Key Points
ABO blood group A transferases possess intrinsic FS activity upon deletion of exon 3 or 4 of A transferase messenger RNAs.
Cointroduction of exon 3 or 4 deletion and GlyGlyAla substitution synergistically confers human A transferases with strong FS activity.
Abstract
Evolutionarily related ABO and GBGT1 genes encode, respectively, A and B glycosyltransferases (AT and BT) and Forssman glycolipid synthase (FS), which catalyze the biosynthesis of A and B, and Forssman (FORS1) oligosaccharide antigens responsible for the ABO and FORS blood group systems. Humans are a Forssman antigen–negative species; however, rare individuals with Apae phenotype express FORS1 on their red blood cells. We previously demonstrated that the replacement of the LeuGlyGly tripeptide sequence at codons 266 to 268 of human AT with GBGT1-encoded FS-specific GlyGlyAla enabled the enzyme to produce FORS1 antigen, although the FS activity was weak. We searched for additional molecular mechanisms that might allow human AT to express FORS1. A variety of derivative expression constructs of human AT were prepared. DNA was transfected into COS1 (B3GALNT1) cells, and cell-surface expression of FORS1 was immunologically monitored. To our surprise, the deletion of exon 3 or 4, but not of exon 2 or 5, of human AT transcripts bestowed moderate FS activity, indicating that the A allele is inherently capable of producing a protein with FS activity. Because RNA splicing is frequently altered in cancer, this mechanism may explain, at least partially, the appearance of FORS1 in human cancer. Furthermore, strong FS activity was attained, in addition to AT and BT activities, by cointroducing 1 of those deletions and the GlyGlyAla substitution, possibly by the synergistic effects of altered intra-Golgi localization/conformation by the former and modified enzyme specificity by the latter.
Introduction
Blood group A and B glycosyltransferases (ATs and BTs), Forssman glycolipid synthases (FSs), isogloboside 3 synthases, and α1,3-galactosyltransferases of the α1,3-Gal (NAc) transferase family are encoded by evolutionarily related ABO (A and B alleles), GBGT1, A3GALT2, and GGTA1 genes, respectively.1 Blood group ATs and BTs transfer an N-acetyl-d-galactosamine (GalNAc) and a d-galactose (Gal) to the same acceptor substrate H substance to synthesize A and B oligosaccharide glycan antigens in that order. These antigens are expressed on red blood cells (RBCs) and also on some epithelial and endothelial cells, depending on an individual’s ABO genotype/phenotype. Because individuals who do not express A and/or B antigens contain anti-A and/or anti-B antibodies in their sera (Landsteiner’s law), the ABO matching is crucial not only for blood transfusion but also for cell/tissue/organ transplantations to avoid potentially fatal immune reactions. Immunodominant structures of A and B antigens and H substance are GalNAcα1-3(Fucα1-2)Gal− and Galα1-3(Fucα1-2)Gal− and Fucα1-2Gal−, respectively.2,3 The GBGT1 gene–encoded FSs catalyze the last step of pentasaccharide Forssman glycolipid (Gb5) biosynthesis from the precursor globoside (Gb4) by transferring a GalNAc.4 Immunodominant structure of Forssman antigen (FORS1) is GalNAcα1-3GalNAcβ1−, which is carried on the Forssman glycolipid.
Homo sapiens is a Forssman antigen–negative species, and most individuals do not express this antigen. However, FORS1 was identified on RBCs from rare individuals exhibiting what is called the Apae phenotype, and the International Society of Blood Transfusion designated the Forssman (FORS) system as the 31st blood group system of RBCs.5,6 In addition to occurring in Apae individuals, FORS1 expression has also been reported in cancer cells/tissues from non-Apae individuals.7-13 However, the molecular mechanisms that induce heterophilic FORS1 expression remained to be determined.
We have been studying various aspects of the ABO genes, ATs and BTs, and A and B oligosaccharide antigens since 1990, when we cloned human A, B, and O allelic complementary DNAs and correlated their nucleotide sequences with blood group A and B antigen expression.14,15 Human AT and BT differ by only 4 of 354 amino acids at codons 176, 235, 266, and 268. They are Arg, Gly, Leu, and Gly in AT and Gly, Ser, Met, and Ala in BT. By analyzing the 14 A-B transferase chimeras that were different at those 4 positions, having either amino acid of AT or BT, we were able to demonstrate that the amino acids at codons 266 and 268 are crucial.16 Later studies concluded that the sizes and charges of side chains of those amino acids play a decisive role in determining both the GalNAc/galactose sugar specificity and transferase activity.17-23
We expanded enzymatic structural analyses to include the FSs encoded by the GBGT1 genes. Human GBGT1 gene is nonfunctional,24 and we demonstrated that 2 amino acid substitutions (Gly230Ser and Gln296Arg) are responsible for the loss of FS activity of the human protein.25 We also showed that human FS protein gained weak FS activity after the reversion of 1 of the 2 substitutions, whereas the reversion of both restored strong FS activity. Recently, we revealed the crosstalk between ABO gene–encoded AT/BT and GBGT1 gene–encoded FS.26 During species evolution, the GlyGlyAla tripeptide sequence has been conserved in a majority of FSs at codons corresponding to codons 266 to 268 of human AT/BT.23 We realized that mouse cis-AB transferase possesses the same GlyGlyAla tripeptide sequence, although it is encoded by the ABO gene.27 We therefore examined whether this murine enzyme might exhibit FS activity. The answer was yes. The mouse cis-AB transferase with GlyGlyAla synthesized FORS1, whereas human cis-AB transferase with LeuGlyAla did not. Additionally, we also detected weak FS activity of the human AT, LeuGlyGly of which was artificially replaced with GlyGlyAla. Those results showed that GlyGlyAla tripeptide is important for FS activity. However, the results also showed that the substitution is not sufficient to confer full FS activity to human AT. Therefore, we initiated a search for additional molecular mechanisms that allow the blood group ABO gene–encoded glycosyltransferases to synthesize FORS1. Here, we present the identification of 1 mechanism that may potentially explain the heterophilic FORS1 expression in human cancer.
Methods
Data extraction of SNPs, disease/trait associations, and splicing alterations in cancer of the human ABO and GBGT1 genes
The single-nucleotide polymorphism (SNP) data of the coding sequences (CDSs) of the human ABO and GBGT1 gene transcripts were retrieved from Ensembl Genome Assembly: GRCh38.p10 (GCA_000001405.25). The nucleotide and deduced amino acid sequences of the ABO gene transcript were modified to encode for an A allele (A101) rather than the original O allele. The splicing pattern of the ABO gene transcript was also corrected, following the GenBank entry AF134412. The National Human Genome Research Institute–European Bioinformatics Institute Genome-Wide Association Studies (GWAS) Catalog and The Cancer Genome Atlas (TCGA) SpliceSeq Database were searched for diseases and traits associated with SNPs and alternative splicing variants in cancer at the ABO and GBGT1 genetic loci.
In vitro mutagenesis to prepare amino acid substitution/exon deletion/exon replacement constructs
The original human AT, mouse cis-AB transferase, and mouse FS expression constructs H_ABO-A, M_ABO-AB, and M_GBGT1, respectively, were previously prepared in the pSG5 eukaryotic expression plasmid vector.23,26 Human AT derivative with GlyGlyAla substitution, H_ABO-A (GlyGlyAla), was also described. In vitro mutagenesis was performed to obtain the remaining 13 constructs. We employed the 2-round primer-mediated polymerase chain reaction strategy using specific primers with nucleotide substitutions to introduce amino acid substitutions as described.23 We used the same strategy to delete a specific exon using primers bridging the 2 exons and deleting the exon in between. In case of exon replacement constructs, primers bridging mouse and human exons were designed. Using DNA from the constructs created as a template, additional constructs were prepared by introducing another substitution or deletion. The names and nucleotide sequences of primers, as well as detailed polymerase chain reaction protocols and preparations of expression constructs, can be found in the supplemental Data.
DNA transfection of COS1 (B3GALNT1) and HeLa (FUT2) cells and immunological detection of Forssman and blood group A and B antigens
COS1 (B3GALNT1) cells expressing globoside, the acceptor substrate for FS, and HeLa (FUT2) cells expressing H substance, the acceptor substrate for AT and BT, were used as recipients of DNA transfection as previously described.23,26 For immunocytochemistry experiments, DNA from the expression constructs and DNA from the pEGFP plasmid vector expressing enhanced green fluorescent protein were cotransfected to the COS1 (B3GALNT1) cells, using Lipofectamine 3000 reagents (Life Technologies). For immunocytometry experiments, COS1 (B3GALNT1) cells were cotransfected with DNA from pLL3.7-mRFP plasmid vector encoding monomer red fluorescent protein using Lipofectamine 2000, whereas HeLa (FUT2) cells were cotransfected with pEGFP DNA. To detect FORS1 expression, DNA-transfected COS1 (B3GALNT1) cells were immunostained with clone FOM-1 rat immunoglobulin M monoclonal antibody (BMA Biomedicals). To detect blood group A and B antigen expression, DNA-transfected HeLa (FUT2) cells were immunostained using a mixture of murine anti-A monoclonal antibodies or mixture of murine anti-B monoclonal antibodies (OrthoDiagnostic Systems). For detailed protocols for DNA transfection, immunocytochemistry, immunocytometry, and fluorescence-activated cell sorter (FACS) analyses, see the supplemental Data.
Results
Numerous SNPs are identified in the human ABO and GBGT1 genes
The cDNA sequences and SNPs of the ABO and GBGT1 genes extracted are shown on ABO-203 (ENST00000611156.4) and GBGT1-203 (ENST00000372040.7) transcripts, respectively, in Figure 1A-B. Many SNPs were identified in each gene (372 SNPs of 1,065 nucleotides, including a termination codon in the CDS of the ABO gene transcript, and 331 SNPs of 1,044 nucleotides in the GBGT1 gene CDS), although data obtained using next-generation sequencing may have some errors. Those SNPs include 9 nonsense and 28 frameshift mutations in the ABO gene and 8 nonsense and 20 frameshift mutations in the GBGT1 gene. The search for disease/trait associations in the GWAS Catalog with the SNPs in the ABO gene identified several diseases, including pancreatic cancer,28 venous thromboembolism,29 and severe brain malaria,30 as well as a few dozen biological parameters,31-33 as previously reviewed.1,34 However, no associations have so far been identified for the SNPs in the GBGT1 genetic locus. Several alternatively spliced GBGT1 gene transcripts in cancer were found in the TCGA SpliceSeq Database; however, no entry was listed for the ABO gene.
SNPs found in the CDSs of the human ABO and GBGT1 gene transcripts. Together with SNPs, cDNA sequences of ABO (ABO-203 ENST0000061156.4) and GBGT1 (GBGT1-203 ENST00000372040.7) genes were retrieved and modified; they are shown in panels A and B, respectively. The SNPs have been color coded by variation, as indicated at the bottom of the figure. The nucleotide and deduced amino acid sequences in the CDSs were numbered; they are shown at the right of each column.
SNPs found in the CDSs of the human ABO and GBGT1 gene transcripts. Together with SNPs, cDNA sequences of ABO (ABO-203 ENST0000061156.4) and GBGT1 (GBGT1-203 ENST00000372040.7) genes were retrieved and modified; they are shown in panels A and B, respectively. The SNPs have been color coded by variation, as indicated at the bottom of the figure. The nucleotide and deduced amino acid sequences in the CDSs were numbered; they are shown at the right of each column.
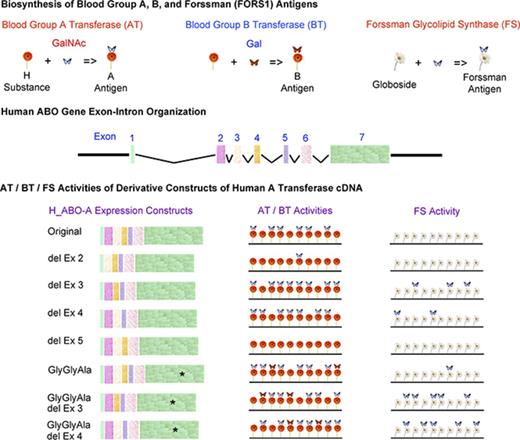
Deletion of exon 3 or 4, but not exon 2 or 5, of human AT mRNA transcripts confers moderate FS activity
The human ABO gene spans >18 kilobase pairs.35,36 The nucleotide sequence encoding the 354–amino acid AT protein is spread over 7 exons: 28 nucleotides in exon 1, 70 in exon 2, 57 in exon 3, 48 in exon 4, 36 in exon 5, 135 in exon 6, and 691 (including the termination codon) in exon 7. Although the functional significance is unclear, human AT/BT messenger RNAs (mRNAs) may take several alternatively spliced patterns, missing 1 or a few exons.37 Accordingly, we questioned whether exon deletions of human AT mRNAs might result in the appearance of FORS1 antigen. For this purpose, we prepared human AT constructs deleting exon 2, 3, 4, or 5. Because the coding sequences in exons 3, 4, and 5 are multiples of 3 nucleotides, the coding frame of translation remains the same. However, exon 2 has 70 nucleotides, which is not a multiple of 3 nucleotides. Accordingly, we designed primers to restore the codon frame after the exon deletion. However, we realized that several clones retained the frameshift. Therefore, we analyzed both constructs with and without the frameshift.
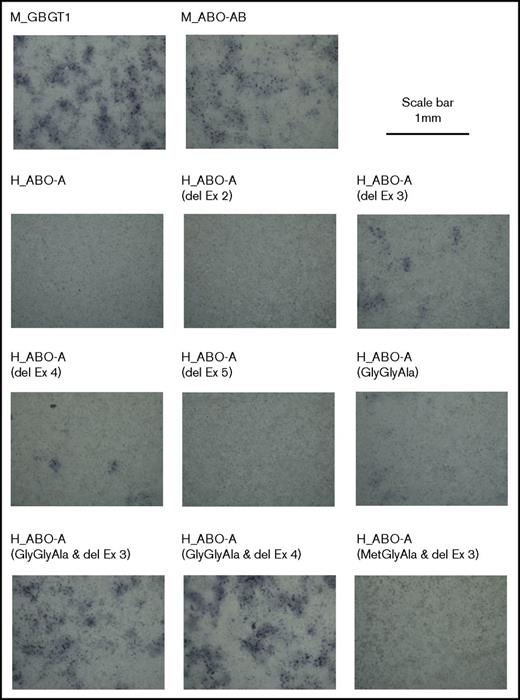
Results of experiments in triplicate are shown in Panel 1 of Table 1. Some FORS1 antigen+ cells were observed in the COS1 (B3GALNT1) cells transfected with human AT constructs with the deletion of exon 3 or 4. However, the constructs deleting exon 2 or 5 did not produce more positive cells than the background. Representative images of the cells transfected and immunostained with anti-FORS1 antibody are shown in Figure 2.
Forssman antigen expression after DNA transfection of a variety of expression constructs into COS1 (B3GALNT1) cells analyzed by immunoperoxidase staining procedure
| Gene name . | Tripeptide sequence . | Exon deletion . | Forssman antigen+ cells, % (adjusted) . | Deduced FS activity . | ||
|---|---|---|---|---|---|---|
| Exp. 1 . | Exp. 2 . | Exp. 3 . | ||||
| 1 | ||||||
| M_GBGT1 | GlyGlyAla | — | 100 | 100 | 100 | +++++ |
| M_ABO-AB | GlyGlyAla | — | 68 | 29 | 66 | ++++ |
| H_ABO-A | LeuGlyGly | — | 0 | 0 | 0 | — |
| Exon 2 | 0 | 0 | 0 | — | ||
| Exon 2 (frameshift) | 0 | 0 | 0 | — | ||
| Exon 3 | 8 | 1 | 11 | +++ | ||
| Exon 4 | 3 | 3 | 17 | +++ | ||
| Exon 5 | 0 | 0 | 0 | — | ||
| 2 | ||||||
| M_GBGT1 | GlyGlyAla | — | 100 | 100 | 100 | +++++ |
| M_ABO-AB | GlyGlyAla | — | 68 | 29 | 66 | ++++ |
| H_ABO-A | LeuGlyGly | — | 0 | 0 | 0 | — |
| GlyGlyAla | 2 | 0 | 3 | + | ||
| Exon 3 | 68 | 123 | 95 | +++++ | ||
| Exon 4 | 60 | 110 | 140 | +++++ | ||
| MetGlyAla | 0 | 0 | 0 | — | ||
| Exon 3 | 0 | 0 | 0 | — | ||
| Exon 4 | 0 | 0 | 0 | — | ||
| 3 | ||||||
| M_GBGT1 | GlyGlyAla | — | 100 | 100 | 100 | +++++ |
| M_ABO-AB | GlyGlyAla | — | 68 | 29 | 66 | ++++ |
| H_ABO-A | LeuGlyGly | — | 0 | 0 | 0 | — |
| H_ABO-A(Ex 1,2)-M_ABO-AB(Ex 3)-H_ABO-A(Ex 5,6,7) | 1 | 1 | 12 | ++ | ||
| M_ABO-AB(Ex 1,2)-H_ABO-A(Ex 3,4)-M_ABO-AB(Ex 4,5,6) | 6 | 0 | 43 | +++ | ||
| Gene name . | Tripeptide sequence . | Exon deletion . | Forssman antigen+ cells, % (adjusted) . | Deduced FS activity . | ||
|---|---|---|---|---|---|---|
| Exp. 1 . | Exp. 2 . | Exp. 3 . | ||||
| 1 | ||||||
| M_GBGT1 | GlyGlyAla | — | 100 | 100 | 100 | +++++ |
| M_ABO-AB | GlyGlyAla | — | 68 | 29 | 66 | ++++ |
| H_ABO-A | LeuGlyGly | — | 0 | 0 | 0 | — |
| Exon 2 | 0 | 0 | 0 | — | ||
| Exon 2 (frameshift) | 0 | 0 | 0 | — | ||
| Exon 3 | 8 | 1 | 11 | +++ | ||
| Exon 4 | 3 | 3 | 17 | +++ | ||
| Exon 5 | 0 | 0 | 0 | — | ||
| 2 | ||||||
| M_GBGT1 | GlyGlyAla | — | 100 | 100 | 100 | +++++ |
| M_ABO-AB | GlyGlyAla | — | 68 | 29 | 66 | ++++ |
| H_ABO-A | LeuGlyGly | — | 0 | 0 | 0 | — |
| GlyGlyAla | 2 | 0 | 3 | + | ||
| Exon 3 | 68 | 123 | 95 | +++++ | ||
| Exon 4 | 60 | 110 | 140 | +++++ | ||
| MetGlyAla | 0 | 0 | 0 | — | ||
| Exon 3 | 0 | 0 | 0 | — | ||
| Exon 4 | 0 | 0 | 0 | — | ||
| 3 | ||||||
| M_GBGT1 | GlyGlyAla | — | 100 | 100 | 100 | +++++ |
| M_ABO-AB | GlyGlyAla | — | 68 | 29 | 66 | ++++ |
| H_ABO-A | LeuGlyGly | — | 0 | 0 | 0 | — |
| H_ABO-A(Ex 1,2)-M_ABO-AB(Ex 3)-H_ABO-A(Ex 5,6,7) | 1 | 1 | 12 | ++ | ||
| M_ABO-AB(Ex 1,2)-H_ABO-A(Ex 3,4)-M_ABO-AB(Ex 4,5,6) | 6 | 0 | 43 | +++ | ||
Three sets of experiments (Exp.) were performed in triplicates, and the results are shown in Panels 1 to 3. The original gene names are listed in the leftmost column. M_GBGT1, M_ABO-AB, and H_ABO-A are mouse GBGT1 gene, mouse ABO gene, and the A allele of the human ABO gene, respectively. The tripeptide sequences at the codons corresponding to codons 266 to 268 of human AT/BT are shown in the second column from left. The exon deletions are indicated in the third column. H_ABO-A(Ex 1,2)-M_ABO-AB(Ex 3)-H_ABO-A(Ex 5,6,7) and M_ABO-AB(Ex 1,2)-H_ABO-A(Ex 3,4)-M_ABO-AB(Ex 4,5,6) are human-mouse chimera constructs. The former encodes human AT with exons 3 and 4 replaced by mouse exon 3, and the latter encodes mouse cis-AB transferase with exon 3 replaced by human exons 3 and 4. The results of immunostaining with anti-FORS1 antibody in the triplicate experiments are shown in the next 3 columns. The numbers of cells positive with green fluorescent protein, the expression vector of which was cotransfected, were used to normalize Forssman antigen positivity. The values are shown in percentage of the expression observed of the original mouse FS, M_GBGT1, construct to be 100. The deduced FS activity is also shown semiquantitatively (—, none; +, very weak; ++, weak; +++, moderate; ++++, strong; +++++, very strong activity).
Representative images of cells that were DNA transfected and immunostained. After fixation, cells were incubated with the anti-FORS1 monoclonal antibody and then with a biotinylated secondary antibody. After washing, they were treated with Vectastain ABC reagents. 3,3′-Diaminobenzidine was then used for peroxidase-mediated color development. Photos were taken of cells transfected with different expression constructs, as indicated at the top of the photos. The scale bar of 1 mm is indicated at the top right. del, deletion; Ex, exon.
Representative images of cells that were DNA transfected and immunostained. After fixation, cells were incubated with the anti-FORS1 monoclonal antibody and then with a biotinylated secondary antibody. After washing, they were treated with Vectastain ABC reagents. 3,3′-Diaminobenzidine was then used for peroxidase-mediated color development. Photos were taken of cells transfected with different expression constructs, as indicated at the top of the photos. The scale bar of 1 mm is indicated at the top right. del, deletion; Ex, exon.
Cointroduction of deletion of exon 3 or 4 with the GlyGlyAla tripeptide substitution enables human AT to express strong FS activity
The amino acid residues in exons 3 and 4 are located in the transmembrane (TM)/stem region of the enzyme, whereas codons 266 to 268 are located in the catalytic domain of the enzyme. We therefore thought that the molecular mechanisms conferring FS activity might be similar between the deletion of exon 3 and the deletion of exon 4 and that they might be different from the mechanism in which the GlyGlyAla substitution might function. Accordingly, we investigated the effects of the combined double mutations on FS activity. We prepared the human AT constructs with the FS-specific GlyGlyAla tripeptide sequence in combination with the deletion of exon 3 or 4. We also prepared similar human AT constructs with the BT-specific MetGlyAla sequence in combination with the deletion of exon 3 or 4.
Results are shown in Panel 2 of Table 1. The double combinations with the GlyGlyAla sequence and exon 3 or 4 deletion examined exhibited an increase in FS activity, suggesting that those 2 alterations, 1 in the TM-to-stem region and another (GlyGlyAla) at the substrate recognition/binding site in the catalytic domain, complemented each other and synergistically conferred strong FS activity. Contrastingly, none of the constructs containing MetGlyAla showed any FS activity. Representative images of the cells after immunostaining are also shown in Figure 2.
Different exon-intron organizations and tripeptide sequences account for differential FS activity between the mouse and human ABO genes
As opposed to the human ABO gene, the coding region of which is carried on 7 exons, the mouse ABO gene CDS is spread over 6 exons. Human AT and BT have 354 amino acids, whereas mouse cis-AB transferase is made up of 332 amino acids. The distribution of the nucleotide sequence encoding the mouse cis-AB transferase (999 nucleotides, including the termination codon) is as follows: 13 nucleotides in exon 1, 70 in exon 2, 57 in exon 3, 36 in exon 4, 135 in exon 5, and 688 (including the termination codon) in exon 6. Except for 1 amino acid that is deleted at the C-terminus of the mouse enzyme, the other differences are present in the N-terminal region. One difference is that exon 1 has different numbers of coding nucleotides, 28 and 13, in the human and mouse genes, respectively. Another and more significant difference is that the exon corresponding to human exon 4 consisting of 48 nucleotides is missing in the mouse ABO gene. This is reflected in the experimental results. Mouse cis-AB transferase with GlyGlyAla tripeptide (inherently missing the human exon 4 equivalent) expressed strong AT/BT and FS activities, whereas human AT with both GlyGlyAla and exon 4 deletion exhibited strong AT/BT and FS activities. Therefore, we prepared chimera constructs between mouse cis-AB transferase and human AT, replacing exon 3 of mouse FS with exons 3 and 4 of human AT and also replacing exons 3 and 4 of human AT with exon 3 of mouse cis-AB transferase.
Results of immunostaining of FORS1 antigen after DNA transfection are shown in Panel 3 of Table 1. The substitution of exon 3 of M_ABO-AB with exons 3 and 4 of H_ABO-A reduced the FS activity of mouse cis-AB transferase, whereas the substitution of exons 3 and 4 of H_ABO-A with exon 3 of M_ABO-AB conferred human AT with weak FS activity, supporting our hypothesis.
FACS immunocytometry quantification reconfirms strong FS activity of the human AT constructs with GlyGlyAla substitution and deletion of exon 3 or 4
Immunocytochemistry provided semiquantitative results because of the cell detachment during immunostaining. The difficulty in counting positive cells that were weakly stained under microscope observation may lead to underestimation, whereas too much signal amplification may lead to overestimation. Therefore, we repeated the DNA transfection experiments with key constructs to investigate FS activity in a more quantitative manner by FACS immunocytometry. We selected the human AT derivative constructs containing deletion of exon 2, 3, 4, or 5, GlyGlyAla substitution alone or in combination with deletion of exon 3 or 4, or MetGlyAla substitution alone or in combination with deletion of exon 3 or 4. These constructs were examined, together with M_GBGT1 and M_ABO-AB as positive controls and H_ABO-A as a negative control.
Results were compared among the constructs and are shown in Table 2. Using FACS immunocytometry, similar FORS1 expression results were obtained, confirming the immunocytochemistry results. High FS activity was again observed in the human AT constructs with double mutations of deletion of exon 3 or 4 and GlyGlyAla substitution, whereas the single-mutant constructs with either exon 3 or 4 deletion or GlyGlyAla substitution exhibited moderate or weak FS activity. We repeated the FACS experiments with several selected constructs, and the cytometry results are shown in Figure 3. Although there were differences in number, the results were similar and clearly showed the appearance of FS activity after the introduction of GlyGlyAla substitution or deletion of exon 4 into human AT expression construct. The introduction of both enhanced FS activity, although not to the level of mouse FS.
Expression of blood group antigens (FORS1, A, and B) after DNA transfection of the selected expression constructs into COS1 (B3GALNT1) and HeLa (FUT2) cells analyzed by FACS cytometry
| Gene name . | Tripeptide sequence . | Exon deletion . | Cells/antigen . | |||||
|---|---|---|---|---|---|---|---|---|
| COS1 (B3GALNT1)/FORS1 antigen . | HeLa (FUT2)/A antigen . | HeLa (FUT2)/B antigen . | ||||||
| Positive cells, % . | MFI value . | Positive cells, % . | MFI value . | Positive cells, % . | MFI value . | |||
| M_GBGT1 | GlyGlyAla | — | 9.3 | 2462 | 0.1 | — | 0.1 | — |
| H_ABO-A | LeuGlyGly | — | 0.4 | — | 4.0 | 6 361 | 0.2 | — |
| Exon 2 | 0.2 | — | 2.3 | 435 | 0.1 | — | ||
| Exon 3 | 5.2 | 464 | 4.9 | 10 068 | 0.0 | — | ||
| Exon 4 | 2.2 | 232 | 5.0 | 9 264 | 0.0 | — | ||
| Exon 5 | 0.2 | — | 0.4 | — | 0.1 | — | ||
| GlyGlyAla | — | 0.6 | 79 | 5.2 | 6 811 | 9.9 | 1307 | |
| Exon 3 | 4.3 | 1315 | 5.2 | 5 863 | 14.4 | 1542 | ||
| Exon 4 | 6.0 | 1256 | 5.7 | 7 383 | 14.2 | 1601 | ||
| MetGlyAla | — | 0.3 | — | 0.5 | — | 33.9 | 1493 | |
| Exon 3 | 0.1 | — | 0.5 | — | 40.4 | 1387 | ||
| Exon 4 | 0.2 | — | 0.4 | — | 38.2 | 1299 | ||
| M_ABO-AB | GlyGlyAla | — | 4.0 | 1158 | 4.0 | 14 349 | 16.5 | 1696 |
| Gene name . | Tripeptide sequence . | Exon deletion . | Cells/antigen . | |||||
|---|---|---|---|---|---|---|---|---|
| COS1 (B3GALNT1)/FORS1 antigen . | HeLa (FUT2)/A antigen . | HeLa (FUT2)/B antigen . | ||||||
| Positive cells, % . | MFI value . | Positive cells, % . | MFI value . | Positive cells, % . | MFI value . | |||
| M_GBGT1 | GlyGlyAla | — | 9.3 | 2462 | 0.1 | — | 0.1 | — |
| H_ABO-A | LeuGlyGly | — | 0.4 | — | 4.0 | 6 361 | 0.2 | — |
| Exon 2 | 0.2 | — | 2.3 | 435 | 0.1 | — | ||
| Exon 3 | 5.2 | 464 | 4.9 | 10 068 | 0.0 | — | ||
| Exon 4 | 2.2 | 232 | 5.0 | 9 264 | 0.0 | — | ||
| Exon 5 | 0.2 | — | 0.4 | — | 0.1 | — | ||
| GlyGlyAla | — | 0.6 | 79 | 5.2 | 6 811 | 9.9 | 1307 | |
| Exon 3 | 4.3 | 1315 | 5.2 | 5 863 | 14.4 | 1542 | ||
| Exon 4 | 6.0 | 1256 | 5.7 | 7 383 | 14.2 | 1601 | ||
| MetGlyAla | — | 0.3 | — | 0.5 | — | 33.9 | 1493 | |
| Exon 3 | 0.1 | — | 0.5 | — | 40.4 | 1387 | ||
| Exon 4 | 0.2 | — | 0.4 | — | 38.2 | 1299 | ||
| M_ABO-AB | GlyGlyAla | — | 4.0 | 1158 | 4.0 | 14 349 | 16.5 | 1696 |
The original construct names, tripeptide sequences, and exon deletions are shown in the 3 leftmost columns. The A antigen+, B antigen+, and Forssman antigen+ cell percentages and the median fluorescence intensity (MFI) values of the antigen+ cell populations are shown in the 6 rightmost columns. The MFI values are shown for samples where the positive cell percentages surpassed the background level of 0.5%.
Cytometric analysis of COS1 (B3GALNT1) cells transfected with M_GBGT1, H_ABO-A, and 3 representative derivative constructs. Cells were cotransfected with DNA from the selected expression construct and DNA from the vector expressing monomer red fluorescent protein (mRFP). Cells were then detached and stained with anti-FORS1 antibody and then with Alexa Fluor 488–linked secondary antibody. The 2 graphs in the top panel correspond to M_GBGT1 transfection. On the left, the selection of cell population by forward- (FSC) and side-scatter (SSC) amplitudes is depicted. Events are dot plotted in pseudocolor, with red representing higher frequency and dark blue the lowest. This selection was maintained for all the other samples. The right graph shows the fluorescence intensity from mRFP, used as transfection control, on the y-axis and the fluorescence corresponding to FORS1 on the x-axis. Both axes are on the logarithmic scale. The upper red rectangle represents the mRFP+ cell population, and the percentage is indicated at the upper left corner. The green rectangle corresponds to Alexa Fluor 488+ cell population, and the percentage is indicated at the lower right corner. The other panels show the results for other constructs, H_ABO-A, H_ABO-A (GlyGlyAla), H_ABO-A (deleted [del] exon 4), and H_ABO-A (GlyGlyAla and del exon 4), using the same gates.
Cytometric analysis of COS1 (B3GALNT1) cells transfected with M_GBGT1, H_ABO-A, and 3 representative derivative constructs. Cells were cotransfected with DNA from the selected expression construct and DNA from the vector expressing monomer red fluorescent protein (mRFP). Cells were then detached and stained with anti-FORS1 antibody and then with Alexa Fluor 488–linked secondary antibody. The 2 graphs in the top panel correspond to M_GBGT1 transfection. On the left, the selection of cell population by forward- (FSC) and side-scatter (SSC) amplitudes is depicted. Events are dot plotted in pseudocolor, with red representing higher frequency and dark blue the lowest. This selection was maintained for all the other samples. The right graph shows the fluorescence intensity from mRFP, used as transfection control, on the y-axis and the fluorescence corresponding to FORS1 on the x-axis. Both axes are on the logarithmic scale. The upper red rectangle represents the mRFP+ cell population, and the percentage is indicated at the upper left corner. The green rectangle corresponds to Alexa Fluor 488+ cell population, and the percentage is indicated at the lower right corner. The other panels show the results for other constructs, H_ABO-A, H_ABO-A (GlyGlyAla), H_ABO-A (deleted [del] exon 4), and H_ABO-A (GlyGlyAla and del exon 4), using the same gates.
A and B antigen expression in the DNA transfected HeLa (FUT2) cells was also examined, and the results are also shown in Table 2. A antigen expression was observed in HeLa (FUT2) cells transfected with the M_ABO-AB or H_ABO-A construct with LeuGlyGly or GlyGlyAla sequence irrespective of the presence or absence of exon deletion, excluding the deletion of exon 5, which abolished AT activity. In contrast, B antigen expression was observed in HeLa (FUT2) cells transfected with the M_ABO-AB or H_ABO-A construct with GlyGlyAla or MetGlyAla sequence irrespective of the presence or absence of exon deletions examined.
Discussion
The presence or absence of α1,3-Gal(NAc) transferase member genes, as well as their functionality or nonfunctionality, is species dependent.23 A birth and death model was proposed to explain the evolution of those genes.38 A recent hypothesis emphasized the role that rearrangements of chromosomal fragments with the ABO and GBGT1 genes at the ends might have played in enhancing evolutionary divergence.39 Clearly, those genes are not indispensable for the survival of individuals in many species. However, they must have some functions. Being expressed on the cell surface, oligosaccharide structures may serve as receptors for bacteria, viruses, and toxins. Because glycan expression varies among species, some of the interactions may exhibit species dependency. As the Landsteiner’s law states, host animals lacking a certain glycan may possess antibodies against the glycan. Therefore, the absence of functional glycosyltransferase genes may not always result in disadvantages. For example, the GGTA1 gene is nonfunctional in humans, and we do not express the α1,3-galactosyl epitope.40,41 Instead, we have a high titer of antibodies against this epitope,42,43 and those antibodies may inhibit interspecies infection of retroviruses produced from murine cells44 and reject porcine organs when xenotransplanted in humans.45,46
In addition to the species variations, glycan polymorphisms may also exist within a species, resulting in differential susceptibility to infection among individuals exhibiting distinct phenotypes. For instance, 1 receptor of Helicobacter pylori, a gram-negative, microaerophilic bacterium found in the stomach, is blood group Lewis b (Leb) antigen.47 Functional AT and BT convert this structure to ALeb and BLeb, respectively, by the transfer of a GalNAc or galactose. As the result, the binding of H pylori to gastric epithelium diminishes, and the susceptibility to gastritis decreases in non-O individuals. In contrast, type A RBCs infected with Plasmodium farciparum, a protozoan parasite that causes malaria, are more adhesive to brain capillaries than similarly parasite-infected type O RBCs, and accordingly, the incidence of severe brain malaria becomes higher with group A children than group O children.48,49 In addition to infectious diseases, ABO-dependent susceptibility has also been reported in other diseases such as pancreatic cancer50 and venous thromboembolism.51,52 Those disease associations have also been observed in nontargeted GWAS studies.28,29
Certain strains of Escherichia coli bacteria are known to preferentially bind to FORS1 (Gb5).53 Therefore, most individuals, without a functional GBGT1 gene, may be less susceptible to the infection, although they may be more susceptible to infection by other strains with an affinity to Gb4. No GWAS associations with diseases/traits have been identified at the GBGT1 gene, despite the fact that numerous SNPs have been identified (Figure 1). High prevalence of 2 inactivating missense mutations in non-Apae individuals may explain this failure.25
A and B antigens appeared as the enzymatic reaction products in amphibians, by the emergence of functional α1,2-fucosyltransferase genes in that class. As the ABO/GBGT1 lineage was separated into the ABO and GBGT1 lineages, the structural divergence of the encoded enzymes increased and the utility and preference of fucosylated and nonfucosylated acceptor substrates were modified. The mouse gene encoding cis-AB transferase is an ABO gene, and there is a separate gene (GBGT1) encoding FS in the mouse genome. The loss of the exon corresponding to human exon 4 is not unique to the mouse ABO gene. It is also deleted in rats (data not shown). Accordingly, the deletion seems to have occurred after the separation of the ABO and GBGT1 genes, rather than the acquisition of the exon in humans and other species. If this is true, humans and other species expressing ATs may be innately capable of producing FORS1 antigen. However, considering the weak FS activity and strong AT activity of the enzymes produced by the deletion of exon 3 or 4 of human AT transcripts, A antigens may be primarily synthesized in the presence of fucosylated acceptor substrates. FORS1 may likely be synthesized only when α1,2-fucosyltransferase is absent and globoside is present.
We previously showed that codons 266 to 268 of human AT/BT interact with acceptor substrates, in addition to donor nucleotide-sugar substrates.26 The results presented here confirm that the specificity of acceptor substrates (H or Gb4) is not as stringent as previously thought, although the gene separation led to the recognition of a novel blood group system, FORS. During the evolution of the α1,3-Gal(NAc) transferase genes, there may have been a period when the enzymatic specificities were loose and overlapping. We apparently observed such a residual activity when certain conditions were met. The fact that the cointroduction of the GlyGlyAla substitution and the deletion of exon 3 or 4 into human AT endorsed strong FS activity indicates that at least 2 critical differences exist between blood group AT/BT and FS: the exon/intron gene organization change, which may have affected the in-Golgi localization/conformation, and the structural alteration in the catalytic domain of enzyme, which may have modified substrate specificity.
Both the A allele– and B allele–encoded AT and BT and the GBGT1 gene–encoded FS catalyze the biosynthesis of oligosaccharides that are at the nonreducing terminal of glycans, and therefore, those transferases may likely be localized in the trans-Golgi rather than cis-Golgi apparatus. We recently examined the role of the amino acids at the N-terminus and the TM region of the human AT in the Golgi localization and found that the in-Golgi residency is vital for the intracellular transferase activity.54 The TM region spans codons 33 to 54 and coincides well with codons 33 to 52 contained in exon 3. Because the codons 53 to 68 present in exon 4 are located in the stem region of the enzymes between the TM region and catalytic domain, conformational change, rather than localization change, may have been induced. The current results show that the deletion of exon 3 or 4 from AT mRNA does not much alter the AT activity of the encoded enzymes, despite the fact that FS activity was acquired.
Forssman antigen expression has been detected in cancers occurring in Forssman antigen–negative individuals.7-13 We previously examined the presence/absence of activating missense mutations at codon 230 or 296 of the human GBGT1 gene in the Catalogue of Somatic Mutations in Cancer Database. However, no such mutations were found, although the database lists 60 mutations in the GBGT1 gene in various cancers. In addition, the allele frequency of the SNP (rs375748588) responsible for the Apae phenotype is extremely low. Accordingly, this SNP does not explain more frequent FORS1 expression in cancer. Human AT and BT may synthesize FORS1 if their tripeptide sequences are replaced with GlyGlyAla. However, neither SNPs nor somatic mutations of that sort have been found in the ABO gene and entered in the database. In conclusion, GlyGlyAla substitution may not account for FORS1 expression in cancer.
Altered splicing is 1 of the hallmarks of cancer. Aberrantly spliced mRNAs may be translated into proteins with altered functions, which could contribute to carcinogenesis. The GWAS studies have revealed more than 15 000 tumor-associated splice variants in a wide variety of cancers.55-57 The TCGA SpliceSeq Database lists the GBGT1 gene with several alternatively spliced transcripts in cancer, but it does not list the ABO gene. However, differential splicing patterns have been experimentally shown in the ABO gene mRNAs in cancerous cells.15,37 In the present study, we show that the A allele may have the capacity to produce AT enzymes with FS activity. However, FORS1 expression is also dependent on the availability of precursor glycans. Therefore, FORS1 appearance is influenced by the polymorphism and expression of additional genes, among which are polymorphic A4GALT and B3GALNT1 genes encoding, respectively, α1,4-galactosyltransferase and β1,3-N-acetylgalactosaminyltransferase 1, which specify the PIPK and GLOB blood group systems.58-61
In addition to glycan changes, there may also be immune reactions in cancer between glycan epitopes and naturally occurring antiglycan antibodies. For instance, anti-FORS1 antibodies may suppress the growth of cancer cells ectopically expressing FORS1. A similar inhibition may also be achieved by anti-A antibodies toward incompatible A antigens expressed in cancers occurring in group B and O individuals and also by anti–T nouvelle antibodies toward the cryptic T-nouvelle (CD175) antigen (GalNAcα1-Ser/Thr), observed frequently in a variety of cancers, including hematological malignancies.62,63 Cross-reactions may also occur as a result of the overlapping structural homology among those antigens. Furthermore, immune cells may also be involved in addition to antibodies. A proposition was recently made to apply controlled transfusion of incompatible blood to enhance immunity.64 It is evident that substantial investigation must occur before the ectopic appearance of those glycan antigens in human cancer and the effects on cancer progression can be explained.
The full-text version of this article includes a data supplement.
Acknowledgments
The authors thank Alba Garcia and David Izquierdo for their technical assistance as well as Marco Antonio Fernández Sanmartín and Gerard Requena Fernández at the Institut d’Investigació Germans Trias i Pujol Cytometry Core Facility for their help with FACS analyses. The majority of work was performed at the now defunct Institut de Medicina Predictiva i Personalitzada del Càncer.
The study was supported by a Spanish Health Research Foundation (Instituto de Salud Carlos III) grant (PI11/00454), funds from Agència de Gestió d’Ajuts Universitaris i de Recerca (2014 SGR 1269), and institutional startup funds from the Institut de Medicina Predictiva i Personalitzada del Càncer and Josep Carreras Leukaemia Research Institute (IJC)–“La Caixa” Foundation to F.Y. The IJC and Institut d’Investigació Germans Trias i Pujol are Centres de Recerca de Catalunya (CERCA) centers supported by CERCA Programme/Generalitat de Catalunya. The IJC is also supported by the Josep Carreras Foundation.
Authorship
Contribution: M.Y. and F.Y. conceived and designed the experiments; M.Y. prepared expression constructs and performed DNA transfection and immunocytochemistry; E.C. performed DNA transfection and FACS immunocytometry; and F.Y. wrote the manuscript with contributions from the other authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Fumiichiro Yamamoto, Josep Carreras Leukaemia Research Institute (IJC), Can Ruti Campus, Ctra. de Can Ruti, Camí de les Escoles s/n, 08916 Badalona, Barcelona, Spain; e-mail: fyamamoto@carrerasresearch.org.
References
Author notes
M.Y. and E.C. contributed equally to this work.


![Figure 3. Cytometric analysis of COS1 (B3GALNT1) cells transfected with M_GBGT1, H_ABO-A, and 3 representative derivative constructs. Cells were cotransfected with DNA from the selected expression construct and DNA from the vector expressing monomer red fluorescent protein (mRFP). Cells were then detached and stained with anti-FORS1 antibody and then with Alexa Fluor 488–linked secondary antibody. The 2 graphs in the top panel correspond to M_GBGT1 transfection. On the left, the selection of cell population by forward- (FSC) and side-scatter (SSC) amplitudes is depicted. Events are dot plotted in pseudocolor, with red representing higher frequency and dark blue the lowest. This selection was maintained for all the other samples. The right graph shows the fluorescence intensity from mRFP, used as transfection control, on the y-axis and the fluorescence corresponding to FORS1 on the x-axis. Both axes are on the logarithmic scale. The upper red rectangle represents the mRFP+ cell population, and the percentage is indicated at the upper left corner. The green rectangle corresponds to Alexa Fluor 488+ cell population, and the percentage is indicated at the lower right corner. The other panels show the results for other constructs, H_ABO-A, H_ABO-A (GlyGlyAla), H_ABO-A (deleted [del] exon 4), and H_ABO-A (GlyGlyAla and del exon 4), using the same gates.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/1/27/10.1182_bloodadvances.2017009795/3/m_advances009795f3.jpeg?Expires=1769230865&Signature=0tgplHihO5JpVPNErvWJOqGCornr4tUQ4VtGtjXjt~i-2LeNVALQBFFcL~6yJVawo48xeyZTb1ilMrGJn0wkCQTB7Hl~JwCuOiEaMxitKbb7Cg162eaHy74MmChhR9xDL9i6CLvFr3qrp1JkENAcF0OWTRMCk3mGZ2fUDb8wpdIQtZfsCFz4RduMFrMouLKE2Fp6QbX~LL6IFi2UlWDiOurk-CVQas1IcsggZRCtb3MCJ0G70dd-tBrUqMYzFBlp9IBvfaGg1Kez3aOQke8rSf3EIxNQoGfXQv~u8Razw0ujkfa3HOcpYyrRaIV59JySaoKCtEzR6SqwHxuLXR4MBg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)