reflected in the preferential expansion of Ebf2+ MPCs in AML mice. These data provide evidence of the molecular mechanisms by which AML cells remodel the BM HSC niche to create a self-reinforcing leukemic niche.
Laminin α4 chain (Lama4) and α5 chain (Lama5) are active chains forming the cell-binding domains in the extracellular matrix proteins laminin-411, laminin-421, laminin-423 and laminin-511 and laminin-521, respectively.43 Previous studies have shown that the interactions between HSC and laminins are critical for adult BM HSC homing and engraftment.44,45 Lama4 upregulation was detected in CML mice treated with placental growth factor, a factor promoting CML proliferation.46 The upregulation of Lama4 and Lama5 in the AML BM niche warrants further investigation of the role of the laminin in AML progression. Furthermore, Sipa1 reduction in AML MSCs and MPCs might contribute to AML progression, since Sipa1 loss could induce BM niche alterations, causing myeloproliferative neoplasms.31
In summary, our findings provide new evidence for MLL-AF9 AML–induced dynamic niche alterations and the temporal roles of BM MSPCs during the development of AML. BM native Ebf2+ cells suppress AML onset and progression. However, these cells can be functionally altered by AML cells to constitute the AML niche. In vivo fate-mapping using Ebf2 reporter mouse models demonstrated the significant contribution of Ebf2+ cells to AML niche formation. The suppressive role of native Ebf2+ cells in AML onset and progression warrants future exploration of the underlying molecular mechanisms involved. Importantly, molecular tools that restore BM MSC function and the molecular niche may be a potentially powerful strategy to suppress AML development.
The full-text version of this article contains a data supplement.
Acknowledgments
Confocal images were obtained at the Live Cell Imaging unit, Department of Biosciences and Nutrition, Karolinska Institute, Huddinge, Sweden. The authors are grateful to Sten Eirik W. Jacobsen at Karolinska Institute for his scientific input on the study. The authors acknowledge the MedH Core Flow Cytometry facility (Karolinska Institute) for providing cell-sorting/analysis services. The authors thank the Nordic Society of Pediatric Hematology and Oncology (http://www.nopho.org) for scientific discussion and providing precious samples from their biobank.
This study was supported by the Swedish Research Council (K2013-99X-22241-01-5), the Swedish Childhood Society (TJ2013-0048, PR2015-0142, and PROJ12/081), the Swedish Cancer Society (CAN 2015/652 and CAN 2012/891), the Åke Olsson Foundation, Radiumhemmets Forskningsfonder, and Karolinska Institute Wallenberg Institute for Regenerative Medicine (H.Q.).
Authorship
Contribution: H.Q. and P.X. designed and performed experiments, analyzed data, and wrote the manuscript; L.S. designed and performed experiments, analyzed data, and participated in manuscript writing; Y.H. generated MLL-AF9 AML cells and helped with transplantation experiments; M.K. performed, analyzed data and participated in the manuscript writing; T.B., M.D., and A.-S.J. assisted in part of the study and with manuscript review; J.W., M.S., and M.E. provided scientific input on the study and reviewed the manuscript; and all authors approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hong Qian, Center for Hematology and Regenerative Medicine, Department of Medicine, Karolinska Institute, Karolinska University Hospital, SE-141 86 Stockholm, Sweden; e-mail: hong.qian@ki.se.

![Figure 3. Primary human AML cells resulted in BM stromal cell alterations in NSG-SGM3 mice. Primary patient AML cells with or without MLL-AF9 mutations were transplanted by intrafemoral injection into sublethally irradiated NSG-SGM3 mice. Femurs and tibias were collected to analyze AML cell engraftment and BM stromal cell phenotype 6 to 20 weeks after transplantation. Engraftment levels (3.8% to 94.6% of total BM cells) in NSG-SGM3 mice were included in the analysis. (A) Representative FACS profile showing the gating strategy used for phenotypic analysis of BM stromal cells, including ECs, MSCs, and MPCs, and human CD45 (hCD45)+ cells in NSG-SGM3 mice transplanted with phosphate-buffered saline (control) or normal BM CD34+ cells or patient AML mononuclear cells (AML). CD44− cells were first gated within CD45−TER119−CD31− cells and then subdivided into SCA1+CD51+ MSCs and SCA1−CD51+ MPCs, as indicated. CD31+ ECs were gated within CD45−TER119− cells. Human CD45+ cells were gated within live (propidium iodide negative [PI−]) mononuclear cells. (B) Engraftment of primary AML cells from patient samples and CD34+ cells from healthy donors in NSG-SGM3 mice. (C) Increased frequencies of BM SCA1+CD51+ MSCs, SCA1−CD51+ MPCs, CD45−TER119−CD31+ ECs, and CD45−TER119−CD31+SCA1+ arteriolar ECs in NSG-SGM3 mice after transplantation. (D) Total BM cell counts in xenografted NSG-SGM3 recipient mice. (E) Absolute counts of ECs (CD45−TER119−CD31+), SCA1+CD51+ MSCs, and SCA1−CD51+ MPCs in NSG-SGM3 recipient mice. The green dots in panels B-E represent the cell counts in recipient mice 8 weeks after transplantation of healthy CD34+ BM cells. The blue dot represents cell count in the NSG mouse with 94.6% AML engraftment. Black dots indicate data from adult AML cells transplantation; red dots, pediatric AML donor; orange dots, pediatric AML donor with MLL-AF9 mutation. The horizontal bars in panels B-C,E show median values. Data are from 4 independent experiments. Differences were determined using an unpaired Student t test or Mann-Whitney U test.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/2/12/10.1182_bloodadvances.2017013870/3/m_advances013870f3.jpeg?Expires=1763461968&Signature=yYEXnyWIEnAhRFKcTji7s8YtsPOuhrRj8hNLs9mxTji4RIFdWEDN3eog3xsw5pr6QRzHPrtIFhfJ-nh5jBZ94NWlH~~pqSNZMiFBQYN-KYT0Zp0Gs~YGiII9vgc6ydfY1hT~Soyg3YbkzK3V0ZMlDT~VIUg1JIGSFBVyVVk~1HVCXKVwaG30LeJyiRxxch4aFcfTTJ10N65~tWm8iHGBKRCkREXBOa8y44Jlm6qgUtbMkYStnmlMPCFA1994ykcAqJIoFr5kM6575FduwADJp8lIb4oaWXxvtpfHPZbJEPHIuVEd8EFVic6MDJMEryPrknDtx40Y6VrofhxIlNvYog__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. Participation of Ebf2+cells and their progeny in leukemic niche formation during AML progression. (A) Experimental setup for analyzing Ebf2+ cells in the AML BM. Normal and MLL-AF9–expressing lineage (LIN) −KIT+ cells were generated from FVB/N mouse BM LIN−KIT+ cells and transplanted to nonirradiated Ebf2-Egfp reporter FVB/N mice. Ebf2+ cells were detected by their GFP expression. BM stromal cells in control and AML mice were collected when the mice developed AML. BM stromal cell subsets were analyzed and sorted by FACS for subsequent functional assays and gene expression analysis. (B) Representative FACS profiles showing abnormal expansion of Ebf2+ MSPCs in AML mice. The gating strategy used for phenotypic analysis and sorting of Ebf2+ cells and MSCs and MPCs within the Ebf2+ cell fraction in the BM of control and AML mice. (C) Increased frequencies and numbers of Ebf2+ MSPCs within total CD45−TER119−CD31− stromal cells in AML mice. Data are from 2 independent experiments. (D) Reduced proportion of MSCs and increased proportion of MPCs within Ebf2+ cells in AML mice. Data are from 2 independent experiments. (E) Frequency of CFU-Fs in sorted Ebf2+ cells from control and AML BM. Each dot in the left panel represents mean values of duplicates or triplicate measurement. Horizontal bars in panels C-E show median values. Data are from 3 independent sorting experiments. (F) qPCR analysis of Cxcl12, Kitl, and Il6 expression in Ebf2+ cells from AML and control mice. Data are from 2 to 4 independent experiments. (G) Experimental setup. Eight- to 10-week old Ebf2-Egfp × Ebf2-CreER × Rosa26-loxpStoploxp-Tomato mice were first injected with TAM to activate expression of tomato in Ebf2+ cells and their progeny. One month later, mice were transplanted either with normal or MLL-AF9–expressing LIN−KIT+ cells without irradiation. The BM stromal cells in the control and AML mice were collected and analyzed by FACS at the onset of AML. (H) FACS profile showing gating of tomato-expressing stromal cells. Tomato+ cells were first gated within CD45−TER119−CD31−7AAD− live cells. Cells were analyzed for CD44− and CD44+ cell fraction, and CD44−Tomato+ cells were then further subdivided into SCA1+PDGFRA/CD140A+ MSCs (PaS), SCA1−CD140A+ MPCs, and SCA1−CD140A/PDGFRA− mature stromal cells. (I) Frequencies of the total tomato-expressing cells and tomato+ MSPCs within the BM stromal cell compartment (CD45−TER119−CD31−) in control and AML mice. (J) Frequencies of the Tomato− MSCs and MPCs within the BM stromal cell compartment (CD45−TER119−CD31−) in control and AML mice. (K) FACS plots showing the distribution of MSCs and MPCs within Tomato− and Tomato+ cells in AML mice. SCA1+PDGFRA/CD140A+ MSCs (PaS) and SCA1−CD140A+ MPCs were first gated within stromal cells (CD45−TER119−CD31−). Cells were further analyzed for their expression of Tomato and GFP. Gates were made based nontransgenic mice and fluorescence-minus-one control. (L) Dot plots represent the gating strategy used for the expression of Tomato and GFP. Differences were determined using an unpaired parametric Student t test (C [right],D-F,I [left],L), unpaired parametric Student t test with Welch’s correction (F), or Mann-Whitney U test (C [left],I). Horizontal bars represent mean values in panels C-F,I-K. Data are from 2 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/2/12/10.1182_bloodadvances.2017013870/3/m_advances013870f6.jpeg?Expires=1763461968&Signature=SvmYzaNF~ry3~eVm0WqhdpyyvswFsNRRP~1qzLv5b4-lXVWE2FzIz32AJHiODT4un0CTPgj34kwedN6K3J~M-lKbENpTWypp3Fg5KXbgEdsIxx2VTLrwozl-URCCO~FuinQC6mJvGM5q-NCfz50xzGQf-pbEFf1GDLMQixDhJuVJrRR47cvNTZlDDqceSUfxNwmG2P1SN0HJRScf-oTFgMuK7ss5umtw~XjaUQDrPgpf3SKPBJznLGwKGBSwP3vi8Bw94M4hditvuD2ukwLw9nmsUoAoRy4jxiTklNuGwMjuOAujENpdEVTR0D4qeLMz16~JtSTlO87efnG0-XqYrA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)