Key Points
The frequency of 20q deletions increases with age and is more common than myeloid disorders.
Mosaic 20q deletions spanning regions deleted in myeloid disorders are found in individuals without diagnosis of myeloid disorders.
Abstract
Deletions on the long-arm of chromosome 20, del(20q), are common karyotypic abnormalities in myeloid disorders. Bioinformatic analyses of the B-allele frequency and log R ratio values from genome-wide association data have identified individuals who are mosaic for large structural abnormalities (>2 Mb). We investigated the most common autosomal event, namely mosaic del(20q), in 46 254 nonhematologic cancer cases and 36 229 cancer-free controls. We detected 91 mosaic del(20q) in leukocytes (80%) and buccal material (20%). The mosaic del(20q) mapped to a well-characterized minimally deleted region (MDR) reported in myeloid disorders. Common breakpoint clusters map to the coordinates of 29.9 to 31.5 Mb on the centromeric side of mosaic del(20q), and 42.0 to 45.4 Mb and 48.1 to 50.7 Mb on the telomeric end (GRCh36). Multivariate analyses suggest del(20q) increases with age, and is more common in males but less common in individuals of African ancestry. No conclusive associations were noted between the presence of mosaic del(20q) and subsequent solid tumor risk. Our observations demonstrate that the MDR of del(20q) is the most common large scale mosaic autosomal abnormality in whole blood and has a frequency of ∼1 in every 1000 adults over the age of 50, which exceeds the expected incidence of myeloid leukemia in the population. Our results indicate that subclonal mosaic events of a region implicated in myeloid disorders on 20q are more frequent than the predicted population-estimated incidence of myeloid diseases, and thus suggest that these events can be tolerated until additional events accumulate that drive myeloid disorders.
Introduction
Deletion of the long arm of chromosome 20, del(20q), is a common chromosomal abnormality in hematologic malignancies, observed in 10% of myeloproliferative neoplasms, 4% of myelodysplastic syndromes, and 2% of acute myeloid leukemias1-5 ; however, del(20q) alone is not a sufficient criterion for clinical diagnosis of a myeloid neoplasm.6 Although del(20q) alone can indicate a favorable prognosis, del(20q) in combination with other karyotypic abnormalities or its appearance later in disease can portend a less favorable outcome.7-9 A minimally deleted region (MDR) of ∼1.7 Mb has been reported in myeloid disorders.10-13 It is hypothesized that haploinsufficiency of one or more tumor suppressors in a cluster of imprinted genes in the MDR could lead to a loss of expression, which may contribute to myeloid disorder development.14-16 Mapping studies have focused on two such candidate tumor suppressors: L3MBTL1 and SGK2. Repression of L3MBTL1 has been shown to increase MYC expression in cell lines and could cooperate with SGK2, a regulator of the SWItch/sucrose nonfermentable-like chromatin-remodeling complex, in the epigenetic regulation of MYC expression.14
Mosaic del(20q) was the most commonly detected autosomal event in previous surveys of large structural mosaic alterations (>2 Mb) in genome-wide association studies conducted with DNA extracted from either leukocytes or buccal material.17-19 Herein, we aim to further characterize mosaic del(20q) in nonhematologic cancer cases and cancer-free controls, and compare mosaic del(20q) to reported del(20q) in myeloid neoplasms. Investigating the clinical and molecular features favoring the development of mosaic del(20q) could provide new insights into del(20q) acquisition, advance our understanding of how some individuals tolerate mosaic del(20q), and determine whether del(20q) could be investigated as a biomarker for early detection of myeloid disorders.
Methods
Study population
The combined genotyped set includes 46 254 nonhematologic cancer cases and 36 229 cancer-free controls drawn from over a dozen cancer genome-wide association studies (GWAS) conducted in the Division of Cancer Epidemiology and Genetics of the US National Cancer Institute (NCI) (dbGap accession numbers: phs000863.v1.p1, phs000351.v1.p1, phs000838.v1.p1, phs000346.v2.p2, phs000093.v2.p2, phs000336.v1.p1, phs000361.v1.p1, phs000652.v1.p1, phs000716.v1.p1, phs000206.v5.p3, phs000734.v1.p1, phs000147.v3.p1, phs000207.v1.p1, and phs000396.v1.p1).19 Scans involving hematologic malignancies (ie, non-Hodgkin lymphoma or chronic lymphocytic leukemia) were excluded from analyses. As described earlier, blood or buccal DNA (20% overall) was previously extracted and genotyped on one or more commercially available Illumina Infinium human single nucleotide polymorphism (SNP) microarrays (Hap300, Hap240, Hap550, Hap610, Hap660, Hap 1, Omni Express, Omni 1, Omni 2.5, and Omni 5). Standard quality control metrics were applied for clustering in batches (based on the specific studies) to optimize accuracy and minimize batch effects. Informed consent was received for each study participant. The protocols for each of the cancer GWAS studies were reviewed by the NCI’s Institutional Review Board and that of all participating study centers, and all participants provided written informed consent.
Mosaic event detection
Two bioinformatics metrics were assessed to detect mosaic events, namely the B-allele frequency (BAF) and log R ratio (LRR). BAF is a measure of allelic imbalance used to determine if a set of linear genotypes deviate from expected genotype clusters. Contiguous runs of heterozygous genotypes with values that deviate from the expected value of 0.5 are evidence for 2 subpopulations of cells that differ by genotype and is known as genetic mosaicism. The LRR of an SNP provides information on copy number state by comparing observed to expected intensity values. LRR values >0 indicate a copy gain and <0 indicate a copy loss. BAF and LRR values were calculated using previously described methods20 and corrected using a framework from our previous work.17
Renormalized BAF and LRR values for each participant were systematically scanned across all chromosomes using a modification of the R GADA package.21 Briefly, chromosomes are segmented using sparse Bayesian learning and backward elimination. Because of the limited marker density on some genotyping arrays and a high false-positive rate associated with events smaller than 2 Mb in size, events smaller than 2 Mb were filtered from the analysis. Gaussian mixture models were fitted to BAF bands to assign event type given the best fitting number of Gaussian components (2-4). Finally, event mosaic proportion was calculated using the deviation from expected BAF given the LRR value. Further details on the methods are available in prior publications.17,19
Statistical analysis
In multivariate analyses, we investigated the relationship of age group (categorized as: <50, 50-54, 55-59, 60-64, 65-69, 70-74, and ≥75), gender (male, female), estimated ancestry (fractions of European, Asian, and African ancestry by SNP genotypes), and nonhematologic cancer (disease free controls, solid tumor cases) with del(20q). Additionally, indicator variables were fitted to adjust for effects from contributing study. Strata-specific multivariate analyses were also performed within strata of nonhematologic cancer type to investigate whether del(20q) was associated with specific nonhematologic cancers. All statistical analyses were performed in R version 3.0.1 (R Foundation for Statistical Computing).
Results
We detected 119 mosaic events >2 Mb on chromosome 20 among 107 individuals (68 nonhematologic cancer cases and 39 cancer-free controls; supplemental Figure 1); 96 (81%) were mosaic losses, 8 (7%) mosaic gains, and 14 (12%) mosaic copy neutral loss of heterozygosity. The high proportion of mosaic chromosome 20 losses in relation to mosaic gains and copy neutral events differs from what is observed overall across all autosomal chromosomes (all autosomal loses = 34%, chromosome 20 loses = 81%; P = 2.2 × 10−16).19 One event was complex in nature and not amenable to distinct copy number state classification. When restricted to events detected on the q arm of chromosome 20, mosaic losses were the primary event detected (n = 91). Of the detected mosaic del(20q), the mean size was 17.0 Mb (range, 34.5-62.4 Mb; supplemental Figure 2A) and the percentage of affected cells differing from a normal karyotype was between 13% and 92% (median = 41%; supplemental Figure 2B), indicating individuals with low-frequency mosaic del(20q) (<13% of cells affected) are not detected or included in our frequency estimates of del(20q). LRR values for detected del(20q) indicated the majority of the detected mosaic losses were monoallelic (supplemental Figure 2C).
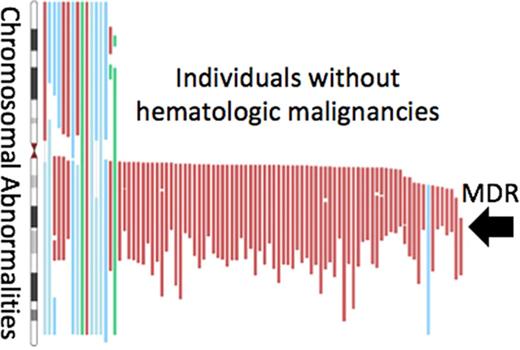
We observed variability in the size but nearly uniform overlap of the known MDR of 1.73 Mb from 40 425 000 to 42 155 000 (GRCh36; Figure 1). Notably, del(20q) spanning the MDR was seen at comparable frequencies in whole blood and buccal material (blood = 0.11%, buccal = 0.10%; P =1.0), suggesting DNA from our buccal samples likely originated from a high proportion of leukocytes. The observed MDR contains the transcripts of 8 genes: PTPRT, SRSF6, L3MBTL1, SGK2, IFT52, MYBL2, GTSF1L, and TOX2. Although no defined breakpoint region could be established, we observed breakpoints on the centromeric end clustering in a 1.6 Mb region spanning from 29.9 to 31.5 Mb, and breakpoint clusters at 42.0 to 45.4 Mb and 48.1 to 50.7 Mb on the telomeric end of del(20q) (NCBI36).
Localization of del(20q) MDR. Upper panel sums detected mosaic losses at each genomic position. Lower panel is a zoomed-in region of 20q (red box) showing genes nearby and in the MDR region from chr20:40 425 000 to 42 155 000 (GRCh36). chr20, chromosome 20.
Localization of del(20q) MDR. Upper panel sums detected mosaic losses at each genomic position. Lower panel is a zoomed-in region of 20q (red box) showing genes nearby and in the MDR region from chr20:40 425 000 to 42 155 000 (GRCh36). chr20, chromosome 20.
We fit logistic regression models to examine the effects of age, gender, ancestry, and overall solid tumor case/control status on the distribution of mosaic del(20)q. We observed a significant association between del(20q) and age (P = 6.82 × 10−4) (Figure 2). When comparing individuals >75 to those <50 years, the odds of a mosaic del(20q) increased substantially (odds ratio [OR], 14.33; 95% CI, 1.86-110.23; P = .01), suggesting a steady age-related acquisition of mosaic del(20q). Gender also displayed an association with del(20q) with males at an elevated risk of del(20q) in comparison with females (OR, 1.97; 95% CI, 1.09-3.57; P = .03). Individuals with African ancestry were observed to have fewer del(20q) events in comparison with those of European ancestry (OR, 0.14; 95% CI, 0.02-0.95; P = .04). No overall association between risk for development of a solid tumor and del(20q) was observed (P = .10). When stratifying on solid tumor subtype, suggestive associations were observed between head/neck cancer (P = 1.12 × 10−4), kidney cancer (P = .04), and ovarian cancer (P = .03), but the estimates are unstable due to small sample sizes. Additionally, the presence of mosaic del(20q) did not correlate with an increased risk of detectible mosaicism on other chromosomes (P = .99).
Frequency of mosaic del(20q) by gender and age category. Males are plotted in blue and females in red. Error bars represent 95% CI around the frequency estimate. CI, confidence interval.
Frequency of mosaic del(20q) by gender and age category. Males are plotted in blue and females in red. Error bars represent 95% CI around the frequency estimate. CI, confidence interval.
Our cross-sectional sampling using case-control and case-cohort GWAS suggests ∼0.1% of nonhematologic cancer cases and cancer-free controls have del(20q) in between 13% and 92% of leukocytes or buccal cells. Within our data set, a small subset of 5 individuals had a longitudinal collection of between 2 and 4 DNA samples. These data showed del(20q) acquired over time, del(20q) increasing in mosaic proportion, or del(20q) remaining at stable mosaic proportions (supplemental Figure 3). Although clinical information on these samples reveals no diagnosis of myeloid malignancies, it is possible that these individuals may harbor preleukemic clones.
Del(20q) is observed in <10% of cases with myeloid disorders.1,2,5 Prevalence estimates for myeloid neoplasms is ∼0.03% in the population of the United States and Europe,22,23 suggesting mosaic del(20q) frequency is approximately threefold higher than the overall population prevalence of myeloid malignancies (0.1% vs 0.03%) and substantially higher than the 0.003% expected frequency, based on combined estimates of myeloid malignancy prevalence (0.03%) and expected cases with del(20) (<10%). Other data sets with longitudinal collection of blood-derived DNA specimens over many years should be conducted to more thoroughly explore the relationship between mosaic del(20q) and the risk of myeloid disorders.
Discussion
Herein, we report that del(20q) is the most common large scale autosomal (>2 Mb) mosaic abnormality observed in DNA derived from leukocytes and buccal cells drawn from 36 229 apparently healthy controls, as well as 46 254 nonhematologic cancer cases. The mosaic MDR in our study includes the transcripts of 8 genes and overlaps the MDR reported in myeloid disorders. Furthermore, we observed elevated frequencies of mosaic del(20q) as age increases, elevated frequencies in men, and reduced frequencies in individuals with predominantly African ancestry. It is notable that myeloid disorder-associated del(20q) is present as a tolerated, mosaic event in subjects without the diagnosis of a myeloid disorder, and the observed frequency of mosaic del(20q) is higher than the predicted prevalence of myeloid disorders with del(20q).
Our observations suggest del(20q) could be an early event, although additional events need to accumulate to drive one or more clones toward myeloid disorders. Several lines of evidence suggest del(20q) may occur early in the development of myeloid disorder. Previous studies have detected del(20q) in stem cells with both myeloid and lymphoid differentiation capabilities.24,25 Several non-neoplastic conditions have also been associated with del(20q), including idiopathic thrombocytopenic purpura,26 pure red blood cell aplasia,27 Schwachman-Diamond syndrome,24 and idiopathic cytopenia.28 In prior studies of cases of myeloid neoplasms, the presence of del(20q) alone or accompanied with one other chromosomal abnormality has been associated with favorable prognosis.7,8
The observation of increased rates of mosaic del(20q) with age suggests that either the genome may have diminished maintenance capacity as age increases or that rare clones harboring del(20q) are in some way selected with age, either due to hematopoietic senescence or gradual age-related loss of stem cells.29,30 The gender (males have elevated del[20q] frequency) and ancestry (African ancestry has a lower del[20q] frequency) associations observed for del(20q) have also been observed with overall mosaicism and may not be specific to del(20q).19 One hypothesis that could partially account for the observed gender and ancestry differences in frequency is gender and ancestry specific differences in recombination rate31,32 ; although, further research is needed to robustly link gender and ancestry specific recombination rate to clonal mosaicism. Interestingly, individuals with high proportions of African ancestry (>30%) were observed to have a smaller mean event size (13.1 vs 17.5 Mb) and on average fewer cells affected (39.3% vs 43.0%), although these differences were not significant (P = .05 and .58, respectively) and no differences were observed in the genomic location of events in individuals with a high proportion of African ancestry. It is also noteworthy that our microarray detection technique detected events with affected cells ranging from 13% to 92%, indicating that individuals with low-frequency mosaic del(20q) (<13% of cells affected) are not detected or included in our frequency estimates of del(20q).
In conclusion, our study demonstrates that apparently healthy individuals and nonhematologic cancer cases harbor mosaic del(20q) covering the same genetic footprint as del(20q) seen associated in myeloid disorders. Our data indicate that del(20q) is age-related and the presence of detectable subpopulations of cells of hematopoietic origin harboring del(20q) do not necessarily commit to frank myeloid disorders. Although we did not observe a conclusive relationship with mosaic del(20q) and solid tumor risk, such mosaic deletions in circulating blood or buccal samples could be investigated as an important biomarker for assessing the future risk of myeloid disorders. Moreover, it is plausible that an increase of large structural mosaic alterations, together with point mosaic mutations, could define a state of genomic instability important for risk for a range of cancers as well as other complex diseases.33-35 At the same time, our study suggests that a small fraction of the population appears to tolerate the presence of somatically acquired del(20q) in a subpopulation of cells.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank the many men and women who contributed samples to make this study possible.
This study was funded by the Intramural Research Program of the National Institutes of Health NCI’s Division of Cancer Epidemiology, and the American Cancer Society.
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the National Institutes of Health.
Authorship
Contribution: S.J.C. and M.Y. conceived the study; M.J.M., W.Z., M.D., M.Y., and S.J.C. participated in the design and analysis of the study; M.J.M., W.Z., N.C., M.D., S.M.G., L.G., N.R., V.L.S., M.Y., and S.J.C. drafted the manuscript; and all authors read and approved the final submitted manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stephen J. Chanock, Division of Cancer Epidemiology and Genetics, National Cancer Institute, 9609 Medical Center Dr, Room 7E412, Bethesda, MD 20892-9776; e-mail: chanocks@mail.nih.gov.