Key Points
Gene-editing correction of the GT deletion in exon 2 of NCF1 pseudogenes corrects p47phox-deficient chronic granulomatous disease.
The nonfunctional pseudogenes NCF1B and NCF1C can be resurrected to produce functional p47phox protein by gene editing.
Abstract
Pseudogenes are duplicated genes with mutations rendering them nonfunctional. For single-gene disorders with homologous pseudogenes, the pseudogene might be a target for genetic correction. Autosomal-recessive p47phox-deficient chronic granulomatous disease (p47-CGD) is a life-threatening immune deficiency caused by mutations in NCF1, a gene with 2 pseudogenes, NCF1B and NCF1C. The most common NCF1 mutation, a GT deletion (ΔGT) at the start of exon 2 (>90% of alleles), is constitutive to NCF1B and NCF1C. NCF1 ΔGT results in premature termination, undetectable protein expression, and defective production of antimicrobial superoxide in neutrophils. We examined strategies for p47-CGD gene correction using engineered zinc-finger nucleases targeting the exon 2 ΔGT in induced pluripotent stem cells or CD34+ hematopoietic stem cells derived from p47-CGD patients. Correction of ΔGT in NCF1 pseudogenes restores oxidase function in p47-CGD, providing the first demonstration that targeted restoration of pseudogene function can correct a monogenic disorder.
Introduction
Chronic granulomatous disease (CGD) is a primary immune deficiency resulting from defects in any 1 of 5 protein subunits that comprise the phagocyte nicotinamide adenine dinucleotide phosphate oxidase (phox) complex, resulting in a failure to generate microbicidal reactive oxygen species. Phenotypically, CGD is characterized by recurrent bacterial and fungal infections, granuloma formation, hyperinflammation, and autoimmunity. Mutations in neutrophil cytosolic factor 1 (NCF1) are responsible for autosomal-recessive p47phox-deficient CGD (p47-CGD), comprising ∼25% of CGD cases. Notably, while mutations span across the genes for all other CGD patient phox genes, >80% of p47-CGD patients are homozygous for a 2-nt deletion (ΔGT) from the GTGT start of exon 2,1-6 resulting in a codon frame shift and premature termination, abrogating p47phox expression. The same ΔGT is constitutive to the start of presumptive exon 2 in the NCF1 pseudogenes NCF1B and NCF1C.2,3,7 Gene conversion resulting from close proximity of NCF1, NCF1B, and NCF1C at chromosome 7q11.23 (Figure 1A)8 likely accounts for the disproportionately high rates of the ΔGT mutation–mediated p47phox-deficient CGD relative to the other 3 genetic types of autosomal-recessive CGD. Recombination events between NCF1 and its pseudogenes may be responsible for the ΔGT mutation defect acquired by the resulting product at the NCF1 locus.2,3,5,7
Schematic diagram of NCF1 and its pseudogenes on chromosome 7 and the corrective donors used. (A) Positions of pseudogenes (NCF1B and NCF1C) relative to NCF1 are shown in the inset of chromosome locus 7q11.23. (B) Distinguishing sequences in normal NCF1 and pseudogenes include unique intronic SNPs, the normal GTGT (or mutant ΔGT) at start of exon 2 in NCF1 vs constitutive ΔGT in both NCF1B and NCF1C as depicted. We do not indicate in this figure that a portion of pseudogene or an entire pseudogene may replace a portion of or the entire NCF1 gene at its NCF1 locus. ZFN targets the start of exon 2, with the recognition sequence (upper case) and the spacer or cutting region (lower case) as shown. The ZFN targets the pseudogene sequence shown: CCCAGGTACATGTTCctggtgAAATGGCAGGAC. The capital letters represent the target sequence, and the lowercase letters denote the space or cutting site. Within the “Donor repair sequence,” the underlined base pair G was changed from a base pair A to avoid recutting but does not change the codon translation and is the codon-optimized choice. (C) Schematic representation of the correction donors used for minigene addition or exon 2 replacement with or without a puromycin (Puro) selection cassette flanked with loxP (A/B or C/D, respectively). The cytomegalovirus (CMV) promoter is used to express the puromycin resistance gene and the polyadenylation signal (pA) is indicated. Shown are left and right homology arms (LHA and RHA, respectively). The inverted terminal repeats (ITRs) in donors B, C, and D allow donor packaging in rAAV vector.
Schematic diagram of NCF1 and its pseudogenes on chromosome 7 and the corrective donors used. (A) Positions of pseudogenes (NCF1B and NCF1C) relative to NCF1 are shown in the inset of chromosome locus 7q11.23. (B) Distinguishing sequences in normal NCF1 and pseudogenes include unique intronic SNPs, the normal GTGT (or mutant ΔGT) at start of exon 2 in NCF1 vs constitutive ΔGT in both NCF1B and NCF1C as depicted. We do not indicate in this figure that a portion of pseudogene or an entire pseudogene may replace a portion of or the entire NCF1 gene at its NCF1 locus. ZFN targets the start of exon 2, with the recognition sequence (upper case) and the spacer or cutting region (lower case) as shown. The ZFN targets the pseudogene sequence shown: CCCAGGTACATGTTCctggtgAAATGGCAGGAC. The capital letters represent the target sequence, and the lowercase letters denote the space or cutting site. Within the “Donor repair sequence,” the underlined base pair G was changed from a base pair A to avoid recutting but does not change the codon translation and is the codon-optimized choice. (C) Schematic representation of the correction donors used for minigene addition or exon 2 replacement with or without a puromycin (Puro) selection cassette flanked with loxP (A/B or C/D, respectively). The cytomegalovirus (CMV) promoter is used to express the puromycin resistance gene and the polyadenylation signal (pA) is indicated. Shown are left and right homology arms (LHA and RHA, respectively). The inverted terminal repeats (ITRs) in donors B, C, and D allow donor packaging in rAAV vector.
Evolutionarily, pseudogenes are nonessential but persistent copies of genes that are dysfunctional through mutation(s) resulting in defective expression or nonfunctional products. By one report,9 at least 19 724 regions in the human genome may encode pseudogenes, although the total number of pseudogenes is expected to be far greater. These changes in the chromosomes due to mutations, insertions, deletions, duplications, and chromosomal rearrangements are driving forces for genome evolution and can be beneficial, detrimental, or inconsequential.
Here, we examined strategies for p47-CGD gene correction using engineered zinc-finger nucleases (ZFNs) targeting the exon 2 ΔGT in induced pluripotent stem cells (iPSCs) derived from p47-CGD patients. We show that production of p47phox protein in association with restoration of oxidase activity can occur following gene-targeted correction of the exon 2 ΔGT at the NCF1 locus or its pseudogenes. This is a first report of functional correction of a monogenic disorder mediated by gene-editing resurrection of pseudogene function.
Methods
Approvals for human blood and animal use
Peripheral blood from healthy volunteer donors and CGD patients was obtained after written informed consent under auspices of National Institute of Allergy and Infectious Diseases (NIAID) institutional review board–approved protocols 05-I-0213 and 94-I-0073. H.L.M. is the principal investigator of these human subjects’ protocols. The conduct of these studies conforms to the Declaration of Helsinki protocols and all US federal regulations required for protection of human subjects.
Use of immunodeficient NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice (The Jackson Laboratory) for teratoma assays of iPSC pluripotency was approved by NIAID Institutional Animal Care and Use Committee under the animal use protocol LHD 3E. H.L.M. is the principal investigator of this animal use protocol. The conduct of these studies conforms to AAALAC International guidelines and all US federal regulations required for protection of research animals.
Human iPSC derivation and maintenance
Peripheral blood mononuclear cells (PBMCs) were purified from non-mobilized subjects’ peripheral blood by Ficoll separation (Lymphocyte Separation Medium; MP Biomedicals). CD34+ cells were isolated from PBMCs using CD34 antibody-conjugated magnetic beads for transduction with STEMCCA-loxP lentivector or Sendai virus to produce iPSCs as described previously.10 P47-02, p47-03, and p47-04 were previously described. P47-05 was reprogrammed with Sendai virus with a method described elsewhere.10 iPSCs were grown on Matrigel (BD Biosciences)–coated plates and cultured with E8 medium (Thermo Fisher Scientific) or Nutristem XF/FF medium (Stemgent, Cambridge, MA). All initial lines and all corrected subclones of iPSCs were rigorously cloned to assure that analyses reflect homogeneity.
Correction donors, CD34+ hematopoietic progenitor cell correction, and iPSC correction
Plasmid donors were synthesized (Genescript). Peripheral blood mobilized CD34+ cells were obtained from the National Institutes of Health Department of Transfusion Medicine, Cell Processing Section.10 CD34+ cells were cultured in Stemspan SFEMII (STEMCELL Technologies) supplemented with stem cell factor, Flt3 ligand, and thrombopoietin (Peprotech; 100 ng/mL each) for 2 days before transfection using Amaxa4D (Lonza Nucleofector) or MaxCyteGT (MaxCyte, Gaithersburg, MD) per the manufacturer’s conditions. Cells were electroporated with the ZFN messenger RNA (mRNA) at 5 μg/mL, immediately followed by recombinant adeno-associated virus serotype 6 (rAAV6) transduction at multiplicities of infection (MOIs) of 1000 to 5000 and incubated overnight.
Two to three million CGD-patient iPSCs grown in Nutristem (Stemgent) were transfected with 5 μg donor plasmid and 5 μg ZFN plasmid DNA (Sigma) or in vitro–transcribed/synthesized mRNA (Sigma, Life Technologies, TriLink) using the AMAXA mouse ES transfection kit (A-023). For correction, fresh medium was used and rAAV-2 (200 to 400 MOI) was added. The next day, iPSCs were transfected with ZFN mRNA. After 3 to 5 days of growth, cells were selected for puromycin resistance at 0.25 to 0.5 μg/mL puromycin. After determining correct insertion specific to NCF1 by polymerase chain reaction (PCR), strongly positive clones were used for additional analysis.
rAAV production and quantitative PCR titer analysis
A map of the correction donors is shown in Figure 1C. The rAAV2 and rAAV6 serotypes were used for generating donors for correction of iPSCs or hematopoietic progenitor cells (HPCs), respectively. rAAV was made by transfecting 293T cells with Polyethyleneimine, linear, M.W. 25,000 (VWR) for the following plasmids, as described previously11 : Ad Helper pRS449B,11 AAV2 RepCap (AAV2 Y730+500+400F), or AAV6 RepCap (pACGr2c6-Y705+731F,12 a gift from Dr. Georgiy Aslanidi), and correction donor with ratios of 2:1:1, respectively. Lysates were mixed with 0.55 g/mL cesium chloride (Sigma) and ultracentrifuged at 38 000 rpm using an SW-41 rotor (Beckman) for 72 hours to form a density gradient. The virus was collected at refractive index 1.372 and screened by real-time PCR. Positive fractions were dialyzed with Slide-A-Lyzer Dialysis Cassette 10 000 MWCO (Thermo Fisher Scientific) in Dulbecco’s modified Eagle medium. Titer was performed using primers DiagPCRP47F 5′-TGCAATCCAGGACAACCGCAA-3′ and R69 5′-TCGGTGAATCTCCGGTACAC-3′ and reconfirmed with puro-specific primers for donor A and donor C using primers Forward(Fwd)-Puro-Sybr 5′-TCACCGAGCTGCAAGAACT-3′ and Reverse(Rev)-Puro-Sybr 5′-GAGGCCTTCCATCTGTTGCT-3′. SYBR Green (Promega) real-time PCR was run obtaining titers of 1011 per milliliter. Plasmid DNA used to generate vector was used to generate the standard curve.
Molecular analysis
Targeted insertion (TI) was tested by PCR analysis using one primer located inside the puromycin selectable cassette. The second primer was located outside of the homology arm of correction donor. For detecting TI, primers Fwd-BGHpA 5′-GAAATTGCATCGCATTGTCTGAGTAGG-3′ and Rev-OuterRHA3 5′- GTCCTCAATCCAGTCGCCAA-3′ were used. Single-nucleotide polymorphism (SNP) analysis determined the correction site NCF1, NCF1B, or NCF1C loci for each corrected iPSC line. This was done by using the puromycin selection cassette as a priming region and second primer either ∼2 kb upstream (intron1) or ∼2 kb downstream (intron 2), both of which are located beyond the homology region of corrective donors, to generate 2 PCR amplification products encompassing genomic DNA containing unique SNPs specific to the correction. Primers for the first PCR were Fwd-1671gNCF1 5′-CTGCTTGTCCTCTGCAGTGA-3′ and RevCMV2 5′-TGGAAAGTCCCTATTGGCGT-3′. Primers for the second PCR were the NCF1-specific primers Fwd-BGHpA 5′- GAAATTGCATCGCATTGTCTGAGTAGG-3′ and Rev5568gNCF1 5′-TTTCTGGCTGTGTGGCTCTG-3′.
Neutrophil differentiation, characterization, and oxidase functional assays
STEMdiff APEL medium (STEMCELL Technologies) differentiation of iPSCs was performed to obtain HPCs, as previously described.13 A stage-3 medium is used for HPC expansion, followed by stage 4 medium for neutrophil differentiation.13,14 Modifications to the procedure are as follows. On day 2 of APEL A, media was changed to fresh APEL A, and APEL B culture differentiation was sometimes extended from day 13 to day 18 depending on differentiation progress. At stage 3 HPC expansion, suspension cells were harvested after 1 week and additional media added. The harvested HPCs were further expanded in stage 3 media or stem cell media (StemSpan SFEMII medium, STEMCELL Technologies) containing 50 ng/mL stem cell factor, FLT-3L, thrombopoietin, 5 ng/mL interleukin 3 (IL-3), and 25 ng/mL IL-6. Neutrophil surface markers were detected using fluorochrome-conjugated antibodies CD13, CD33, and CD45 (BD Biosciences). Intracellular p47phox stain, dihydrorhodamine (DHR), and chemiluminescence assays were performed as previously described.13
DNA fingerprint, FISH, and G-band karyotype analysis
For identity testing, genomic DNA from PBMC (uncorrected) and iPSC lines were analyzed using a multiplex standard tandem repeat method, PowerPlex 16 HS system (Promega). To determine site-specific correction, fluorescent in situ hybridization (FISH) was used for colocalization. Metaphase spreads were prepared and analyzed as previously described.15 NCF1 probe (Empire Genomics) shown in red, recognizes the 3′ end of NCF1 and its downstream sequences. We used a 3.5-kb probe for the puromycin selection cassette. Primers used for the probe were Fwd-FISH-Probe 3.5 kb 5′-TTGTGTATCCAGGGCAGTGG-3′ and Rev-FISH-Probe 3.5 kb 5′-AGGAAAAGGAGCCAGGTGTG-3′.
Southern blot
Southern blot analysis was commercially performed by Lofstrand Labs (Gaithersburg, MD). Fifteen micrograms of genomic DNA was digested with Fast digest BamH1 (Fermentas) for 2.5 hours to detect random vector insertion and Cre-mediated excision of the puromycin resistance selectable marker cassette.
Results
ZFN-targeted gene correction of p47-CGD
We engineered ZFNs to target the shared region at the start of exon 2 containing ΔGT at the NCF1 locus responsible for p47-CGD in most patients (Figure 1A-B). We designed 2 pairs of donor constructs to achieve correction following homologous recombination targeted insertion (Figure 1C). Donors A and B were designed to achieve correction with an abbreviated NCF1 codon-optimized cDNA beginning with exon 2 and continuing to the stop codon plus a polyadenylation signal (minigene). Donors C and D were designed to repair the ΔGT in mutated NCF1 by targeted replacement of only exon 2 with codon-optimized corrected sequence. As shown in Figure 1C, the donor pairs were designed with or without a puromycin-resistance selection cassette. We initiated our studies with cells from p47-CGD patients known to be homozygous for the common ΔGT mutation at the NCF1 locus.
Gene-edited restoration of GTGT at sites of ΔGT in p47-CGD iPSCs
Our initial studies were with the P47-04 iPSC line derived from subject 4, who is a p47-CGD patient homozygous for ΔGT (ΔGT/ΔGT) at the NCF1 locus. We cotransfected with ZFN mRNAs and Donor A plasmid (Figure 1) to achieve minigene insertion correction, generating the P47-04-C1M line following puromycin selection. Gene correction was assessed by PCR to identify specific targeted insertion using one primer to the puromycin selection cassette and a second primer matching sequences in intron 1 or intron 2 common to NCF1, NCF1B and NCF1C located outside the homology arms of the donor. The minigene-corrected iPSC line P47-04-C1M was differentiated into myeloid cells as previously described13 and tested for p47phox protein expression. In Figure 2A (left 3 panels), only the minigene-corrected myeloid cells derived from P47-04-C1M showed restored p47phox expression (Figure 2A) similar to myeloid cells differentiated from healthy control iPSCs. It is important to note in the right 3 panels of Figure 2A that undifferentiated iPSCs, whether uncorrected, corrected, or healthy control, do not express p47phox.
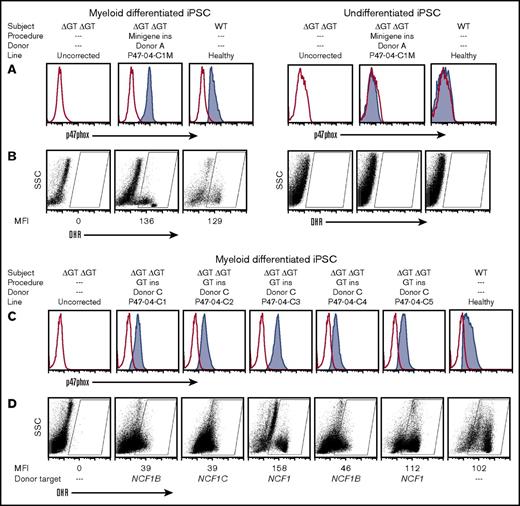
Functional analysis of myeloid cells generated from corrected homozygous exon 2 ΔGT NCF1 p47-CGD iPSCs. The cell treatment procedure is summarized above the graphs, with minigene insertion (ins) mediated by donor A or exon 2 replacement (GT ins) mediated by donor C. (A) Flow cytometric analysis of p47phox expression in myeloid-differentiated iPSC lines, P47-04 (uncorrected) vs minigene-corrected, or healthy control (left 3 panels, respectively) and in undifferentiated iPSCs (right 3 panels). (B) Oxidase function of the uncorrected, minigene-corrected, and healthy control line (left 3 panels, respectively) or the same iPSC lines without differentiation (right 3 panels, respectively) evaluated by dihydrorhodamine (DHR) flow cytometry analysis. (C) Flow cytometric analysis of p47phox expression in myeloid-differentiated iPSC lines, P47-04 (uncorrected) vs five exon 2–corrected, and healthy control, respectively. (D) Oxidase function of the uncorrected, 5 exon 2–corrected, and healthy control lines, respectively, or the same iPSC lines without differentiation, respectively, was evaluated by DHR. Also indicated below these graphs (Donor target) is where the correction occurred (NCF1 gene or the NCF1B or NCF1C pseudogene) as determined by sequence analysis (Figure 5). rAAV2 vector was used between 200 and 400 MOI of viruses per cell. SSC, side scatter; WT, wild-type.
Functional analysis of myeloid cells generated from corrected homozygous exon 2 ΔGT NCF1 p47-CGD iPSCs. The cell treatment procedure is summarized above the graphs, with minigene insertion (ins) mediated by donor A or exon 2 replacement (GT ins) mediated by donor C. (A) Flow cytometric analysis of p47phox expression in myeloid-differentiated iPSC lines, P47-04 (uncorrected) vs minigene-corrected, or healthy control (left 3 panels, respectively) and in undifferentiated iPSCs (right 3 panels). (B) Oxidase function of the uncorrected, minigene-corrected, and healthy control line (left 3 panels, respectively) or the same iPSC lines without differentiation (right 3 panels, respectively) evaluated by dihydrorhodamine (DHR) flow cytometry analysis. (C) Flow cytometric analysis of p47phox expression in myeloid-differentiated iPSC lines, P47-04 (uncorrected) vs five exon 2–corrected, and healthy control, respectively. (D) Oxidase function of the uncorrected, 5 exon 2–corrected, and healthy control lines, respectively, or the same iPSC lines without differentiation, respectively, was evaluated by DHR. Also indicated below these graphs (Donor target) is where the correction occurred (NCF1 gene or the NCF1B or NCF1C pseudogene) as determined by sequence analysis (Figure 5). rAAV2 vector was used between 200 and 400 MOI of viruses per cell. SSC, side scatter; WT, wild-type.
The DHR oxidase assay (Figure 2B) was used to detect superoxide production by myeloid differentiated iPSC cultures (left 3 panels) or by undifferentiated iPSC cultures (right 3 panels). Myeloid differentiation of iPSCs generally leads to a mixture of immature and mature myeloid cells, where only the more mature myeloid cells express significant oxidase activity when stimulated. In Figure 2B, we gated on the portion of the culture population in the oxidase-positive region. Since the number of mature cells in a myeloid differentiated iPSC culture is variable, by gating the oxidase-positive region, it is possible to assess the mean level of oxidase activity in the mature myeloid cells as measured by the mean fluorescence intensity (MFI). The MFI of these mature myeloid cells in uncorrected, minigene-corrected, and healthy iPSCs are 0, 136, and 129, respectively (Figure 2B, left 3 panels). As expected, undifferentiated iPSCs for all lines contained no mature myeloid cells and were therefore devoid of cells within the oxidase-positive gated region. This demonstrates that minigene expression is regulated by the endogenous p47phox promoter, which is active only after myeloid differentiation.
We next conducted studies for correction of the iPSC line P47-04 in which exon 2 was replaced to correct ΔGT using ZFN mRNA plus donor C packaged in rAAV2, achieving TI of donor C in 16 of 18 (90%) puromycin-selected clones. We myeloid differentiated 5 of these clones to demonstrate restored p47phox expression and oxidase function (Figure 2C-D). There was some variability in both average p47phox expression and in oxidase activity (MFI) in the corrected clones compared with the healthy control. No p47phox protein or oxidase activity was seen in myeloid cells derived from the uncorrected line.
Similar studies were performed on the P47-02 iPSC line, which was derived from subject 2, another p47-CGD patient homozygous for ΔGT (ΔGT/ΔGT) at the NCF1 locus. Specifically, we corrected the exon 2 ΔGT in the P47-02 iPSC line. Analysis of 4 corrected iPSC subclone lines derived from P47-02 is shown in supplemental Figure 1, where oxidase function is restored in myeloid cells differentiated from these gene-corrected clones.
Characterization of corrected iPSCs by FISH, karyotype, Cre-mediated excision of puromycin selection cassette, and microsatellite
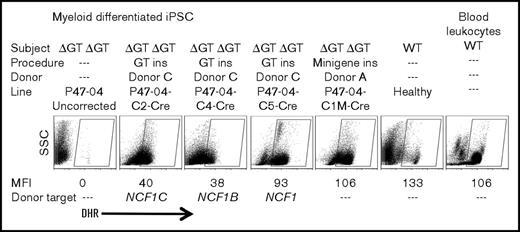
Some corrected iPSC lines were analyzed by FISH, which showed that a probe to the puromycin resistance selection cassette colocalized with the NCF1 probe at chromosome 7 (supplemental Figure 2A), confirming TI at an NCF1 locus. A corrected iPSC line retained a normal karyotype (supplemental Figure 2B). Southern blot analysis using a probe to the puromycin resistance gene was performed on the minigene-corrected iPSC derived from P47-04 (characterized in Figure 2A-B) and on the GT exon 2–corrected iPSC lines derived from P47-04 (characterized in Figure 2C-D) and P47-02 (characterized in supplemental Figure 1), confirming presence of a single TI band with no evidence of off target inserts. Cre-mediated excision in these lines resulted in the expected loss of the puromycin resistance gene (supplemental Figure 3A). To test if oxidase function remained after Cre-mediated excision of the loxP-flanked puromycin selection cassette, several of the GT exon 2–corrected iPSC lines were tested for oxidase function after excision. Myeloid cells generated from the Cre-excised lines continued to express oxidase function by DHR assay with MFIs from some of the lines similar to MFI of oxidase-positive cells from the myeloid differentiated healthy control iPSC line or even compared with healthy control peripheral blood cells (Figure 3). The expression of CD13, CD33, and CD45 cell surface markers was comparable between mature myeloid cells in these cultures and healthy peripheral blood neutrophils (supplemental Figure 3B), confirming the phenotype of the cells generated, and validating the neutrophil differentiation process. Patient donor origin of the corrected iPSC lines was confirmed using DNA fingerprinting analysis of microsatellite short tandem repeats among genomic DNA isolated from PBMCs, uncorrected iPSCs, and corrected iPSCs from the same subject (supplemental Figure 4).
Oxidase activity following Cre excision. Analysis of DHR oxidase activity in myeloid differentiated iPSC lines following Cre-mediated recombination excision of the puromycin cassette. Shown are analyses of corrected iPSC lines (P47-04C2,4,5-Cre GT exon 2 corrected; P47-04-C1M-Cre minigene corrected) compared with uncorrected (P47-04), healthy control iPSCs, or healthy control peripheral blood. The DHR assay, gating for MFI assessment, and donor target assessment are as described in the Figure 2D legend.
Oxidase activity following Cre excision. Analysis of DHR oxidase activity in myeloid differentiated iPSC lines following Cre-mediated recombination excision of the puromycin cassette. Shown are analyses of corrected iPSC lines (P47-04C2,4,5-Cre GT exon 2 corrected; P47-04-C1M-Cre minigene corrected) compared with uncorrected (P47-04), healthy control iPSCs, or healthy control peripheral blood. The DHR assay, gating for MFI assessment, and donor target assessment are as described in the Figure 2D legend.
Gene editing of ΔGT in a pseudogene restores oxidase function in iPSCs from a p47-CGD patient with homozygous 734 to 748 deletion in NCF1
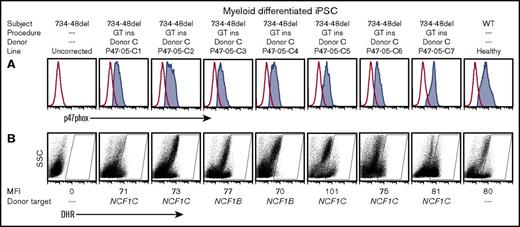
We generated an iPSC line, P47-05, from a unique p47-CGD patient (subject 5) known to have the normal GTGT at the start of exon 2 of NCF1 and a homozygous 15-bp deletion at 734 to 748 (Δ734-748) in exon 8. This patient retains the normal SNPs associated with intron 1 and intron 2 of the native NCF1 gene at both NCF1 loci. We used our ZFNs designed to target ΔGT exon 2 together with donor C rAAV2 (Figure 1C) to correct iPSC P47-05, generating 7 corrected lines following puromycin selection. Any restored p47phox protein expression or oxidase function in myeloid cells differentiated from these corrected lines must arise from correction of one or both pseudogenes, NCF1B or NCF1C, since replacement of exon 2 in NCF1 in this patient’s iPSC would not repair the exon 8 mutation. All 7 lines, when differentiated to myeloid cells, showed restored p47phox protein expression (Figure 4A) and restored oxidase function by DHR assay (Figure 4B). This strongly suggests that a strategy of repair of ΔGT in exon 2 could restore p47phox expression and oxidase function by resurrection of one of the pseudogenes. This presumption was confirmed by sequencing described below (Figure 5), with results for each clone also indicated in the row marked “Donor target” in Figure 4B.
Functional analysis of myeloid cells generated from homozygous Δ734-748 NCF1 p47-CGD iPSCs corrected by exon 2 replacement. (A) p47phox expression in myeloid cells differentiated from iPSCs derived from the homozygous Δ734-748 NCF1 p47-CGD subject, including the uncorrected line, P47-05 (first panel on the left), 7 exon 2 replacement (GT ins) clones, P47-05-C1 to C7 (middle 7 panels), and the healthy control iPSCs (last panel on the right). (B) Oxidase function in corresponding clones is shown by DHR assay. The DHR assay, gating for MFI assessment, and donor target assessment are as described in the Figure 2D legend.
Functional analysis of myeloid cells generated from homozygous Δ734-748 NCF1 p47-CGD iPSCs corrected by exon 2 replacement. (A) p47phox expression in myeloid cells differentiated from iPSCs derived from the homozygous Δ734-748 NCF1 p47-CGD subject, including the uncorrected line, P47-05 (first panel on the left), 7 exon 2 replacement (GT ins) clones, P47-05-C1 to C7 (middle 7 panels), and the healthy control iPSCs (last panel on the right). (B) Oxidase function in corresponding clones is shown by DHR assay. The DHR assay, gating for MFI assessment, and donor target assessment are as described in the Figure 2D legend.
Schematic diagram of sequencing determined SNP detection at TI sites. Forward and reverse sequencing in duplicate was performed on the ∼2-kb PCR products amplified from genomic DNA of donor C TI gene-corrected p47-CGD iPSCs. PCR products were generated from 2 primer sets, one amplifying from 5′ of the first distinguishing SNP in intron 1 to the puromycin cassette and the other from the puromycin cassette to 3′ of the last distinguishing SNP in intron 2 of NCF1 or its pseudogenes. The highlighted regions are color-coded based on the SNP’s gene identity with blue as NCF1 (1), orange as NCF1B pseudogene (1B), and yellow as NCF1C pseudogene (1C). Some SNPs represent only NCF1, NCF1B, or NCF1C, while others can represent 2 different potential combinations. The single-color rows for 16 of 18 clones indicate unambiguous concordance across all SNPs in both primer products, allowing single-locus assignment of the TI. Crosshatch rows for 2 clones represent discordance of SNPs across or between the primer products, suggestive of the presence of TI at 2 loci. The first PCR product contained 3 potential SNPs, and the second PCR product contained exon 2 and multiple SNPs.
Schematic diagram of sequencing determined SNP detection at TI sites. Forward and reverse sequencing in duplicate was performed on the ∼2-kb PCR products amplified from genomic DNA of donor C TI gene-corrected p47-CGD iPSCs. PCR products were generated from 2 primer sets, one amplifying from 5′ of the first distinguishing SNP in intron 1 to the puromycin cassette and the other from the puromycin cassette to 3′ of the last distinguishing SNP in intron 2 of NCF1 or its pseudogenes. The highlighted regions are color-coded based on the SNP’s gene identity with blue as NCF1 (1), orange as NCF1B pseudogene (1B), and yellow as NCF1C pseudogene (1C). Some SNPs represent only NCF1, NCF1B, or NCF1C, while others can represent 2 different potential combinations. The single-color rows for 16 of 18 clones indicate unambiguous concordance across all SNPs in both primer products, allowing single-locus assignment of the TI. Crosshatch rows for 2 clones represent discordance of SNPs across or between the primer products, suggestive of the presence of TI at 2 loci. The first PCR product contained 3 potential SNPs, and the second PCR product contained exon 2 and multiple SNPs.
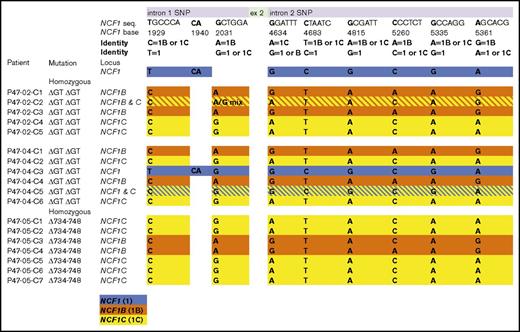
Analyses of the location of the donor C targeted insertion
Figure 1B shows the SNP distribution defining sequence flanking exon 2 as deriving from NCF1 or its pseudogenes (NCF1B or NCF1C). Using PCR-based sequence analysis, these SNPs were used to determine the context of the exon 2 ΔGT repair by donor C (Figure 5). PCR primer sets are shown in “Methods.” One set amplified from 5′ upstream of the first SNP of intron 1 to the beginning of the puromycin cassette. The second set amplified from the end of the puromycin cassette to downstream of the last SNP in intron 2 (Figure 1B). PCR products were fully sequenced forward and reverse in duplicate. In most cases, identification by presence of multiple SNPs was unambiguous. Unequivocal assignment of locus by presence of a distinguishing complete set of linked SNPs throughout the entire sequence of both primer products is indicated by single-color rows in Figure 5. The 2 clones with hatched dual-color rows have discordant SNPs identified that suggest the possibility of inserts at 2 different loci. One example of the discordance can be appreciated from sequence data shown in supplemental Figure 5 for clone P47-02-C2, indicating this clone may have TI targeted to both NCF1B and NCF1C. A second example is P47-04-C5, indicating that this subclone has a TI targeted to NCF1C, but there are also SNPs identified that are characteristic of NCF1 itself. This initially caused some confusion, because the report by Hayrapetyan et al had previously published data to show that for the great majority of p47phox-deficient CGD patients, the ΔGT at the NCF1 as the basis for their CGD likely had arisen either from complete replacement of NCF1 by one of its pseudogenes at the NCF1 locus or by a variable-length replacement of the 5′ portion of NCF1 by the equivalent region of one of its pseudogenes.16 In general, this should replace the flanking SNPs characteristic of NCF1 by those of one of its pseudogenes. There was also the sequence analysis of subclone P47-04-C3, which indicated the TI is flanked by NCF1 SNPs. Upon sequencing the primary iPSC P47-04 uncorrected clone from this patient, we found at least one NCF1 locus retained detectable NCF1-associated SNPs in the region of intron 1 and intron 2 flanking exon 2, indicating that the origin of the ΔGT at this NCF1 locus in this patient derived from an atypical crossover event. This clarified the source of NCF1 SNPs that we detected flanking the TI in some of the corrected clones from this patient (subject 4).
It is of note that for 16 of 18 lines repaired by donor C shown in Figure 5, there is full concordance of all SNPs found in those clones to allow unequivocal assignment of the TI to a sequence associated only one putative locus. It does not eliminate the possibility of homozygous correction at more than one location in these subclones. It is also of note that a majority of all TIs occurred in one of the pseudogenes, and for the functionally corrective TIs found in the homozygous Δ734-748 NCF1 clones derived from P47-05, all TIs are found in one of the pseudogenes. None of the sequenced PCR products for SNP analysis detected inverted terminal repeat sequences from the rAAV donor, indicating that seamless homologous recombination occurred instead of nonhomologous viral sequence integration.
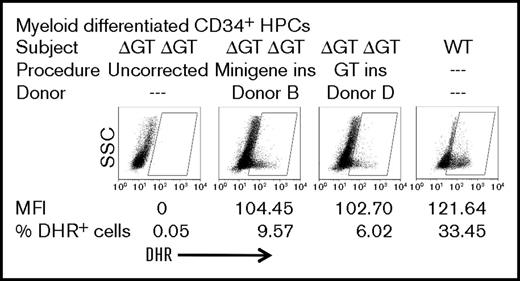
Gene-editing correction of exon 2 ΔGT in peripheral blood mobilized CD34+ HPCs from a homozygous ΔGT NCF1 p47-CGD patient
To test the feasibility of pseudogene correction in HPC peripheral blood, mobilized CD34+ HPCs were used, which were purified from apheresis product from subject 3, an adult patient with p47-CGD with homozygous ΔGT at the NCF1 locus and lacking any of the SNPs characteristic of native NCF1 intron 1 and intron 2. Thus, any level of correction at all observed in these CD34+ HPCs from this p47-CGD patient must target pseudogene sequence. We used the same ZFN pair mRNA as used for the iPSC studies noted above but then used rAAV6 donor B (minigene without puromycin selection cassette) or rAAV6 donor D (exon 2 without puromycin selection cassette) for targeted insertion. Since we did not use puromycin selection, the resulting correction as measured is a reflection of the direct-targeted insertion correction rates. Following gene targeting of patient HPCs at day 2 of culture, cells were cultured for an additional week in conditions resulting in myeloid differentiation as indicted in “Methods.” The myeloid cells generated were tested for oxidase function by DHR flow cytometry. The results with donor B insertion (minigene) or donor D insertion (exon 2) are shown in Figure 6. Since significant oxidase activity by DHR assay can only be detected from sufficiently mature myeloid cells arising in the differentiating culture, it should be noted that even using differentiating HPCs from a healthy adult donor, only ∼33% of cells in the culture fall into the oxidase-positive gate (Figure 6, right-most graph). In the myeloid differentiated culture of uncorrected p47-CGD patient HPCs, there is no significant detection of cells in the oxidase-positive gate (Figure 6, left-most graph). Donor B (minigene) or donor D (exon 2 replacement) TI correction of patient HPCs results in 9% or 6% of cells in the myeloid differentiating culture in the oxidase-positive gate, respectively (Figure 6, middle graphs). It is of note that for the 2 gene-targeted corrections, oxidase-positive myeloid cells have an MFI that is 85% of that seen in the healthy control, indicating a high level of correction per cell, of which a major portion likely derives from pseudogene correction if the proportions of locus corrections observed in the iPSCs hold true for HPCs.
Functional analysis of gene-targeting correction of p47-CGD patient HPCs. HPCs from p47-CGD subject 3 (homozygous exon 2 ΔGT NCF1) were cultured in Stemspan SFEMII supplemented with stem cell factor, Flt3 ligand, and thrombopoietin and gene targeted with ZFN mRNA and donor B or donor D rAAV6 at day 2 of culture and analyzed at day 10 of culture under conditions inducing myeloid differentiation with 50 ng/mL granulocyte colony-stimulating factor. Shown first on the left are uncorrected myeloid differentiating HPCs from the patient in whom no oxidase-positive cells were detected. Shown last on the right are myeloid differentiating HPCs from a healthy control, where 33% of cells fall into the oxidase-positive gate. Shown on the middle-left and on the middle-right panels, respectively, are myeloid differentiating minigene (donor B) or exon 2 replacement (donor D) corrected HPCs from the patient where significant numbers of oxidase-positive cells are detected with MFI approaching that of a healthy control subject.
Functional analysis of gene-targeting correction of p47-CGD patient HPCs. HPCs from p47-CGD subject 3 (homozygous exon 2 ΔGT NCF1) were cultured in Stemspan SFEMII supplemented with stem cell factor, Flt3 ligand, and thrombopoietin and gene targeted with ZFN mRNA and donor B or donor D rAAV6 at day 2 of culture and analyzed at day 10 of culture under conditions inducing myeloid differentiation with 50 ng/mL granulocyte colony-stimulating factor. Shown first on the left are uncorrected myeloid differentiating HPCs from the patient in whom no oxidase-positive cells were detected. Shown last on the right are myeloid differentiating HPCs from a healthy control, where 33% of cells fall into the oxidase-positive gate. Shown on the middle-left and on the middle-right panels, respectively, are myeloid differentiating minigene (donor B) or exon 2 replacement (donor D) corrected HPCs from the patient where significant numbers of oxidase-positive cells are detected with MFI approaching that of a healthy control subject.
Discussion
The genetics of autosomal-recessive p47-CGD is noteworthy in that >80% of patients are homozygous for a ΔGT mutation at the start of exon 2 at the NCF1 locus. Furthermore, the same ΔGT is constitutively present in the 2 pseudogenes, NCF1B and NCF1C. The predominance of this mutation in NCF1 as a cause of p47-CGD is likely due to gene conversion from the pseudogenes, where the result is that for the great majority of patients either a 5′ region of the NCF1 gene is derived from one of its pseudogenes or the pseudogene replaces the entire NCF1 gene at its NCF1 locus. We explored the feasibility of targeted gene correction at the exon 2 ΔGT using ZFNs designed to cut this site, plus donors that either replace exon 2 with a minigene or that replace only exon 2. Our studies demonstrate that the exon 2 replacement gene-targeting strategy will target pseudogenes, resulting in resurrection of functional production of p47phox protein and oxidase activity.
We have previously shown that we can reprogram iPSCs from the low number of CD34+ HPCs present in small volumes of peripheral blood or a repository sample of cryopreserved blood mononuclear cells from nonmobilized patients.10 This allowed us to conduct our gene-targeting studies on an iPSC line generated from a repository sample of cryopreserved blood mononuclear cells from a p47-CGD patient previously shown to have normal GTGT at the start of exon 2 in NCF1 but homozygous for Δ734-748 mutation in exon 8. Use of this line from this patient allowed unequivocal demonstration that ΔGT correction of the putative exon 2 of pseudogenes NCF1B or NCF1C is sufficient to achieve production of p47phox and correction of oxidase activity in myeloid cells differentiated from the corrected iPSC clones. While natural reactivation of human pseudogenes can occur and has been demonstrated in human embryonic ζ-globin locus gene conversion of the ψ ζ gene17 and human IRGM gene,18 a gene editing approach to restore pseudogene function has not been previously demonstrated.
Our molecular analysis to delineate correction at the NCF1, NCF1B, or NCF1C loci using SNPs confirmed that correction at NCF1 or its pseudogenes, NCF1B and NCF1C, could restore levels of oxidase function comparable to those of healthy mature myeloid cells as measured by DHR flow cytometry assay. We noticed differences in average per-cell p47phox protein production and per-cell oxidase activity (MFI) in myeloid cells differentiating from the gene-targeting–corrected p47-CGD iPSCs. The differences were not consistent enough, nor were enough individual iPSC clones analyzed to be conclusive as to whether pseudogene correction could result in equivalent restoration of function as correction of mutated NCF1 or as seen from healthy donor iPSC
Our work with iPSCs allowed cloning and detailed analysis of the outcome of our gene-targeting strategy, but given the current impasse in generating long-term repopulating HPCs from human iPSCs, it was important for us to also show that we could use our gene-targeting strategy to correct primary HPCs from a patient. Furthermore, correction of autologous HPCs holds the future potential for clinical therapeutic application, if, with additional work, we can optimize and significantly improve the level of correction we observed in p47-CGD CD34+ HPCs. Nonetheless, we were able to demonstrate a level of correction of oxidase activity using either exon 2 replacement or minigene strategies for correction of p47-CGD in peripheral blood mobilized CD34+ HPCs to provide proof of principle that the same gene targeting of ΔGT that worked in iPSCs also works to correct primary HPCs. This is inline with recent reports that have described efficient CD34+ HPC targeted corrections or insertions.19-22
We demonstrated our gene-targeting correction in iPSCs from 3 subjects and in HPCs from a fourth subject with p47-CGD. Three of these patients had the common homozygous ΔGT mutation at the NCF1 locus while one patient (subject 5; the source of iPSC of the P47-05 correction series) had normal GTGT at the start of exon 2 of NCF1 and instead had a homozygous Δ734-748 mutation in exon 8 of NCF1. This allows us to conclude that our gene-targeting strategy appears to successfully restore function in cells from several patients, including one with an uncommon mutation, and that correction may occur via targeting of the NCF1 locus or in the case of the exon 8 deletion exclusively via resurrection of either of its pseudogenes at their loci. Of note is that correction of P47-05 must have occurred with TI correction of pseudogenes NCF1B or NCF1C in their normal locus, with mRNA transcription driven by the pseudogene promoter. p47phox was only detected following myeloid differentiation of the corrected P47-05 series subclones, which demonstrates that the pseudogene promoters are regulated similar to that of the NCF1 gene.
Since mutated NCF1 at its locus and its 2 pseudogenes are all appropriate targets for gene correction using our strategy, each cell has 6 potential targets for TI correction of p47-CGD. It is possible to speculate that the increased number of targets per cell might increase efficiency of targeted correction. However, we did not determine whether this is the case from our studies, and it may be that gene-targeting efficiency is more highly correlated with other cell physiology factors that make a particular cell more susceptible to targeted insertion than simply the number of genomic targets in the cell.
In summary, we demonstrate for the first time that apparently nonfunctional pseudogenes can be resurrected to functionality by gene editing, providing an approach to achieve functional correction of p47-CGD regardless of the mutation in NCF1. The broader ramification is that other human pseudogenes, such as ζ-globin, IRGM, human T-cell receptor γ variable pseudogene V10,23 may be potential targets for gene editing with potential both for defining novel genetic and evolutionary aspects of such pseudogenes and for clinical application.
The full-text version of this article contains a data supplement.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health under intramural project numbers Z01-Al-00644 and Z01-Al-00988. This project has also been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Authorship
Contribution: R.K.M. and S.S.D.R. designed experiments, performed experiments and wrote the paper: D.B.K., C.L.S., and E.M.K. contributed reagents and wrote the paper; X.W., S.B., J.C., J.L., and S.K. performed experiments: G.D.P., S.A.A., J.A.C., and U.C. contributed reagents and critical methodology; and H.L.M. designed experiments and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Harry L. Malech, LHD/NIAID/NIH, Building 10, Room 5-3750, 10 Center Dr, MSC-1456, Bethesda, MD 20892-1456; e-mail: hmalech@nih.gov.