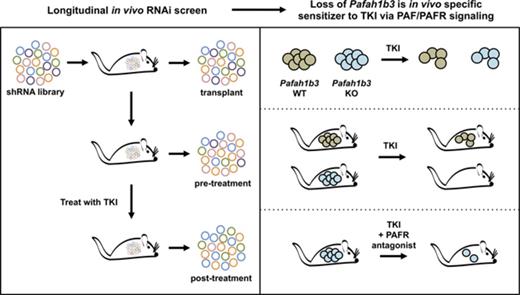
Key Points
Functional in vivo screening reveals distinct genetic dependencies in a mouse model of BCR-ABL1+ BCP-ALL after dasatinib treatment.
Disruption of the PAF/PAFR/PAFAH1B axis sensitizes leukemia cells to TKIs; this pathway may be a novel target for combination therapy.
Abstract
Despite the addition of tyrosine kinase inhibitors (TKIs) to the treatment of patients with BCR-ABL1+ B-cell precursor acute lymphoblastic leukemia (BCR-ABL1+ BCP-ALL), relapse both with and without BCR-ABL1 mutations is a persistent clinical problem. To identify BCR-ABL1–independent genetic mediators of response to the TKI dasatinib, we performed in vivo and in vitro RNA interference (RNAi) screens in a transplantable syngeneic mouse model of BCR-ABL1+ BCP-ALL. By using a novel combination of a longitudinal screen design and independent component analysis of screening data, we identified hairpins that have distinct behavior in different therapeutic contexts as well as in the in vivo vs in vitro settings. In the set of genes whose loss sensitized BCR-ABL1+ BCP-ALL cells to dasatinib, we identified Pafah1b3, which regulates intracellular levels of platelet-activating factor (PAF), as an in vivo–specific mediator of therapeutic response. Pafah1b3 loss significantly sensitized leukemia cells to the multiple TKIs, indicating that inhibition of PAFAH1B3 in combination with TKI treatment may be an effective therapeutic strategy for BCR-ABL1+ BCP-ALL patients. PAF-induced cell death as well as surface levels of PAF receptor (PAFR) in our model are altered upon dasatinib treatment and depend on the local leukemia microenvironment; the response of Pafah1b3 KO vs overexpressing cells to dasatinib is also dependent on microenvironmental context. Antagonism of the PAFR partially reverses the observed sensitization to TKI treatment upon Pafah1b3 loss in vivo, suggesting that signaling via the PAF/PAFR pathway is at least partially responsible for this effect.
Introduction
Expression of the oncogenic tyrosine kinase Bcr-Abl is associated with chronic myelogenous leukemia (CML) and BCR-ABL1+ B-cell precursor acute lymphoblastic leukemia (BCR-ABL1+ BCP-ALL), the latter of which is characterized by additional genetic changes including activation of SRC kinases and loss of tumor suppressors like ARF and IKZF1.1,2 Tyrosine kinase inhibitors (TKIs) that inhibit BCR-ABL and related kinases are effective and well-tolerated treatments for BCR-ABL1+ leukemia, but relapse after therapy is a persistent clinical problem.1,3,4 Relapse without alterations to BCR-ABL1 occurs in BCR-ABL1+ BCP-ALL patients after treatment with all currently approved TKIs and can be fueled by alternative intracellular signaling, inhibition of apoptosis, or interaction with the leukemic microenvironment.1,4-10 Identification of specific pathways that can alter TKI response is crucial in understanding and treating these relapses and improving front-line therapeutic strategies.10
Using a transplantable mouse model of p19Arf−/− p185BCR-ABL+ BCP-ALL, we performed an unbiased, pooled short hairpin RNA (shRNA) screen for genetic mediators of response to dasatinib in the native leukemic microenvironment.8,9 This model is suitable for large-scale screens and has distinct genetic dependencies in vivo vs in vitro.11 Here, we perform a longitudinal screen by collecting serial peripheral blood (PB) or culture samples, thus allowing us to compare hairpin representation before and after therapy in a single mouse or cell culture dish. We used independent component analysis (ICA)12 to remove noise from the RNA interference (RNAi) dataset and identify high-value candidate shRNAs that promote resistance or sensitivity to dasatinib in specific environmental contexts.
Our screen revealed Pafah1b3 loss as an in vivo–specific sensitizer of BCR-ABL1+ BCP-ALL cells to dasatinib. Pafah1b3 encodes a catalytic subunit of platelet-activating factor (PAF) acetyl hydrolase Ib (Pafah1b), a phospholipase that hydrolyzes PAF to regulate intracellular PAF levels and prevent apoptosis caused by high intracellular PAF concentrations.13,14 Pafah1b3 loss extends the lifespan of leukemia-bearing mice after multiple TKIs and differentially sensitizes to dasatinib based on microenvironmental context. The PAF/PAF receptor (PAFR) pathway in our model is also sensitive to dasatinib treatment and local microenvironment, and blocking this pathway can partially reverse the phenotype of Pafah1b3 loss.
Methods
Cell culture
Murine p19Arf−/− p185BCR-ABL+ BCP-ALL cells8,9 and mouse spleen (SPL) and bone marrow (BM) stromal cells were cultured in RPMI 1640 media supplemented with 10% fetal bovine serum (FBS), 4 mM l-glutamine, and 55 μM β-mercaptoethanol. Stromal cells were isolated from nontumor-bearing C57BL/6J mice by culturing BM aspirate or homogenized SPL and removing nonadherent cells every 24 hours for 7 to 10 days. Stromal cells for were passaged once in culture.
Vectors
shRNAs were expressed in the mir30 context; shRNA cloning and design were performed as described.15 For complementary DNA (cDNA) expression, we replaced the puromycin-resistance cassette from pMSCVpuro (Clontech) with tdTomato or E2Crimson fluorescent proteins. Single-guide RNAs (sgRNAs) were designed using the Zhang laboratory web tool (crispr.mit.edu) and cloned into the pX458 vector (Addgene plasmid 48138) according to the referenced protocol.16
Mice
Mice were housed at the Koch Institute for Integrative Cancer Research at MIT in facilities accredited by the American Association of Laboratory Animal Care and Use Committee. Procedures were performed in accordance with Institutional Animal Care and Use Committee and National Institutes of Health guidelines. All mouse experiments except survival studies were performed using 6- to 10-week-old female C57BL6/J mice (The Jackson Laboratory).
RNAi screening and validation
One hundred million p19Arf−/− p185BCR-ABL+ BCP-ALL cells cultured for >1 week were infected in 10 plates with a library of 25 206 shRNAs (14 703 genetic loci) tagged with green fluorescent protein (GFP) at a multiplicity of infection (MOI) <1. Forty-eight hours posttransduction, 5 × 106 shRNA-expressing cells were isolated using fluorescence-activated cell sorting (FACS), expanded in culture for 3 to 4 days, and 106 cells were plated into 3 replicate cultures or transplanted into 8 recipient mice; 3 input samples (2 × 106 cells) were taken at transplant/plating. Five days postplating, 2 × 106 cells from each culture were taken as a pretreatment sample; cultures were treated with 0.6 nM dasatinib, a dose lethal to approximately 30% of the test population (∼LD30), for 3 days, then 2 × 106 cells were taken from each culture as posttreatment samples. For the in vivo screen, a pretreatment blood sample (70 μL) was taken via retro-orbital bleed at the presentation of overt symptoms of leukemia 11 days posttransplant, and mice were treated with 10 mg/kg dasatinib once per day for 3 days. At morbidity (14-16 days), >106 leukemia cells were collected from PB for posttreatment samples.
Sequencing was performed on an Illumina HiSequation 2000 instrument to obtain single-end 51-nt reads. Polymerase chain reaction (PCR) primer sequences are provided in supplemental Table 1. GFP competition assays repeat the screening procedure with a single hairpin or cDNA infected at 40% to 60% and use flow cytometry to assess enrichment or depletion.11 Clustered regularly interspaced short palindromic repeats (Crispr) knockouts (KOs) were generated by transfection with pX458 vector using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocol; 24 hours later, single GFP+ cells were sorted into individual wells of a 96-well plate. Clones were expanded (∼2-3 weeks), frozen down, and checked for Pafah1b3 loss; confirmed clones were thawed and used within 3 weeks. All flow data were collected on LSR analyzers and analyzed using FACSDiva software (BD Biosciences). Pafah1b3 levels were assessed via western blot. PAFR levels were assessed by flow cytometry using a fluorescein isothiocyanate (FITC)-conjugated antibody (Bioss bs-1478R-FITC). Lifespan analysis and analysis of associated imaging data were performed blinded to genotype of transplanted leukemic cells. Further details of screening and GFP competition procedures are in supplemental Methods.
Western blotting
BCR-ABL1+ BCP-ALL cells were infected with constructs coexpressing a fluorescent marker or a puromycin cassette and were sorted based on marker expression or isolated via puromycin selection; cell lysates were isolated from purified populations. The following antibodies were used for protein detection: Pafah1b3 (1:100 or 1:250 in Tris-buffered saline with Tween 20 (TBS-T) with 5% milk; Santa Cruz Biotechnology), Heat shock protein 90 (Hsp90) (1:5000 in TBS-T with 5% milk; BD Transduction Laboratories).
Therapeutics
Dasatinib (LC Laboratories) was dissolved in dimethyl sulfoxide (DMSO) and diluted into cell culture media. For in vivo use, dasatinib and ponatinib (LC Laboratories) were dissolved in 80 mM citric acid at pH 2.3; imatinib (LC Laboratories) was dissolved in water. TKI doses varied and are noted in figure legends; doses are similar to that used in previous studies but for shorter time periods to avoid emergence of BCR-ABL1–resistance mutations and to prevent loss of complexity in screen/GFP competition assays.8-10,17 WEB-2086 (Tocris) was dissolved in phosphate-buffered saline (PBS) plus 0.3% DMSO and was administered intraperitoneally at 5 mg/kg 3 times per week starting at 9 days post injection of tumor cells. PAF (Tocris) was dissolved in water and diluted into cell culture media. All drugs were administered to mice at 0.01 mL/g.
Imaging
Bioluminescent imaging was performed using albino B6 mice (strain B6(Cg)-Tyrc-2J/J; The Jackson Laboratory ) on a Xenogen VIS system and analyzed using Living Image version 4.4 software (Caliper Life Sciences). Mice were injected intraperitoneally with 4.5 mg of D-Luciferin and anesthetized with 2.5% isofluorane delivered at 1 L per minute in O2. Animals were imaged 10 minutes after injection at multiple exposures lasting 5 to 60 seconds with small binning. Images were normalized to the same color scale for figure generation.
Computational methods
All analyses were conducted in the R Statistical Programming language (http://www.r-project.org/), including ICA signature analysis, hierarchical clustering, and principal component analysis (PCA)-based sample clustering. Gene set enrichment analysis (GSEA) was carried out using the preranked mode with default settings.18 Reads were mapped to library sequences using the BWA aligner allowing at most 1 mismatch.19 Read counts were upper-quartile normalized to a count of 4000 (A.B., I. Blatz, P. L. Boutz, E.R.C.F., P. Y. Chen, S. Chen, R. Feretti, A. M. Gurtan, and M. D. Muzumdar, manuscript in preparation). Hairpins with normalized upper-quartile values <100 for across all in vivo samples were eliminated from downstream analyses. Hierarchical clustering was performed with Pearson correlation–based distance and average linkage. For signature analysis, an unsupervised blind source separation strategy using ICA was applied to elucidate statistically independent hairpin representation signatures11,20 (A.B., I. Blatz, P. L. Boutz, E.R.C.F., P. Y. Chen, S. Chen, R. Feretti, A. M. Gurtan, and M. D. Muzumdar, manuscript in preparation). The R implementation of the core Join Approximate Diagonalization of Eigenmatrices (JADE) algorithm20-22 was used along with custom R utilities. A high-resolution signature discovery approach was used to characterize global hairpin representation profiles. ICA was used on this discrete count-based dataset to elucidate statistically independent and biologically relevant signatures (A.B., I. Blatz, P. L. Boutz, E.R.C.F., P. Y. Chen, S. Chen, R. Feretti, A. M. Gurtan, and M. D. Muzumdar, manuscript in preparation). Further details of ICA are in supplemental Methods.
Results
Longitudinal RNAi screening reveals a set of hairpins with context-specific effects
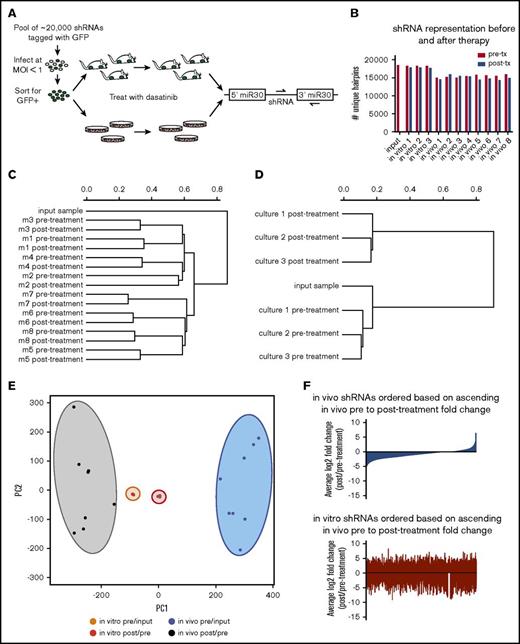
To identify genes that confer resistance or sensitivity to dasatinib in vivo in an unbiased manner, we performed pooled longitudinal shRNA screens in mice and in culture (Figure 1A). Fifteen thousand to 18 000 unique hairpins were identified in our screen; the median number of reads/shRNA were 91 in vivo and 374 in vitro (Figure 1B; supplemental Figure 1). Hierarchical clustering of normalized representation of shRNAs revealed that while in vivo samples cluster based on sample origin, in vitro samples cluster based on therapeutic context (Figure 1C-D). If we instead measure fold change of shRNA representation over time, samples cluster based on therapeutic context and microenvironmental setting by both PCA and hierarchical clustering (Figure 1E; supplemental Figure 2). Behavior of shRNAs in vivo does not predict their behavior in vitro or vice versa (Figure 1F; supplemental Figure 3). This is not due to random hairpin behavior; as fold changes in replicate samples are more correlated than nonreplicates (supplemental Figure 4).
RNAi screening for genetic mediators of response to dasatinib in vivo using a transplantable BCR-ABL1+BCP-ALL model. (A) Schematic representation of the longitudinal screening strategy. BCR-ABL1+ BCP-ALL cells were infected with a pool of 25 206 shRNAs tagged with GFP at MOI <1. GFP+ cells were isolated 48 hours postinfection by FACS, and cultured for 3 days before plating in 3 replicate cultures for in vitro screen and 4 days before injection into 8 nonirradiated syngeneic mice for in vivo screen. Input samples were taken at beginning of screens; pretreatment samples were taken after disease developed from cultures or blood of recipient mice, then mice/cultures were treated with dasatinib, allowed to relapse, and then posttreatment samples were taken from cultures or blood of recipient mice. Dasatinib was used in vitro at 0.6 nM (∼LD30) for 3 days, and in vivo at 10 mg/kg for 3 days once per day (q.d.) starting at 11 days postinjection. Hairpin representation was determined by high-throughput sequencing. (B) Bar graph showing the number of unique hairpins (1 or more reads) detected in each in vivo and in vitro screen sample both before (pre-tx) and after treatment (post-tx), as well as at transplant/plating (input). (C-D) Dendrograms generated by hierarchical clustering of normalized hairpin representation in vivo (C) and in vitro (D). (E) Principal component analysis of log2 fold changes before (pre/input) and after treatment (post/pre) in vivo and in vitro. (F) Waterfall plots representing the log2 fold changes after dasatinib therapy of all shRNAs in the library in vivo (blue) and in vitro (red), with shRNAs arranged in rank ascending order based on their log2 fold change in vivo.
RNAi screening for genetic mediators of response to dasatinib in vivo using a transplantable BCR-ABL1+BCP-ALL model. (A) Schematic representation of the longitudinal screening strategy. BCR-ABL1+ BCP-ALL cells were infected with a pool of 25 206 shRNAs tagged with GFP at MOI <1. GFP+ cells were isolated 48 hours postinfection by FACS, and cultured for 3 days before plating in 3 replicate cultures for in vitro screen and 4 days before injection into 8 nonirradiated syngeneic mice for in vivo screen. Input samples were taken at beginning of screens; pretreatment samples were taken after disease developed from cultures or blood of recipient mice, then mice/cultures were treated with dasatinib, allowed to relapse, and then posttreatment samples were taken from cultures or blood of recipient mice. Dasatinib was used in vitro at 0.6 nM (∼LD30) for 3 days, and in vivo at 10 mg/kg for 3 days once per day (q.d.) starting at 11 days postinjection. Hairpin representation was determined by high-throughput sequencing. (B) Bar graph showing the number of unique hairpins (1 or more reads) detected in each in vivo and in vitro screen sample both before (pre-tx) and after treatment (post-tx), as well as at transplant/plating (input). (C-D) Dendrograms generated by hierarchical clustering of normalized hairpin representation in vivo (C) and in vitro (D). (E) Principal component analysis of log2 fold changes before (pre/input) and after treatment (post/pre) in vivo and in vitro. (F) Waterfall plots representing the log2 fold changes after dasatinib therapy of all shRNAs in the library in vivo (blue) and in vitro (red), with shRNAs arranged in rank ascending order based on their log2 fold change in vivo.
ICA identifies candidate shRNAs with distinct behavior after therapy in vivo
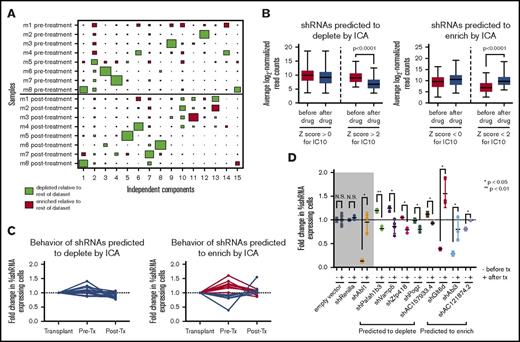
The biological noise and strong sample effects observed in our in vivo dataset indicate that basic statistical hypothesis testing (eg, Student t test) may not be sufficient to identify all of the hairpins that have distinct behavior after as compared with before therapy. To avoid this issue, we used ICA, a form of blind-source separation that has been successfully used with expression data to reduce clonality and identify latent variables by decomposing the overall dataset into (n − 1) independent components that represent distinct signatures.12,23,24 The signature for each component consists of the correlation of every shRNA in the dataset to the hairpin representation pattern defined by that component across all samples, which is expressed as a Z score. High-ranking positively and negatively correlated hairpins in each signature, identified by the high absolute value of their Z score, represent sets of shRNAs that drive the corresponding hairpin representation pattern. These signatures can be visualized using the sample-to-signature correspondence schematic afforded by Hinton plots; ICA identifies both sample (mouse)-dependent and -independent sources of variation in our screen (Figure 2A; supplemental Figure 5).
ICA of in vivo RNAi screening data identifies a set of hairpins with distinct behavior after therapy. (A) A Hinton plot of the independent components generated by ICA shows the signature generated by each component across all in vivo samples before and after dasatinib treatment. Colors represent directionality of hairpin representation (red enriched, green depleted) defined by that component and the size of each rectangle quantifies the strength of the signature for that component (column) in a given sample (row). Each component identifies a 2-sided signature, such that there are enriched and depleted hairpins within each sample for each signature; only 1 side of the component is depicted here. IC10 identifies a signature of hairpins that have distinct representation before and after therapy (P = .0078, Mann-Whitney U test). (B) Box-and-whisper plots showing the normalized representation before and after therapy from the original RNAi screen of those shRNAs predicted to deplete (left) or enrich (right) after therapy by IC10. Hairpins with a Z score <−2 or >2 are considered significant by this analysis. (C) Graphs showing normalized hairpin representation over time from individual GFP competition assays of 16 shRNAs predicted by IC10 to deplete after therapy (left) and 13 shRNAs predicted by IC10 to enrich after therapy (right). Blue lines denote shRNAs that behave as predicted by IC10; red lines denote shRNAs that do not behave as predicted. (D) Scatterplots showing normalized fold change of shRNA-expressing cells before and after dasatinib treatment in vivo in GFP competition assays of a set of 16 shRNAs that are predicted to deplete or enrich after therapy by IC10 and have at least a 1.5-fold difference in representation after therapy as compared with before. − denotes the fold change is before therapy (pre/input); + signifies fold change after therapy (post/pre). Controls are in the gray box: an empty hairpin vector and a hairpin targeting Renilla luciferase, which these cells do not express, are negative controls expected to lack significant change in representation, and a hairpin targeting ABL1 is included as a positive control that is expected to behave significantly differently after therapy vs before. Fold changes are normalized to an empty vector or a hairpin targeting Renilla luciferase. At least 3 mice were used per hairpin. Error bars represent standard deviation; P values were calculated using the Student t test. N.S., not significant.
ICA of in vivo RNAi screening data identifies a set of hairpins with distinct behavior after therapy. (A) A Hinton plot of the independent components generated by ICA shows the signature generated by each component across all in vivo samples before and after dasatinib treatment. Colors represent directionality of hairpin representation (red enriched, green depleted) defined by that component and the size of each rectangle quantifies the strength of the signature for that component (column) in a given sample (row). Each component identifies a 2-sided signature, such that there are enriched and depleted hairpins within each sample for each signature; only 1 side of the component is depicted here. IC10 identifies a signature of hairpins that have distinct representation before and after therapy (P = .0078, Mann-Whitney U test). (B) Box-and-whisper plots showing the normalized representation before and after therapy from the original RNAi screen of those shRNAs predicted to deplete (left) or enrich (right) after therapy by IC10. Hairpins with a Z score <−2 or >2 are considered significant by this analysis. (C) Graphs showing normalized hairpin representation over time from individual GFP competition assays of 16 shRNAs predicted by IC10 to deplete after therapy (left) and 13 shRNAs predicted by IC10 to enrich after therapy (right). Blue lines denote shRNAs that behave as predicted by IC10; red lines denote shRNAs that do not behave as predicted. (D) Scatterplots showing normalized fold change of shRNA-expressing cells before and after dasatinib treatment in vivo in GFP competition assays of a set of 16 shRNAs that are predicted to deplete or enrich after therapy by IC10 and have at least a 1.5-fold difference in representation after therapy as compared with before. − denotes the fold change is before therapy (pre/input); + signifies fold change after therapy (post/pre). Controls are in the gray box: an empty hairpin vector and a hairpin targeting Renilla luciferase, which these cells do not express, are negative controls expected to lack significant change in representation, and a hairpin targeting ABL1 is included as a positive control that is expected to behave significantly differently after therapy vs before. Fold changes are normalized to an empty vector or a hairpin targeting Renilla luciferase. At least 3 mice were used per hairpin. Error bars represent standard deviation; P values were calculated using the Student t test. N.S., not significant.
Independent component 10 (IC10) identified a statistically independent signature of shRNAs that have different representation in the mice after as compared with before therapy (Figure 2A; P = .0078 by Mann-Whitney U test). Hairpins with a Z score >2 (n = 389) or < −2 (n = 398) for IC10 significantly deplete or enrich after as compared with before therapy, respectively (Figure 2B; supplemental Tables 2 and 3). From this set of scoring shRNAs, we generated a validation set of 17 random shRNAs plus an additional 12 shRNAs that were previously predicted to have in vivo–specific behavior.11 Seventy-two percent behaved as predicted by IC10 in independent GFP competition assays as compared with negative control shRNA vectors, with hairpins predicted to deplete being more likely to validate (Figure 2C; supplemental Figure 6). If we require that shRNAs have at least 1.5-fold difference between the average fold changes before vs after therapy of all the mice in the screen, we reduce our validation set to 16 hairpins, 8 of which validate as having distinct behavior before vs after therapy; an shRNA against Abl1 was used as a positive control (Figure 2D; supplemental Figure 7).
Pafah1b3 loss sensitizes leukemia cells to dasatinib in vivo
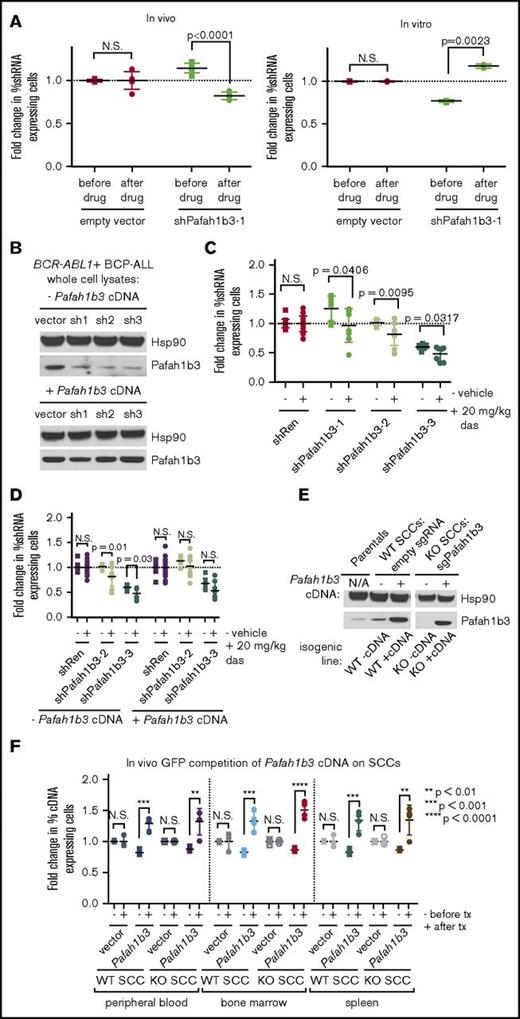
The hairpin that validated with the most significant depletion after dasatinib targets the acetylhydrolase and PAF/PAFR pathway component Pafah1b3; it is the only shRNA against Pafah1b3 in our screen. This shRNA depletes significantly after dasatinib in vivo but not in vitro in multiple independent GFP competition assays, indicating Pafah1b3 may be a viable target for combination therapy (Figure 3A). Additional shRNAs that knock down Pafah1b3 also deplete in the presence of dasatinib specifically in vivo, and rescue with a Pafah1b3 cDNA reverses this effect (Figure 3B-D; supplemental Figure 8). Coculturing leukemia cells with BM-derived stromal cells (BMSCs) to model aspects of the in vivo setting failed to recapitulate the observed in vivo phenotype (supplemental Figure 8).
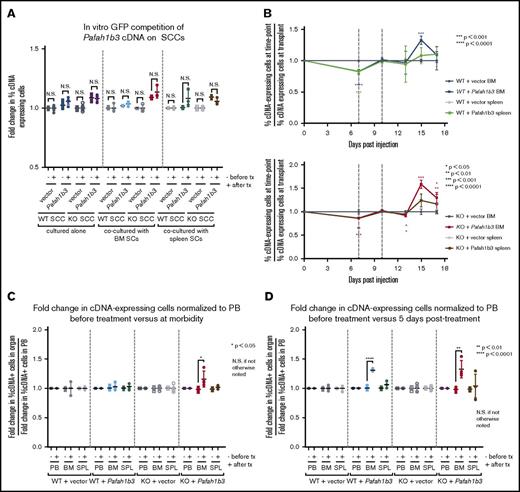
Pafah1b3 loss confers sensitivity to dasatinib specifically in vivo. (A) Scatterplots showing that the initial hairpin against Pafah1b3 from the 20K screen depletes significantly after dasatinib (das) treatment (20 mg/kg q.d. for 3 days starting 11 days postinjection of 106BCR-ABL1+ BCP-ALL cells; 1 nM in culture for 3 days) in vivo (left) but not in vitro (right) in GFP competition assays performed as described in supplemental Figure 6; fold changes are normalized to empty vector. Values are an average of at least 7 mice per condition. (B) Western blot showing Pafah1b3 knockdown by shRNAs in the presence of absence of a Pafah1b3 cDNA in cultured BCR-ABL1+ BCP-ALL cells infected with cDNA and/or shRNA and sorted by flow cytometry or selected by antibiotic resistance for pure populations. shPafah1b3-1 is the initial shRNA identified in 20K screen that targets an exon; shPafah1b3-2 and shPafah1b3-3 are additional shRNAs targeting the 3′UTR of the Pafah1b3 gene. Hsp90 is used as a loading control; Hsp90 and Pafah1b3 panels of the same lysates are different exposures of the same blot. −/+ cDNA lysates were run on distinct blots. (C) Scatterplot showing fold change in percentage of shPafah1b3-expressing cells from transplant to morbidity in both untreated and dasatinib-treated (20 mg/kg, 3 days q.d. starting 11 days postinjection of 106BCR-ABL1+ BCP-ALL cells) mice. GFP competition is performed as described in supplemental Figure 6, except that some mice are treated with vehicle instead of dasatinib, and no pretreatment sample is taken. − indicates vehicle-treated mice (morbidity/transplant); + indicates dasatinib-treated mice (morbidity/transplant). Fold changes are normalized to shRenilla; values are an average of at least 7 mice per condition. (D) Scatterplot showing fold change in percentage of shPafah1b3-expressing cells that coexpress either the Pafah1b3 cDNA or an empty GFP vector from transplant to morbidity in both untreated and dasatinib-treated (20 mg/kg, 3 days q.d. starting 11 days postinjection of 106BCR-ABL1+ BCP-ALL cells) mice. − indicates vehicle-treated mice (morbidity/transplant); + indicates dasatinib-treated mice (morbidity/transplant); the presence or absence of the Pafah1b3 cDNA is noted below the x-axis. Fold changes are normalized to shRenilla vector for matching cDNA condition. GFP competition assays are performed as in panel C. Values are an average of at least 7 mice per condition. (E) Western blot showing Pafah1b3 expression in single-cell clones (SCCs) generated by using CRISPR/Cas9 empty constructs (WT −cDNA, WT +cDNA) or CRISPR/Cas9 constructs targeting the Pafah1b3 gene (KO −cDNA, KO +cDNA), with or without rescue of Pafah1b3 protein levels via expression of the Pafah1b3 cDNA. Each pair of isogenic lines was created from a single SCC. Hsp90 is used as a loading control. Hsp90 and Pafah1b3 panels of the same lysates are different exposures of the same blot; WT vs KO SCCs are from the same exposure(s) of the same blot, but are edited to remove clones that were not used in subsequent experiments. (F) Scatterplot showing fold change in cDNA-expressing cells on either Pafah1b3 WT or KO backgrounds before and after dasatinib treatment (20 mg/kg, 3 days q.d. starting 7 days postinjection of 106BCR-ABL1+ BCP-ALL cells) in mouse PB, BM, and SPL, as normalized to an empty vector control. Pre- and posttreatment samples are taken from distinct mice rather than longitudinally sampled. − indicates fold change before treatment (pre/transplant); + indicates fold change after treatment (post/pre). Fold changes are normalized to empty vector control within each organ for the same SCC (WT or KO). GFP competition assays are performed as described in supplemental Figure 6, except timing of treatment is changed as noted, and mice are not longitudinally sampled. Values are an average of at least 3 mice per condition and time point. Error bars in all panels indicate standard deviation; P values were calculated using the Student t test.
Pafah1b3 loss confers sensitivity to dasatinib specifically in vivo. (A) Scatterplots showing that the initial hairpin against Pafah1b3 from the 20K screen depletes significantly after dasatinib (das) treatment (20 mg/kg q.d. for 3 days starting 11 days postinjection of 106BCR-ABL1+ BCP-ALL cells; 1 nM in culture for 3 days) in vivo (left) but not in vitro (right) in GFP competition assays performed as described in supplemental Figure 6; fold changes are normalized to empty vector. Values are an average of at least 7 mice per condition. (B) Western blot showing Pafah1b3 knockdown by shRNAs in the presence of absence of a Pafah1b3 cDNA in cultured BCR-ABL1+ BCP-ALL cells infected with cDNA and/or shRNA and sorted by flow cytometry or selected by antibiotic resistance for pure populations. shPafah1b3-1 is the initial shRNA identified in 20K screen that targets an exon; shPafah1b3-2 and shPafah1b3-3 are additional shRNAs targeting the 3′UTR of the Pafah1b3 gene. Hsp90 is used as a loading control; Hsp90 and Pafah1b3 panels of the same lysates are different exposures of the same blot. −/+ cDNA lysates were run on distinct blots. (C) Scatterplot showing fold change in percentage of shPafah1b3-expressing cells from transplant to morbidity in both untreated and dasatinib-treated (20 mg/kg, 3 days q.d. starting 11 days postinjection of 106BCR-ABL1+ BCP-ALL cells) mice. GFP competition is performed as described in supplemental Figure 6, except that some mice are treated with vehicle instead of dasatinib, and no pretreatment sample is taken. − indicates vehicle-treated mice (morbidity/transplant); + indicates dasatinib-treated mice (morbidity/transplant). Fold changes are normalized to shRenilla; values are an average of at least 7 mice per condition. (D) Scatterplot showing fold change in percentage of shPafah1b3-expressing cells that coexpress either the Pafah1b3 cDNA or an empty GFP vector from transplant to morbidity in both untreated and dasatinib-treated (20 mg/kg, 3 days q.d. starting 11 days postinjection of 106BCR-ABL1+ BCP-ALL cells) mice. − indicates vehicle-treated mice (morbidity/transplant); + indicates dasatinib-treated mice (morbidity/transplant); the presence or absence of the Pafah1b3 cDNA is noted below the x-axis. Fold changes are normalized to shRenilla vector for matching cDNA condition. GFP competition assays are performed as in panel C. Values are an average of at least 7 mice per condition. (E) Western blot showing Pafah1b3 expression in single-cell clones (SCCs) generated by using CRISPR/Cas9 empty constructs (WT −cDNA, WT +cDNA) or CRISPR/Cas9 constructs targeting the Pafah1b3 gene (KO −cDNA, KO +cDNA), with or without rescue of Pafah1b3 protein levels via expression of the Pafah1b3 cDNA. Each pair of isogenic lines was created from a single SCC. Hsp90 is used as a loading control. Hsp90 and Pafah1b3 panels of the same lysates are different exposures of the same blot; WT vs KO SCCs are from the same exposure(s) of the same blot, but are edited to remove clones that were not used in subsequent experiments. (F) Scatterplot showing fold change in cDNA-expressing cells on either Pafah1b3 WT or KO backgrounds before and after dasatinib treatment (20 mg/kg, 3 days q.d. starting 7 days postinjection of 106BCR-ABL1+ BCP-ALL cells) in mouse PB, BM, and SPL, as normalized to an empty vector control. Pre- and posttreatment samples are taken from distinct mice rather than longitudinally sampled. − indicates fold change before treatment (pre/transplant); + indicates fold change after treatment (post/pre). Fold changes are normalized to empty vector control within each organ for the same SCC (WT or KO). GFP competition assays are performed as described in supplemental Figure 6, except timing of treatment is changed as noted, and mice are not longitudinally sampled. Values are an average of at least 3 mice per condition and time point. Error bars in all panels indicate standard deviation; P values were calculated using the Student t test.
To control for off-target effects, as evidenced by hairpin behavior that cannot be rescued by Pafah1b3 cDNA, we used CRISPR/Cas9 as an orthogonal approach, generating leukemic single cell clones (SCCs) that either express wild-type (WT) levels of Pafah1b3 or are Pafah1b3 KOs (Figure 3E; supplemental Figure 9). By partially transducing WT or KO SCCs with a Pafah1b3 cDNA or an empty vector control, we can perform GFP competition assays in which Pafah1b3 cDNA-expressing cells replace shRNA-expressing cells, and compete with an internal control of Pafah1b3 KO or WT cells. As expected from our shRNA data, Pafah1b3 cDNA-expressing cells, which have higher levels of Pafah1b3 than either WT or KO cells, enrich in multiple leukemic organs of dasatinib-treated mice (Figure 3F).
Changes to Pafah1b3 levels affect survival of leukemia cells after dasatinib in a microenvironment-dependent manner
In experiments with WT cells in our model, we found the BM to be protective during dasatinib treatment; coculture with BMSCs protects against dasatinib-mediated killing in vitro as compared with leukemia cells alone or in coculture with SPL-derived stromal cells (SPL SCs) (supplemental Figures 10 and 11). However, coculturing with stromal cells did not replicate the in vivo phenotype of enrichment of Pafah1b3 cDNA-expressing cells in vitro, nor was there an increase in cell death after dasatinib in vitro in Pafah1b3 KO cells in coculture (Figure 4A; supplemental Figure 12). To determine if Pafah1b3 levels differentially affect leukemia cell survival in distinct microenvironments, we performed in vivo GFP competitions of Pafah1b3-cDNA expressing cells on WT and KO backgrounds and assessed the percentage of cDNA-expressing cells in PB, SPL, and BM at multiple time points. To lengthen the window between cessation of therapy and morbidity, we treated mice earlier than in GFP competition assays with shRNAs. While Pafah1b3 cDNA-expressing cells enriched vs at transplant at various time points after dasatinib in both SPL and BM, they were more consistently enriched in the BM vs SPL, and on the KO background vs WT, potentially due to relative depletion of the KO internal control cells (Figure 4B). To control for potential blood contamination in the SPL and BM, as cDNA-expressing cells do enrich in PB, we normalized fold changes in cDNA-expressing cells of each organ to the changes in cDNA-expressing cells in the PB at the same time point (supplemental Figure 13). Using this measure of relative enrichment, Pafah1b3 cDNA-expressing cells were enriched solely in the BM, and the enrichment was only maintained at multiple time points on the KO background (Figure 4C-D).
Changes to Pafah1b3 levels alter distribution of leukemia cells in vivo after dasatinib therapy. (A) Scatterplots showing fold change in cDNA-expressing cells on either Pafah1b3 WT or KO backgrounds before and after dasatinib treatment (1 nM for 3 days) in vitro when BCR-ABL+ BCP-ALL cells are cultured alone, with BM–derived stromal cells, or with SPL-derived stromal cells. − indicates fold change before treatment (pre/plating); + indicates fold change after treatment (post/pre). Fold changes are normalized to an empty vector control from the same culture condition and SCC. (B) Plots showing the fold change in Pafah1b3 cDNA-expressing cells, as compared with percentage of cDNA-expressing cells at transplant, over time in the BM and SPL on both Pafah1b3 WT (top) and Pafah1b3 KO (bottom) backgrounds over the course of dasatinib treatment (20 mg/kg, 3 days q.d. starting 7 days postinjection of 106BCR-ABL1+ BCP-ALL cells; vertical gray lines indicate time period when dasatinib is present). Asterisks denoting significance are color-matched to the organs; changes are not significant if not otherwise noted. (C-D) Scatterplots show relative enrichment of Pafah1b3 cDNA-expressing cells, or cells expressing an empty vector control, on a Pafah1b3 WT or KO background from transplant to the start of treatment (pre/transplant) vs relative enrichment from (C) transplant to morbidity (morbidity/transplant) or (D) transplant to 5 days posttreatment (5 days post/transplant) from GFP competition assays shown in panel B in PB, BM, and SPL. − indicates fold change before treatment; + indicates fold change after treatment. Fold changes are normalized to empty vector control from the same organ and SCC. Relative enrichment is defined as the fold change of percentage of cDNA+ cells in each organ normalized to the fold change of percentage of cDNA+ cells in the blood of the same mouse. For all panels, values are an average of at least 3 mice per genotype at each time point. Individual mice are used for each time point. GFP competition assays are performed as described in “Methods”/supplemental Figure 6, except for timing of treatment as detailed here. Error bars represent standard deviation; P values were calculated using the Student t test.
Changes to Pafah1b3 levels alter distribution of leukemia cells in vivo after dasatinib therapy. (A) Scatterplots showing fold change in cDNA-expressing cells on either Pafah1b3 WT or KO backgrounds before and after dasatinib treatment (1 nM for 3 days) in vitro when BCR-ABL+ BCP-ALL cells are cultured alone, with BM–derived stromal cells, or with SPL-derived stromal cells. − indicates fold change before treatment (pre/plating); + indicates fold change after treatment (post/pre). Fold changes are normalized to an empty vector control from the same culture condition and SCC. (B) Plots showing the fold change in Pafah1b3 cDNA-expressing cells, as compared with percentage of cDNA-expressing cells at transplant, over time in the BM and SPL on both Pafah1b3 WT (top) and Pafah1b3 KO (bottom) backgrounds over the course of dasatinib treatment (20 mg/kg, 3 days q.d. starting 7 days postinjection of 106BCR-ABL1+ BCP-ALL cells; vertical gray lines indicate time period when dasatinib is present). Asterisks denoting significance are color-matched to the organs; changes are not significant if not otherwise noted. (C-D) Scatterplots show relative enrichment of Pafah1b3 cDNA-expressing cells, or cells expressing an empty vector control, on a Pafah1b3 WT or KO background from transplant to the start of treatment (pre/transplant) vs relative enrichment from (C) transplant to morbidity (morbidity/transplant) or (D) transplant to 5 days posttreatment (5 days post/transplant) from GFP competition assays shown in panel B in PB, BM, and SPL. − indicates fold change before treatment; + indicates fold change after treatment. Fold changes are normalized to empty vector control from the same organ and SCC. Relative enrichment is defined as the fold change of percentage of cDNA+ cells in each organ normalized to the fold change of percentage of cDNA+ cells in the blood of the same mouse. For all panels, values are an average of at least 3 mice per genotype at each time point. Individual mice are used for each time point. GFP competition assays are performed as described in “Methods”/supplemental Figure 6, except for timing of treatment as detailed here. Error bars represent standard deviation; P values were calculated using the Student t test.
Pafah1b3 KO results in increased survival of TKI-treated mice
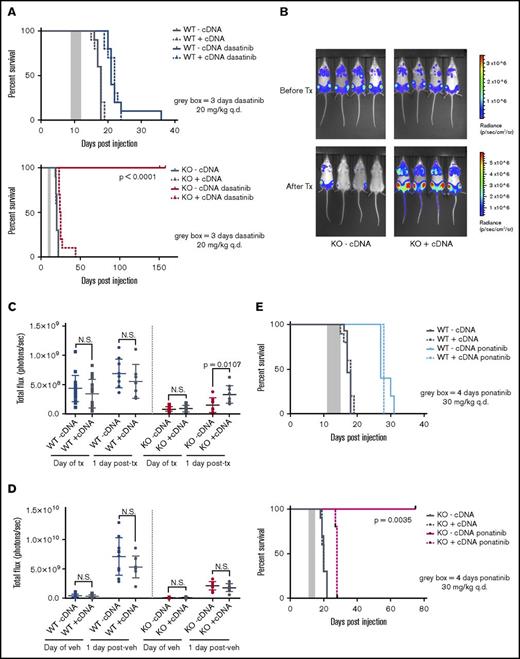
To compare pure populations of Pafah1b3 KO and WT cells while controlling for clonal effects, we used isogenic lines of WT or KO SCCs expressing either a Pafah1b3 cDNA (WT/KO +cDNA) or an empty vector (WT/KO −cDNA) (Figure 3E). Mice transplanted with Pafah1b3 KO cells have significantly extended lifespans when treated with dasatinib as compared with vehicle, and expression of a Pafah1b3 cDNA on the KO background completely reverses this effect; this is not the case for mice receiving Pafah1b3 WT cells (Figure 5A). In vivo bioluminescent imaging of these mice revealed significantly lower burden in mice with KO −cDNA leukemia after treatment than mice with KO +cDNA leukemia; this is not the case in mice transplanted with WT cells or in vehicle treated mice (Figure 5B-D).
CRISPR/Cas9-mediated KO of Pafah1b3 in BCR-ABL1+BCP-ALL cells results in increased survival of leukemia-bearing mice after TKI treatment. (A) Survival analysis of dasatinib-treated mice receiving 104 WT-cDNA or WT+cDNA cells (top) or KO −cDNA or KO +cDNA cells (bottom). Significance was calculated using the Mantel-Cox test; the gray rectangle indicates the time period (3 days q.d. starting at 9 days postinjection) over which dasatinib was administered at 20 mg/kg. Ten mice per condition were used in 2 independent experiments. (B) In vivo luminescent imaging of representative mice before and after dasatinib treatment of mice from survival curve shown in panel A that received KO −cDNA or KO +cDNA cells. Different color scales are used in before treatment and after treatment images to for visualization purposes, but images at each time point use the same color scale and duration of exposure. Bioluminescent images were collected using a Xenogen IVIS system and analyzed using Living Image version 4.4 software (Caliper Life Sciences). (C-D) Scatterplots quantifying in vivo luminescent imaging data (total flux = photons per second) before and after dasatinib treatment (C) or vehicle treatment (D) of mice from survival curves shown in panel A. Values are an average of at least 8 mice per genotype. Error bars indicate standard deviation; P values were calculated using the Student t test. (E) Survival analysis of ponatinib-treated mice receiving 104 WT −cDNA or WT +cDNA cells (top) or KO −cDNA or KO +cDNA cells (bottom). Significance was calculated using the Mantel-Cox test; the gray rectangle indicates the time period (4 days q.d, starting at 11 days postinjection) over which ponatinib was administered at 30 mg/kg. Five mice were used per condition.
CRISPR/Cas9-mediated KO of Pafah1b3 in BCR-ABL1+BCP-ALL cells results in increased survival of leukemia-bearing mice after TKI treatment. (A) Survival analysis of dasatinib-treated mice receiving 104 WT-cDNA or WT+cDNA cells (top) or KO −cDNA or KO +cDNA cells (bottom). Significance was calculated using the Mantel-Cox test; the gray rectangle indicates the time period (3 days q.d. starting at 9 days postinjection) over which dasatinib was administered at 20 mg/kg. Ten mice per condition were used in 2 independent experiments. (B) In vivo luminescent imaging of representative mice before and after dasatinib treatment of mice from survival curve shown in panel A that received KO −cDNA or KO +cDNA cells. Different color scales are used in before treatment and after treatment images to for visualization purposes, but images at each time point use the same color scale and duration of exposure. Bioluminescent images were collected using a Xenogen IVIS system and analyzed using Living Image version 4.4 software (Caliper Life Sciences). (C-D) Scatterplots quantifying in vivo luminescent imaging data (total flux = photons per second) before and after dasatinib treatment (C) or vehicle treatment (D) of mice from survival curves shown in panel A. Values are an average of at least 8 mice per genotype. Error bars indicate standard deviation; P values were calculated using the Student t test. (E) Survival analysis of ponatinib-treated mice receiving 104 WT −cDNA or WT +cDNA cells (top) or KO −cDNA or KO +cDNA cells (bottom). Significance was calculated using the Mantel-Cox test; the gray rectangle indicates the time period (4 days q.d, starting at 11 days postinjection) over which ponatinib was administered at 30 mg/kg. Five mice were used per condition.
We repeated survival experiments on Pafah1b3 WT −/+cDNA and KO −/+cDNA isogenic lines with other TKIs to see if this effect is specific to dasatinib. Imatinib shows little extension of life in this model unless treatment is given continuously, but does confer a survival advantage to mice transplanted with KO −cDNA vs KO +cDNA cells (supplemental Figure 14).8,9 Ponatinib treatment significantly extended lifespan in this model, and KO −cDNA cells but not WT −/+cDNA or KO +cDNA cells were sensitized to ponatinib treatment in vivo as they were to dasatinib (Figure 5E).
Sensitization of leukemia cells to dasatinib upon Pafah1b3 loss is partially due to PAF/PAFR signaling
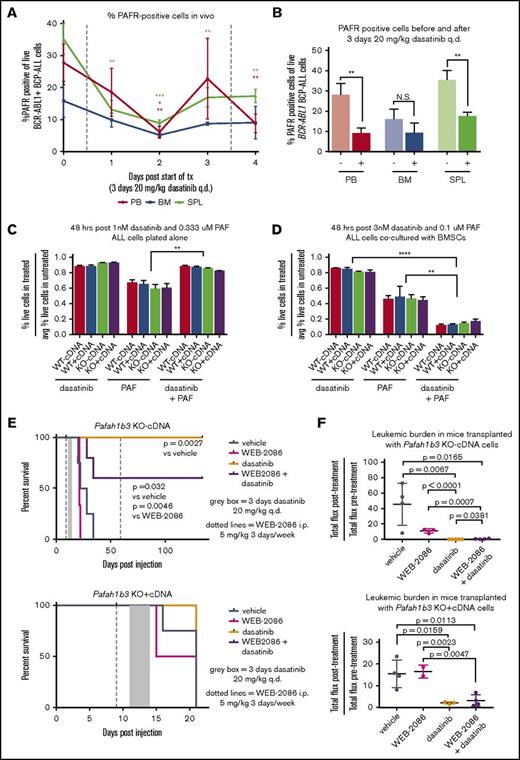
The ability of Pafah1b3 to reduce intracellular PAF levels allows it to protect cells from PAF-mediated death; accumulation of intracellular PAF can occur via both PAFR-dependent and -independent pathways.13,14 To determine if our cell express PAFR, we used an FITC-conjugated antibody that binds to PAFR. We found that PAFR is expressed on the surface of BCR-ABL1+ BCP-ALL cells in culture, and PAFR levels significantly decrease after dasatinib treatment only when leukemia cells are cocultured with stroma (supplemental Figure 15). The percentage of PAFR-expressing leukemia cells in vivo also decreases after dasatinib exposure, but less in BM than PB or SPL (Figure 6A-B). To determine what the effect of dasatinib is on PAF response in our model, we treated cells cultured either alone or with BMSCs with dasatinib, PAF, or dasatinib plus PAF, with higher doses used in coculture to achieve the same extent of cell death. Dasatinib and PAF cotreatment caused increased leukemia cell death only when stroma was present (Figure 6C-D).
PAF/PAFR signaling pathway is partially responsible for in vivo sensitization to TKI treatment upon Pafah1b3 loss. (A) Line graph showing the percentage of PAFR-expressing cells, of total leukemia cells, in mouse PB, BM, and SPL during dasatinib treatment. Mice were injected with 106BCR-ABL1+ BCP-ALL cells expressing tdtomato fluorescent protein, and were then treated with dasatinib (20 mg/kg q.d. for 3 days, starting at 11 days postinjection). Samples of mouse blood, BM, and SPL were taken from 3 mice/day starting on day 11 postinjection prior to dasatinib treatment, and were assessed for percentage of cells expressing PAFR on their surface via flow cytometry by staining cells with an anti-PAFR antibody conjugated to FITC and measuring the percentage of FITC+ cells of tdtomato+ cells. (B) Bar graph summarizing total percentage of leukemia cells expressing surface PAFR from before treatment (−) and 1 day after dasatinib treatment (+) time points from experiment shown in panel A. At least 3 mice were used per time point. (C-D) Bar graphs showing cell viability of BCR-ABL1+ BCP-ALL cells cultured in vitro either alone (C) or with BMSCs (D) after treatment with dasatinib alone, PAF alone, or a combination or dasatinib + PAF. Viability is normalized to untreated cells cultured in parallel in all cases, and is measured via flow cytometry after 48 hours of treatment by staining with 4′,6-diamidino-2-phenylindole (DAPI) as a cell exclusion stain. Drugs were used at an ∼LD85 for dasatinib and ∼LD55 for PAF when dosed alone, with the same dose of each drug used for the combination treatment; higher doses are needed to achieve equivalent killing when leukemia cells are cocultured with BMSCs, so dosage is adjusted accordingly. Three culture plates are used per condition. (E) Survival analysis of dasatinib-treated mice receiving 104 KO −cDNA (top) or KO +cDNA (bottom) cells in the presence of absence of the PAFR antagonist WEB-2086. WEB-2086 was given 3 times per week intraperitoneally (i.p.) at 5 mg/kg starting 9 days postinjection; dasatinib was given at 20 mg/kg q.d. for 3 days starting 11 days postinjection. Significance was calculated using the Mantel-Cox test; 3 to 5 mice were used per condition and genotype. (F) Scatterplots showing fold change in leukemic burden of mice from survival curves in depicted in panel E of mice receiving transplants of KO −cDNA (top) or KO +cDNA (bottom) cells as measured by in vivo bioluminescent imaging (total flux = photons per second) from start of treatment to 1 day post end of treatment. The posttreatment burden for each mouse is normalized to the pretreatment burden of that same mouse. Bioluminescent images were collected using a Xenogen IVIS system and analyzed using Living Image version 4.4 software (Caliper Life Sciences). Error bars represent standard deviation; P values were calculated using the Student t test.
PAF/PAFR signaling pathway is partially responsible for in vivo sensitization to TKI treatment upon Pafah1b3 loss. (A) Line graph showing the percentage of PAFR-expressing cells, of total leukemia cells, in mouse PB, BM, and SPL during dasatinib treatment. Mice were injected with 106BCR-ABL1+ BCP-ALL cells expressing tdtomato fluorescent protein, and were then treated with dasatinib (20 mg/kg q.d. for 3 days, starting at 11 days postinjection). Samples of mouse blood, BM, and SPL were taken from 3 mice/day starting on day 11 postinjection prior to dasatinib treatment, and were assessed for percentage of cells expressing PAFR on their surface via flow cytometry by staining cells with an anti-PAFR antibody conjugated to FITC and measuring the percentage of FITC+ cells of tdtomato+ cells. (B) Bar graph summarizing total percentage of leukemia cells expressing surface PAFR from before treatment (−) and 1 day after dasatinib treatment (+) time points from experiment shown in panel A. At least 3 mice were used per time point. (C-D) Bar graphs showing cell viability of BCR-ABL1+ BCP-ALL cells cultured in vitro either alone (C) or with BMSCs (D) after treatment with dasatinib alone, PAF alone, or a combination or dasatinib + PAF. Viability is normalized to untreated cells cultured in parallel in all cases, and is measured via flow cytometry after 48 hours of treatment by staining with 4′,6-diamidino-2-phenylindole (DAPI) as a cell exclusion stain. Drugs were used at an ∼LD85 for dasatinib and ∼LD55 for PAF when dosed alone, with the same dose of each drug used for the combination treatment; higher doses are needed to achieve equivalent killing when leukemia cells are cocultured with BMSCs, so dosage is adjusted accordingly. Three culture plates are used per condition. (E) Survival analysis of dasatinib-treated mice receiving 104 KO −cDNA (top) or KO +cDNA (bottom) cells in the presence of absence of the PAFR antagonist WEB-2086. WEB-2086 was given 3 times per week intraperitoneally (i.p.) at 5 mg/kg starting 9 days postinjection; dasatinib was given at 20 mg/kg q.d. for 3 days starting 11 days postinjection. Significance was calculated using the Mantel-Cox test; 3 to 5 mice were used per condition and genotype. (F) Scatterplots showing fold change in leukemic burden of mice from survival curves in depicted in panel E of mice receiving transplants of KO −cDNA (top) or KO +cDNA (bottom) cells as measured by in vivo bioluminescent imaging (total flux = photons per second) from start of treatment to 1 day post end of treatment. The posttreatment burden for each mouse is normalized to the pretreatment burden of that same mouse. Bioluminescent images were collected using a Xenogen IVIS system and analyzed using Living Image version 4.4 software (Caliper Life Sciences). Error bars represent standard deviation; P values were calculated using the Student t test.
Levels of surface PAFR expression and PAF-mediated cell death in this leukemia model change in response to dasatinib, suggesting that PAF/PAFR signaling may play a role in leukemia cell sensitization to dasatinib upon loss of Pafah1b3. In an in vivo survival experiment with KO −/+cDNA cells we found that cotreatment with the PAFR antagonist WEB-2086 blocked the dasatinib-mediated lifespan extension of some KO −cDNA mice (Figure 6E). In vivo bioluminescent imaging showed that mice transplanted with KO-cDNA cells, but not KO +cDNA cells, and treated with dasatinib plus WEB-2086 had a significantly higher posttreatment burden than those treated with dasatinib alone (Figure 6F; supplemental Figure 16).
Discussion
Large-scale pooled in vivo RNAi screens suffer from high variation between biological replicates and recovery of a relatively low number of candidate hits.11,25,26 Therapy screens have the additional challenge of differentiating the effects of drug response from those of normal cellular growth; we were able to separate those effects by performing a longitudinal screen and clustering the data based on changes in shRNA representation over time. ICA of hairpin fold changes successfully isolated sources of variation in our dataset and identified a significant shRNA representation signature of enrichment or depletion after treatment as compared with before. With additional filtering, we identified a set of 16 shRNAs predicted to have distinct behavior before vs after dasatinib, 50% of which validated in independent assays. These data suggest that in in vivo screens, ability to control for the mouse-specific signals that dominate hairpin behavior is likely to be essential for identification of candidate hits. Our screen is limited by low representation in some samples; utilization of a smaller library or a library with more shRNAs per gene could improve validation rates.
In a small validation set, ICA predicted the direction of shRNA behavior 72% of the time, even when the change was not biologically significant. In RNAi or CRISPR screens in which some constructs do not fully knock down or KO the target gene, being able to identify small but consistent effects that are maintained across multiple samples is important for defining genes of interest. Our data show that ICA analysis is capable of detecting even small biological effects in screening data, potentially reducing the incidence of false negatives in large-scale screens. We did find that shRNAs predicted to deplete are more likely to validate; this is likely due to the aggressive nature of the leukemia model. The challenge of identifying a genetic change that consistently confers resistance to drug is seen later in our Pafah1b3 results; while Pafah1b3 loss consistently sensitized leukemia cells to dasatinib, Pafah1b3 overexpression in GFP competition assays only conferred resistance in certain contexts. In survival assays, Pafah1b3 overexpression alone did not decrease lifespan at all, likely because the small amount of resistance it conferred was not sufficient to speed up an already fast-growing disease.
The initial shRNA targeting Pafah1b3 was the strongest depletor after dasatinib treatment in our initial validation set, and it only sensitized to drug in vivo. One of the main functions of the Pafah1b enzymatic complex is to reduce intracellular levels of PAF, which can activate caspase-dependent apoptosis in PAFR-dependent and -independent manners.13,14 PAF/PAFR signaling normally functions to activate leukocytes and promote their accumulation at sites of inflammation.27 Though PAF/PAFR/Pafah1b signaling has been linked to growth and metastasis in a number of solid tumors, and PAFR is expressed on both normal B lymphocytes and ALL patient blasts, a role for this pathway in leukemia has not been described.28-35 Here, we show that Pafah1b3 loss results in dramatic in vivo sensitization of BCR-ABL1+ BCP-ALL cells to dasatinib that cannot be recapitulated by coculture with stromal cells in vitro, and that the sensitization is more pronounced in the BM than SPL or blood. Dasatinib also decreased PAFR surface expression in vivo, although less in BM than SPL or blood, and when leukemia cells are cocultured with BMSCs, cotreatment of dasatinib and PAF results in more cell death than either drug is alone. Finally, we showed that cotreatment with the PAFR antagonist WEB-2086 partially reversed the in vivo sensitization to dasatinib in Pafah1b3 KO cells, indicating that this effect is at least somewhat due to signaling via the PAFR.
Our inability to use coculture to reproduce the in vivo phenotype of Pafah1b3 loss is not entirely surprising, as the cytokine milieu, to which PAF/PAFR signaling is responsive, should be different in leukemia cells cultured with stroma vs in vivo.13,27 Similarly, the specific in vivo leukemia microenvironment (eg, BM vs SPL) affects the PAF/PAFR/Pafah1b axis at both the level of PAFR surface expression and the degree of sensitization of cells with low/no vs high Pafah1b3 expression. The BM is an established protective site for leukemia cells after therapy, and our data seems to suggest that cells with higher levels of Pafah1b3 either traffic to or survive better in the BM vs SPL or blood after dasatinib treatment.36,37 The decrease in PAFR surface expression after dasatinib is also less pronounced in the BM, perhaps due to altered levels of PAF in this organ. Our data suggest that changes to dasatinib response upon modulation of the PAF/PAFR/Pafah1b pathway is distinct in different organs, with BM showing unique properties as compared with blood or SPL; it is unclear to what extent this is due to inherent microenvironmental differences vs alterations to leukemia cell trafficking between organs.
Overall, the in vivo and coculture data suggest that PAF binding to PAFR in the presence of dasatinib may sensitize cells to dasatinib treatment, potentially explaining the increased sensitivity of Pafah1b3 KO leukemia cells to TKI treatment as an effect of the KO cells’ inability to deal with accumulation of intracellular PAF. Our finding that pharmacologically blocking PAFR in vivo somewhat reverses the sensitization of Pafah1b3 KO cells to dasatinib further supports this idea. Further experiments modifying the PAF/PAFR/Pafah1b pathway, such as knocking down or knocking out PAFR in leukemia cells or treating mice directly with PAF should elucidate the role of this pathway in TKI response in BCR-ABL1+ BCP-ALL. Repeating our experiments in human cell lines or PDX models would also clarify whether our findings might be relevant in human disease.
Our data show that Pafah1b3 loss in an Arf null model of BCR-ABL1+ BCP-ALL results in dramatic sensitization to multiple TKIs in vivo, including the most recently developed TKI ponatinib. Nonspecific inhibitors of Pafah1b do exist, and a specific inhibitor of Pafah1b2/3 has recently been shown to have in vitro efficacy against cancer cell lines but is not suitable for in vivo use.38,39 Our results indicate that combination of Pafah1b inhibition with TKI therapy might be efficacious in BCR-ABL1+ BCP-ALL, particularly in those patients that lack expression of p14ARF. Additionally, these data highlight the role of the microenvironment in the development of minimal residual disease in BCR-ABL1+ BCP-ALL and reinforce the need for preclinical studies in this disease to be performed in vivo whenever possible.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank the Koch Institute Swanson Biotechnology Center for technical support, specifically the Biopolymer and Proteomics, Flow Cytometry, Bioinformatics, and Animal Imaging and Preclinical Testing cores. The authors also thank the Sherr laboratory for providing BCR-ABL1+ BCP-ALL murine cells, Corbin Meacham for help with the screening protocols, Yadira Soto-Feliciano for tail vein injections of mice, Yadira Soto-Feliciano and Jordan Bartlebaugh for monitoring mice when authors were unavailable, Luke Gilbert for advice on coculture experiments, Holly Criscione for experimental support, and members of the Hemann laboratory for advice, technical assistance, and critical examination of the manuscript.
The Biopolymer and Proteomics, Flow Cytometry, Bioinformatics, and Animal Imaging and Preclinical Testing cores were supported by Koch Institute Support (core) grant P30-CA14051 from the National Institutes of Health, National Cancer Institute. This work was also supported by the Ludwig Foundation for Cancer Research and an MIT School of Science Fellowship (E.R.C.F.).
Authorship
Contribution: E.R.C.F. and M.T.H. designed the experiments, except for the ICA and associated normalization and clustering of data, which was designed by A.B. in collaboration with E.R.C.F.; E.R.C.F. and M.T.H. wrote the manuscript except for the computational methods, which were written by A.B. and edited by E.R.C.F.; E.R.C.F. performed the screen; E.R.C.F. and E.L. performed subsequent validation experiments, with the exception of the WEB-2086 experiments, which were performed by R.B.; and A.B. performed ICA, associated clustering of screening data, and GSEA and created Hinton plots, dendrograms, and performed PCA.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael T. Hemann, Massachusetts Institute of Technology, 77 Massachusetts Ave, 76-301, Cambridge, MA 02139; e-mail: hemann@mit.edu.