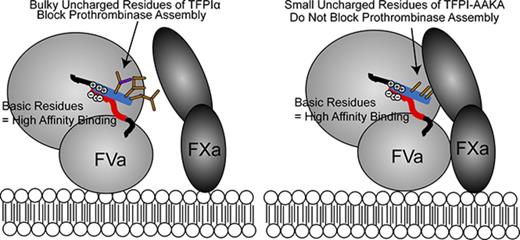
Key Points
TFPIα inhibits prothrombinase through interactions with the FXa active site and B-domain acidic region and heavy chain of FXa-activated FVa.
Leu252-Thr255 of the TFPIα basic region is necessary for inhibitory activity but not for binding the FXa-activated FVa acidic region.
Abstract
Tissue factor pathway inhibitor α (TFPIα) inhibits prothrombinase, the thrombin-generating complex of factor Xa (FXa) and factor Va (FVa), during the initiation of coagulation. This inhibition requires binding of a conserved basic region within TFPIα to a conserved acidic region in FXa-activated and platelet-released FVa. In this study, the contribution of interactions between TFPIα and the FXa active site and FVa heavy chain to prothrombinase inhibition were examined to further define the inhibitory biochemistry. Removal of FXa active site binding by mutation or by deletion of the second Kunitz domain (K2) of TFPIα produced 17- or 34-fold weaker prothrombinase inhibition, respectively, establishing that K2 binding to the FXa active site is required for efficient inhibition. Substitution of the TFPIα basic region uncharged residues (Leu252, Ile253, Thr255) with Ala (TFPI-AAKA) produced 5.8-fold decreased inhibition. This finding was confirmed using a basic region peptide (Leu252-Lys261) and Ala substitution peptides, which established that the uncharged residues are required for prothrombinase inhibitory activity but not for binding the FVa acidic region. This suggests that the uncharged residues mediate a secondary interaction with FVa subsequent to acidic region binding. This secondary interaction seems to be with the FVa heavy chain, because the FV Leiden mutation weakened prothrombinase inhibition by TFPIα but did not alter TFPI-AAKA inhibitory activity. Thus, efficient inhibition of prothrombinase by TFPIα requires at least 3 intermolecular interactions: (1) the TFPIα basic region binds the FVa acidic region, (2) K2 binds the FXa active site, and (3) Leu252-Thr255 binds the FVa heavy chain.
Introduction
Prothrombinase, the complex of the serine protease factor Xa (FXa) and its cofactor factor Va (FVa), generates the thrombin needed to form a blood clot.1,2 FVa is generated through proteolytic activation of its precursor, FV, which is maintained as an inactive procofactor through interactions between basic and acidic regions within its activation peptide (B-domain).3-5 Removal of either the acidic or basic region activates FV to FVa.3,4 FVa generated through limited proteolysis by FXa6 and forms of FVa released from activated platelets7,8 retain the acidic region, which remains only during the initiation phase of coagulation, as thrombin catalyzes rapid removal of the entire B-domain.6,9
Forms of prothrombinase containing the FVa acidic region are inhibited by tissue factor pathway inhibitor α (TFPIα), a trivalent Kunitz-type serine protease inhibitor that regulates the initiation of coagulation.10-13 However, the biochemical mechanism of this inhibition remains unclear. TFPIα contains a 9-residue basic sequence in its C-terminal region (LIKTKRKRK) that is nearly identical to the FV B-domain basic region (LIKTRKKKK).12 This sequence is conserved in both proteins across mammals.12,14 The TFPIα basic region tightly binds to the FVa acidic region12,15,16 in a charge-dependent interaction that is blocked by negatively charged polymers, such as heparins17,18 and polyphosphates.12,19,20 TFPIα does not effectively inhibit thrombin generation by prothrombinase if either the FVa acidic region or TFPIα basic region is missing.10-12 Because a peptide mimicking the homologous FV basic region competes against FXa for FVa binding, it has been proposed that this peptide, and by extension TFPIα, inhibits thrombin generation by displacing FXa from FVa, thereby preventing prothrombinase assembly.21
The ability of the FV basic region peptide to compete against FXa for FVa binding suggests that inhibition by the peptide involves at least 2 binding sites: (1) a high-affinity interaction with the FVa acidic region, and (2) a low-affinity interaction with the FVa heavy chain, which does not occur in the absence of acidic region binding.12,21 Consistent with this supposition, the FV Leiden (FVL) mutation (Arg506→Gln in the FVa heavy chain) causes an approximately twofold decrease in the ability of either TFPIα or the TFPIα basic region peptide to inhibit prothrombinase containing the FVa acidic region.22 This small change in inhibitory kinetics suggests that TFPIα may not bind directly to Arg506, but rather that its binding site is close enough to Arg506 that accessibility is affected by this mutation. In addition, poly-L-lysine does not inhibit prothrombinase activity, demonstrating that the basic residues, although required, are not sufficient for inhibition.21
Even though the TFPIα and FV basic region peptides bind the FVa acidic region with low nanomolar affinity, inhibition of thrombin production requires 2 orders of magnitude higher concentrations of either peptide than TFPIα.12,21 This discrepancy suggests that the efficient inhibition achieved by TFPIα requires more than simply the interaction of the basic C-terminus with FVa. Because the second Kunitz domain (K2) of TFPIα is a well-described inhibitor of the FXa active site,23 we hypothesized that this interaction is also required for prothrombinase inhibition. In the present study, full-length forms of TFPIα with amino acid substitutions in K2 that prevent binding to the FXa active site or within the uncharged amino acids of the basic region, truncated forms of TFPIα, and basic region peptides were used in purified protein– and plasma-based assays to define the interactions required for prothrombinase inhibition.
Materials and methods
Proteins
Human FXa, thrombin, prothrombin, and FX were from Enzyme Research Laboratories (South Bend, IN). Human FV was from Paula B. Tracy or Kathleen M. Brummel-Ziedins (University of Vermont, Burlington, VT) and activated with FXa (FVaXa) or thrombin (FVaIIa).12 FV810, a recombinant form of FV that retains the TFPIα-binding acidic region,3,5 and FV810 containing the Leiden Arg506→Gln substitution (FVL810) were from Rodney M. Camire (University of Pennsylvania, Philadelphia, PA). TF (Dade Innovin) was from Siemens (Washington, DC). Human FVIIa and a mouse antibody against the TFPI K2 domain (anti-K2) were obtained as described.24 A rabbit polyclonal antibody that recognizes the final 12 amino acids of TFPIα (anti-CTP) was produced by Abcam (Burlingame, CA). Goat anti-mouse IRDye680 and goat anti-rabbit IRDye800CW were from LI-COR Biosciences (Lincoln, NE).
Expression and purification of altered TFPIα proteins
The DNA sequences of TFPI were synthesized with CD33 signal peptide and cloned into vector pJSV002 for mammalian expression. The genes of TFPIΔK2 (Arg107Ala) and TFPI-AAKA (Leu252Ala/Ile253Ala/Thr255Ala) were constructed based on wild-type TFPIα by site-directed mutagenesis. Plasmids encoding TFPI variants were transfected into HEK293-6E cells (1.0 × 106 cells per mL), with DNA molar ratio of 1:1. Transfection was performed following the FreeStyle 293 expression manual (Invitrogen). Cells were harvested for purification 5 days posttransfection.
For TFPIΔK2, the supernatant was applied to an anti-TFPI sepharose affinity column (anti-TFPI antibody coupled to the HiTrap NHS-activated HP column; GE Healthcare) and equilibrated in phosphate-buffered saline (PBS). The bound protein was eluted with 0.1 M of glycine-hydrochloride (HCl), pH 2.8. Fractions were collected and neutralized immediately with 1/20 volume 2 M of Tris-HCl, pH 9.0, for further purification. For TFPI-AAKA, the supernatant was applied to a Heparin Sepharose 6 Fast Flow affinity column (GE Healthcare), equilibrated in 20 mM of Tris-HCl, pH 7.5. The bound protein was eluted with a 0.2- to 1-M linear gradient of sodium chloride. Fractions containing the TFPI variants were collected. The pooled fractions were further purified by size-exclusion chromatography on a Superdex 75 prep grade column (GE Healthcare) in PBS.
His-thioredoxin (Trx)-3C-K3C (TFPIα residues 185-271) was synthesized and cloned into the pET32a vector. The plasmid was transformed into Escherichia coli BL21(DE3) host strain and cultured in terrific broth medium. Expression was induced when optical density measured at a wavelength of 600 nm reached 0.1, with 0.1 mM of isopropyl-β-D-thiogalactoside. Protein was expressed overnight at 18°C, and the bacterial pellets were then harvested for purification. The cell pellets were resuspended in lysis buffer (50 mM of Tris, pH 8.0, 0.1% Tween-20) and disrupted by French press. Cell debris was removed by centrifugation. Supernatant was applied to an Ni-NTA Superflow column (QIAGEN), equilibrated in 50 mM of Tris, pH 8.0; 4 M of urea; 300 mM of sodium chloride; and 20 mM of imidazole. The bound protein was eluted with a 20- to 400-mM linear gradient of imidazole. The pooled fractions were further purified by cation exchange chromatography on an SP-HP column (GE Healthcare). Eluted protein was digested by 3C protease to remove the His-Trx-3C tag. Then, the TFPI K3C fragment was further purified using an SP-HP column (GE Healthcare) and buffer exchanged to PBS.
All purified proteins were sterilized by filtration through a 0.2-μm filter unit (Sartorius). The purity was analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and size-exclusion high-performance liquid chromatography. The protein identity was confirmed by mass spectrometry.
Peptides
Peptides were made by the Protein Chemistry Core Laboratory (BloodCenter of Wisconsin, Milwaukee, WI) or Peptide 2.0 (Chantilly, VA). Peptide concentrations were determined by measurement at 205 nm25 and comparison with a standard curve generated using a peptide of similar molecular weight and isoelectric point (GPYVKQNTLKLAT). The concentration of the peptide standard was measured at 280 nm (E1%, 10.4). The wild-type peptide (LIKTKRKRKK) was also synthesized with 2 N-terminal aminohexanoic acid spacers and 5(6)carboxy-X-rhodamine (Life Technologies, Grand Island, NY) coupled to the N-terminus (Rhod-LIKTKRKRKK). The concentration of Rhod-LIKTKRKRKK was determined by measuring the absorbance at 575 nm (carboxy-X-rhodamine; E1M, 82 000) and at 205 nm, which yielded similar results (190 and 215 µM, respectively).
Phospholipid vesicles
Phospholipid vesicles containing 20% phosphatidylserine, 20% phosphatidylethanolamine, and 60% phosphatidylcholine were prepared according to the method of Morrissey.26
Prothrombinase activity assays
FVaXa, FVaIIa, FV810, or FVL810 (0.5 nM) was incubated with phospholipid vesicles (20 μM), the thrombin inhibitor dansylarginine N-(3-ethyl-1,5-pentanediyl)amide (DAPA; 3 μM; Haematologic Technologies, Essex Junction, VT), and varying concentrations of TFPIα protein or peptide. DAPA was included in these reactions to limit thrombin-mediated removal of the FVa B-domain acidic region. Nesheim et al27 demonstrated that the inclusion of DAPA does not alter the rate of thrombin generation. Reactions were initiated by addition of prothrombin (1.4 μM) and FXa (5 nM). Aliquots were removed at timed intervals and quenched with EDTA (33 mM), diluting DAPA to a final concentration of 0.2 μM. Thrombin was measured using the chromogenic substrate Spectrozyme TH (0.32 mM; Sekisui Diagnostics, Lexington, MA). GraphPad Prism v.6 (GraphPad Software, La Jolla, CA) was used to determine 50% inhibitory concentrations (IC50s).12 Assuming Ki = 0.15 μM for thrombin inhibition by DAPA,28 and Km = 2.5 μM for Spectrozyme TH cleavage by thrombin,29 thrombin is expected to be 99% active under these conditions.
Calibrated automated thrombography
All experiments using humans were approved by the BloodCenter of Wisconsin Institutional Review Board. Platelet-rich plasma (PRP) was prepared from whole blood collected into citrate and corn trypsin inhibitor (50 µg/mL; Haematologic Technologies). TFPI-depleted plasma was from Sekisui Diagnostics. Calibrated automated thrombography assays were performed using a Fluoroskan Ascent microplate fluorometer (Thermo Scientific), as described.12 Assays using PRP were initiated with FXa (0.1 nM) and collagen (15 μg/mL). Assays using TFPI-depleted plasma were initiated with FXa and phospholipid vesicles (4 μM).
FXa activity assays
FXa (0.2 nM) was incubated with varying concentrations of the altered TFPIα proteins, and cleavage of the chromogenic substrate Spectrozyme Xa (0.5 mM; Sekisui Diagnostics) was monitored at 405 nm.
TF-FVIIa activity assays
TF (6 pM) was incubated with phospholipid vesicles (20 μM), FVIIa (20 pM), and varying concentrations of the altered TFPIα proteins. Reactions were initiated by addition of FX (20 nM), and cleavage of Spectrozyme Xa was monitored at 405 nm.
Fluorescence anisotropy
Steady-state fluorescence anisotropy was measured in a QuantaMaster spectrafluorometer (Photon Technology International, Edison, NJ).12 Emission and excitation wavelengths were 580 and 605 nm, respectively. For direct binding measurements, Rhod-LIKTKRKRKK (30 nM) was incubated with increasing concentrations of FV810. For displacement experiments, Rhod-LIKTKRKRKK (30 nM) and FV810 (30 nM) were incubated with increasing concentrations of unlabeled peptide. Distribution coefficient (Kd) values were obtained from the direct binding30 and displacement experiments.31
Western blotting
Recombinant TFPIα proteins were diluted into sample preparation buffer (62.5 mM of Tris, pH 6.8; 10% glycerol; 1% SDS; 0.0005% Bromphenol Blue), separated by SDS-PAGE on a 4% to 20% acrylamide gel,32 and transferred to nitrocellulose.33 The blot was probed with mouse anti-K2 and rabbit anti-CTP (5 μg/mL each), followed by detection with goat anti-mouse IRDye680 and goat anti-rabbit IRDye800CW secondary antibodies (1:10 000 dilution each). The blot was developed using Odyssey Infrared Imager Running Image Studio v.4.0 (LI-COR Biosciences).
Results
Characterization of recombinant TFPIα proteins
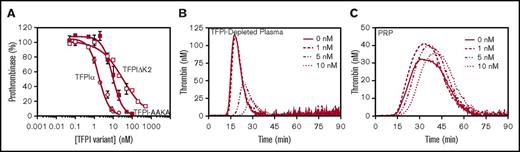
The roles of the TFPIα K2 domain and the uncharged residues within the TFPIα basic region in prothrombinase inhibition were examined using altered recombinant TFPIα proteins (Figure 1A). These proteins included: (1) wild-type TFPIα; (2) 2 altered TFPIα proteins incapable of binding the FXa active site, TFPIΔK2 (Arg107 of K2 mutated to Ala) and K3C, which lacks the K1 and K2 domains; and (3) TFPIα in which the uncharged residues of the basic region (Leu252, Ile253, Thr255) were substituted with Ala (TFPI-AAKA). Because the TFPIα C-terminus is susceptible to proteolysis, the presence of an intact C-terminus was verified by western blotting with an antibody directed against the final 12 amino acids of TFPIα (Figure 1B). This antibody recognized all of the recombinant proteins used in these studies, confirming the presence of an intact C-terminus in each. A recombinant TFPI containing only the K1 and K2 domains, previously shown to be a poor inhibitor of prothrombinase,12 was included as a negative control for the western blot. Similarly, an antibody against the K2 domain (anti-K2) recognized all of the recombinant proteins except K3C. TFPIα and TFPI-AAKA directly inhibited FXa and also inhibited TF-FVIIa in assays using purified proteins, confirming the presence of active K1 and K2 domains (supplemental Figures 1 and 2). K3C did not inhibit TF-FVIIa or FXa, and TFPIΔK2 only weakly inhibited FXa. The weak FXa inhibition by TFPIΔK2 was likely caused by the functional K1 domain, as demonstrated by Petersen et al.34
TFPIα variant proteins used in these studies. (A) Shown are the domain structures of TFPIα, K1K2 (amino acids 1-160), TFPIΔK2 (R107A mutation), TFPI-AAKA (L252A, I253A, T255A mutations), and K3C (amino acids 185-271). The 3 Kunitz domains are shown in gray. The C-terminal basic region is blue. (B) Western blot of the TFPI variant proteins, probed with antibodies against the K2 domain (red) and the last 12 amino acids of the C-terminus (green), with overlay indicated in yellow. The K1K2 sample contains some dimerized protein.
TFPIα variant proteins used in these studies. (A) Shown are the domain structures of TFPIα, K1K2 (amino acids 1-160), TFPIΔK2 (R107A mutation), TFPI-AAKA (L252A, I253A, T255A mutations), and K3C (amino acids 185-271). The 3 Kunitz domains are shown in gray. The C-terminal basic region is blue. (B) Western blot of the TFPI variant proteins, probed with antibodies against the K2 domain (red) and the last 12 amino acids of the C-terminus (green), with overlay indicated in yellow. The K1K2 sample contains some dimerized protein.
Binding of the FXa active site is required for efficient prothrombinase inhibition
The role of the K2 domain in prothrombinase inhibition was evaluated using TFPIΔK2 and K3C. K3C is incapable of inhibiting the FXa active site, and TFPIΔK2 is a poor FXa inhibitor (supplemental Figure 1). In purified prothrombinase assays, TFPIΔK2 was a 17-fold weaker inhibitor than wild-type TFPIα (IC50, 30.6vs 1.8 nM), and K3C was even weaker than TFPIΔK2 (IC50, 61.4 nM; Figure 2A). Consistent with the prothrombinase assays using purified proteins, TFPIα dose dependently inhibited FXa-initiated thrombin generation in TFPI-depleted plasma, increasing the lag time by 79.9% ± 4.3% at 5 nM and 214.6% ± 20.6% at 10 nM (Figure 2B). TFPIΔK2 had significantly less effect, only increasing the lag time by 45.5% ± 8.1% at 10 nM (P < .0001; Figure 2C). Because activated platelets release forms of FVa that bind TFPIα and provide a surface for prothrombinase assembly, the inhibitory activity of TFPIα variants was also evaluated in PRP. Although this system is complicated by the presence of endogenous TFPIα, a similar inhibitory pattern was apparent. In PRP, 5 nM of TFPIα inhibited FXa-initiated thrombin generation, delaying the lag time by 31% ± 10% (Figure 2D). In contrast, thrombin generation was not inhibited by 10 nM of TFPIΔK2 (Figure 2E) or K3C (Figure 2F). These results demonstrate that binding of TFPIα to the FXa active site is required for efficient prothrombinase inhibition in assays performed using purified proteins and in assays performed using TFPI-depleted plasma or PRP.
The K2 domain of TFPIα enhances prothrombinase inhibition. (A) FVaXa (0.5 nM) was incubated with phospholipid vesicles (20 μM), the thrombin inhibitor DAPA (3 μM), and TFPIα (open circle), TFPIΔK2 (open square), or K3C (filled square). Reactions were initiated by addition of prothrombin (1.4 μM) and FXa (5 nM). After dilution to reduce the effect of DAPA, thrombin was quantified by the rate of cleavage of a chromogenic substrate (0.32 mM). The initial rate of thrombin generation is shown as a percentage of control (mean ± standard deviation; n ≥ 3). Lines represent best-fit inhibition curves. (B-F) Thrombin generation was measured in TFPI-depleted plasma (B-C) or PRP (D-F). Reactions were initiated with a mixture of FXa (0.1 nM) and phospholipid vesicles (4 μM) (B-C) or FXa and collagen (15 µg/mL) (D-F), in the presence of the indicated concentrations of TFPIα (B,D), TFPIΔK2 (C,E), or K3C (F). Shown are the average thrombin generation curves from experiments performed in triplicate using TFPI-depleted plasma (B-C) or using PRP from 4 different donors (D-F).
The K2 domain of TFPIα enhances prothrombinase inhibition. (A) FVaXa (0.5 nM) was incubated with phospholipid vesicles (20 μM), the thrombin inhibitor DAPA (3 μM), and TFPIα (open circle), TFPIΔK2 (open square), or K3C (filled square). Reactions were initiated by addition of prothrombin (1.4 μM) and FXa (5 nM). After dilution to reduce the effect of DAPA, thrombin was quantified by the rate of cleavage of a chromogenic substrate (0.32 mM). The initial rate of thrombin generation is shown as a percentage of control (mean ± standard deviation; n ≥ 3). Lines represent best-fit inhibition curves. (B-F) Thrombin generation was measured in TFPI-depleted plasma (B-C) or PRP (D-F). Reactions were initiated with a mixture of FXa (0.1 nM) and phospholipid vesicles (4 μM) (B-C) or FXa and collagen (15 µg/mL) (D-F), in the presence of the indicated concentrations of TFPIα (B,D), TFPIΔK2 (C,E), or K3C (F). Shown are the average thrombin generation curves from experiments performed in triplicate using TFPI-depleted plasma (B-C) or using PRP from 4 different donors (D-F).
Uncharged residues of the TFPIα basic region are required for efficient prothrombinase inhibition
We previously showed that the basic residues of the TFPIα basic region are required for prothrombinase inhibition, because this activity is blocked by negatively charged molecules, including heparins and polyphosphates.12,18 However, basic residues are not sufficient for inhibition, because poly-L-lysine does not block prothrombinase activity.21 Interestingly, the basic regions of both TFPIα and FV contain 3 uncharged residues (Leu252, Ile253, and Thr255 in human TFPIα). These uncharged residues are conserved in both proteins in mammals, birds, and reptiles (supplemental Figures 3-6). TFPI-AAKA, in which Leu252, Ile253, and Thr255 are substituted with Ala, was an intermediate inhibitor of purified prothrombinase (IC50, 10.4 nM) compared with TFPIα (1.8 nM) and TFPIΔK2 (30.6 nM; Figure 3A). Consistent with the prothrombinase assays using purified proteins, TFPI-AAKA was a weaker inhibitor of FXa-initiated thrombin generation in TFPI-depleted plasma (Figure 3B) or PRP (Figure 3C) than TFPIα. TFPI-AAKA (10 nM) was similar to 5 nM of TFPIα in delaying the lag time by 34% ± 4%, whereas 5 nM of TFPI-AAKA delayed the lag time by 17% ± 2%.
The basic region uncharged residues enhance prothrombinase inhibition. (A) Prothrombinase activity was measured as in Figure 2A, in the presence of the indicated concentrations of TFPI-AAKA (filled square). The initial rate of thrombin generation is shown as a percentage of control (mean ± standard deviation; n ≥ 3). Lines represent best-fit inhibition curves. Inhibition of prothrombinase by TFPIα (open circle) and TFPIΔK2 (open square) is reproduced from Figure 2 for reference. (B-C) Thrombin generation was measured in TFPI-depleted plasma (B) or PRP (C) as in Figure 2, in the presence of the indicated concentrations of TFPI-AAKA. Shown are average thrombin generation curves from experiments performed in triplicate using TFPI-depleted plasma (B) or PRP from 4 donors (C).
The basic region uncharged residues enhance prothrombinase inhibition. (A) Prothrombinase activity was measured as in Figure 2A, in the presence of the indicated concentrations of TFPI-AAKA (filled square). The initial rate of thrombin generation is shown as a percentage of control (mean ± standard deviation; n ≥ 3). Lines represent best-fit inhibition curves. Inhibition of prothrombinase by TFPIα (open circle) and TFPIΔK2 (open square) is reproduced from Figure 2 for reference. (B-C) Thrombin generation was measured in TFPI-depleted plasma (B) or PRP (C) as in Figure 2, in the presence of the indicated concentrations of TFPI-AAKA. Shown are average thrombin generation curves from experiments performed in triplicate using TFPI-depleted plasma (B) or PRP from 4 donors (C).
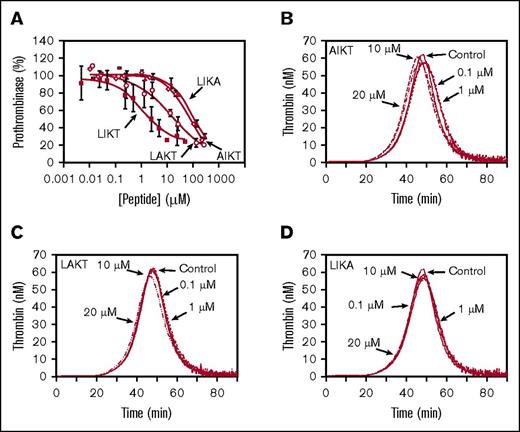
The importance of Leu252, Ile253, and Thr255 for prothrombinase inhibition was more apparent in studies using a TFPIα basic region peptide (LIKTKRKRKK). Similar to a 26-residue C-terminal peptide previously characterized,12 LIKTKRKRKK inhibited prothrombinase assembled with FVaXa (IC50, 1.0 µM) but did not inhibit prothrombinase assembled with FVaIIa (Figure 4A; Table 1) and prolonged the lag time of FXa-initiated thrombin generation in PRP (Figure 4B). These results are consistent with inhibition requiring the FVa acidic region,12,16 which is present in FVaXa but absent from FVaIIa.6 Substitution of all 3 uncharged residues with Ala (AAKAKRKRKK) essentially abolished inhibitory activity (Figure 4C; Table 1). Similarly, AAKAKRKRKK did not inhibit thrombin generation in PRP (Figure 4D). These results demonstrate that the uncharged Leu, Ile, and Thr amino acids within the TFPIα basic region are required for efficient prothrombinase inhibition in assays performed using purified proteins and in assays performed using PRP.
The uncharged residues are required for prothrombinase inhibition by a peptide mimicking the TFPIα basic region. (A) Prothrombinase activity assays were performed as in Figure 2A using either FVaXa (filled square) or FVaIIa (open circle) and the indicated concentrations of LIKTKRKRKK (LIKT). The initial rate of thrombin generation is shown as a percentage of control (mean ± standard deviation; n ≥ 3). Lines represent best-fit inhibition curves. (B,D) Thrombin generation was measured in platelet-rich plasma as in Figure 2D, in the presence of the indicated concentrations of LIKT (B) or AAKAKRKRKK (AAKA) (D). Shown are average thrombin generation curves from experiments using at least 3 donors. (C) Prothrombinase activity assays were performed using FVaXa and the indicated concentrations of AAKA (open circle). Inhibition of FVaXa prothrombinase by LIKT (filled square) is reproduced from panel A for reference.
The uncharged residues are required for prothrombinase inhibition by a peptide mimicking the TFPIα basic region. (A) Prothrombinase activity assays were performed as in Figure 2A using either FVaXa (filled square) or FVaIIa (open circle) and the indicated concentrations of LIKTKRKRKK (LIKT). The initial rate of thrombin generation is shown as a percentage of control (mean ± standard deviation; n ≥ 3). Lines represent best-fit inhibition curves. (B,D) Thrombin generation was measured in platelet-rich plasma as in Figure 2D, in the presence of the indicated concentrations of LIKT (B) or AAKAKRKRKK (AAKA) (D). Shown are average thrombin generation curves from experiments using at least 3 donors. (C) Prothrombinase activity assays were performed using FVaXa and the indicated concentrations of AAKA (open circle). Inhibition of FVaXa prothrombinase by LIKT (filled square) is reproduced from panel A for reference.
Prothrombinase inhibition by peptide variants
| Peptide . | IC50, µM (95% CI)* . | Kd (nM ± SEM)† . |
|---|---|---|
| LIKTKRKRKK | 1.0 (0.4-2.7) | 6.0 ± 2.5 |
| AAKAKRKRKK | >308‡ | 5.9 ± 2.4 |
| AIKTKRKRKK | 70 (60-82) | 10.8 ± 3.9 |
| LAKTKRKRKK | 17 (7.7-36) | 16.2 ± 6.5 |
| LIKAKRKRKK | 110 (89-130) | 9.3 ± 3.6 |
| Peptide . | IC50, µM (95% CI)* . | Kd (nM ± SEM)† . |
|---|---|---|
| LIKTKRKRKK | 1.0 (0.4-2.7) | 6.0 ± 2.5 |
| AAKAKRKRKK | >308‡ | 5.9 ± 2.4 |
| AIKTKRKRKK | 70 (60-82) | 10.8 ± 3.9 |
| LAKTKRKRKK | 17 (7.7-36) | 16.2 ± 6.5 |
| LIKAKRKRKK | 110 (89-130) | 9.3 ± 3.6 |
CI, confidence interval; SEM, standard error of the mean.
IC50 values determined by fitting the data presented in Figures 4 and 5 to a 4-parameter variable slope equation using GraphPad Prism v.6.
Binding affinities determined by fitting the data presented in Figure 6A, as described by Marchand et al.31
30% inhibition observed at 308 µM of peptide.
Leu252, Ile253, and Thr255 each contribute to inhibitory activity
Single Ala substitution peptides were used to examine the individual contributions of Leu252, Ile253, and Thr255 to the inhibition of prothrombinase by the TFPIα basic region. Substitution of any of the 3 uncharged residues resulted in decreased prothrombinase inhibitory activity (Figure 5A). The Leu252 (AIKTKRKRKK) and Thr255 (LIKAKRKRKK) substitution peptides had the least inhibitory activity (IC50, 70and 110µM, respectively), whereas the Ile253 substitution peptide (LAKTKRKRKK) retained greater inhibitory activity (IC50, 17 µM). None of the single Ala peptides inhibited thrombin generation in PRP, at concentrations up to 20 µM (Figure 5B-D). Thus, each of these residues individually contributes to the inhibitory activity of the basic region peptide.
Leu252, Ile253, and Thr255 each contribute to prothrombinase inhibition. (A) Prothrombinase activity was measured as in Figure 2A, in the presence of the indicated concentrations of AIKTKRKRKK (AIKT; open diamond), LAKTKRKRKK (LAKT; open circle), or LIKAKRKRKK (LIKA; filled triangle). The initial rate of thrombin generation is shown as a percentage of control (no peptide; mean ± standard deviation; n ≥ 3). Lines represent best-fit inhibition curves. Inhibition of prothrombinase by LIKTKRKRKK (LIKT; filled square) is reproduced from Figure 3 for reference. (B-D) Thrombin generation was measured in PRP as in Figure 2D, using the indicated concentrations of AIKT (B), LAKT (C), or LIKA (D). Shown are the average thrombin generation curves from experiments using 3 donors.
Leu252, Ile253, and Thr255 each contribute to prothrombinase inhibition. (A) Prothrombinase activity was measured as in Figure 2A, in the presence of the indicated concentrations of AIKTKRKRKK (AIKT; open diamond), LAKTKRKRKK (LAKT; open circle), or LIKAKRKRKK (LIKA; filled triangle). The initial rate of thrombin generation is shown as a percentage of control (no peptide; mean ± standard deviation; n ≥ 3). Lines represent best-fit inhibition curves. Inhibition of prothrombinase by LIKTKRKRKK (LIKT; filled square) is reproduced from Figure 3 for reference. (B-D) Thrombin generation was measured in PRP as in Figure 2D, using the indicated concentrations of AIKT (B), LAKT (C), or LIKA (D). Shown are the average thrombin generation curves from experiments using 3 donors.
Conserved basic residues mediate FVa binding
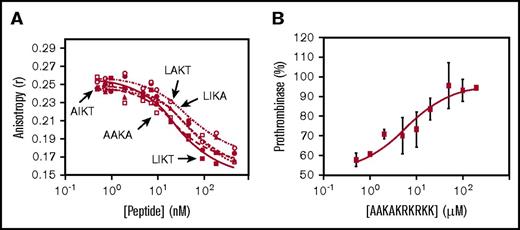
Fluorescence anisotropy was used to measure peptide binding to FV810, a recombinant form of FVa that contains the B-domain acidic region3,5 and tightly binds TFPIα.12 The Kd of each peptide for binding FV810 was determined by measuring its ability to compete against Rhod-LIKTKRKRKK (Kd, 8.6 nM). All of the altered peptides had similar affinity for FV810, which was not different than that of LIKTKRKRKK (Figure 6A; Table 1). Thus, the uncharged residues do not contribute significantly to the binding affinity for FVa.
The basic amino acids, but not the uncharged amino acids, mediate binding of the TFPIα basic region to the FVa acidic region. (A) Rhod-LIKTKRKRKK (30 nM) was incubated with FV810 (30 nM) and the indicated concentrations of LIKTKRKRKK (LIKT; filled square), AAKAKRKRKK (AAKA; open square), AIKTKRKRKK (AIKT; filled circle), LAKTKRKRKK (LAKT; open circle), or LIKAKRKRKK (LIKA; filled triangle). Fluorescence anisotropy was measured and curve fits generated as described in Materials and methods. (B) FV810 (0.5 nM), phospholipid vesicles (20 µM), and the thrombin inhibitor DAPA (3 µM) were incubated with LIKTKRKRKK (LIKT; 3 µM) and varying concentrations of AAKA, and thrombin generation was measured as in Figure 2A. The initial rate of thrombin generation is shown as a percentage of control (no peptide; mean ± standard deviation; n = 3).
The basic amino acids, but not the uncharged amino acids, mediate binding of the TFPIα basic region to the FVa acidic region. (A) Rhod-LIKTKRKRKK (30 nM) was incubated with FV810 (30 nM) and the indicated concentrations of LIKTKRKRKK (LIKT; filled square), AAKAKRKRKK (AAKA; open square), AIKTKRKRKK (AIKT; filled circle), LAKTKRKRKK (LAKT; open circle), or LIKAKRKRKK (LIKA; filled triangle). Fluorescence anisotropy was measured and curve fits generated as described in Materials and methods. (B) FV810 (0.5 nM), phospholipid vesicles (20 µM), and the thrombin inhibitor DAPA (3 µM) were incubated with LIKTKRKRKK (LIKT; 3 µM) and varying concentrations of AAKA, and thrombin generation was measured as in Figure 2A. The initial rate of thrombin generation is shown as a percentage of control (no peptide; mean ± standard deviation; n = 3).
Because AAKAKRKRKK binds FV810 similarly to LIKTKRKRKK (Figure 6A) but does not inhibit prothrombinase activity (Figure 4D), we hypothesized that AAKAKRKRKK would compete against LIKTKRKRKK for binding to the FVa acidic region and prevent prothrombinase inhibition. Purified prothrombinase assays were performed in the presence of LIKTKRKRKK (3 µM) and increasing concentrations of AAKAKRKRKK, which reversed prothrombinase inhibition in a dose-dependent manner (IC50, 6.0 µM; Figure 6B).
Residues Leu252-Thr255 are disrupted by FVL
Because residues Leu252-Thr255 were not necessary for binding the FVa acidic region but were required for optimal inhibitory activity, we hypothesized that they may be involved in an interaction with the FVa heavy chain and mediate displacement of FXa. Consistent with this hypothesis, prothrombinase assembled with FVa Leiden (Arg506Gln) is somewhat less susceptible to inhibition by TFPIα than wild-type FVa.22 We compared the ability of TFPI-AAKA to inhibit prothrombinase assembled with either FV810 or FVL810. Consistent with the experiments using FXa-activated FVa, TFPI-AAKA was a weak inhibitor of prothrombinase containing FV810 (IC50, 16.9 nM; 95% CI, 14.9-20.5 nM; Figure 7A). However, unlike TFPIα, TFPI-AAKA inhibited prothrombinase containing FVL810 identically to prothrombinase containing FV810 (IC50, 17.5 nM; 95% CI, 14.6-19.5 nM). In contrast, the interaction of TFPIα with FXa is unrelated to the effect of FVL, because TFPIΔK2 was a 1.6-fold weaker inhibitor of prothrombinase containing FVL810 (IC50, 46.7 nM; 95% CI, 41.3-52.9 nM) compared with FV810 (IC50, 27.9 nM; 95% CI, 21.7-35.9 nM), similar to the 1.7-fold shift observed with TFPIα (Figure 7B).22
The Leu252-Thr254 region is responsible for the effect of FVL on prothrombinase inhibition by TFPIα. FV810 (filled circle) or FVL810 (open circle; 0.5 nM) was incubated with phospholipid vesicles (20 μM), the thrombin inhibitor DAPA (3 μM), and the indicated concentrations of TFPI-AAKA (A) or TFPIΔK2 (B), and thrombin generation was measured as in Figure 2A. The initial rate of thrombin generation is shown as a percentage of control (mean ± standard deviation; n ≥ 3). Lines represent best-fit inhibition curves.
The Leu252-Thr254 region is responsible for the effect of FVL on prothrombinase inhibition by TFPIα. FV810 (filled circle) or FVL810 (open circle; 0.5 nM) was incubated with phospholipid vesicles (20 μM), the thrombin inhibitor DAPA (3 μM), and the indicated concentrations of TFPI-AAKA (A) or TFPIΔK2 (B), and thrombin generation was measured as in Figure 2A. The initial rate of thrombin generation is shown as a percentage of control (mean ± standard deviation; n ≥ 3). Lines represent best-fit inhibition curves.
Discussion
Inhibition of prothrombinase by TFPIα requires an exosite interaction between the TFPIα basic region and the FVa acidic region. Multiple lines of evidence support this assertion: (1) in vitro studies have shown that the TFPIα basic C-terminus binds to forms of FVa containing the acidic region with high affinity (Kd, ∼90 pM) but does not bind to forms of FVa lacking the acidic region12,15 ; (2) 2 FV mutations producing a truncated B-domain that lacks the basic region but retains the acidic region have been identified16,35 ; these forms of FV circulate bound to TFPIα; and (3) plasma concentrations of TFPIα (∼1 nM) do not inhibit thrombin generation by prothrombinase when the TFPIα basic region or the FVa acidic region is absent.12 In addition, there is evidence suggesting the TFPIα basic region may bind FVa outside of the B-domain acidic region. For example, a peptide mimicking the FV B-domain basic region, which is homologous to the TFPIα basic region, competes with FXa for binding to FV810.21 Because FXa binds the FVa heavy chain,36,37 it has been proposed that the basic peptide binds the FVa heavy chain and inhibits prothrombinase assembly.21 This mechanism has since been extrapolated to full-length TFPIα, which is being described as an inhibitor of prothrombinase assembly.38 However, basic region peptides inhibit prothrombinase only at micromolar concentrations, whereas TFPIα inhibits at low nanomolar concentrations.12 This is a striking discrepancy, particularly because the basic region peptides bind the FVa acidic region with low nanomolar affinity,12 suggesting that structural features of TFPIα beyond its basic region mediate prothrombinase inhibition. FXa-activated FVa (used in the activity assays) and FV810 (used for binding measurements) are chemically dissimilar forms of FV/FVa. The structural differences between these forms were considered as an explanation for the discrepancy between the binding affinity and inhibitory activity observed. This does not seem likely. The major difference between FXa-activated FVa and FV810 is that the former contains the B-domain basic region peptide at 0.5 nM (equimolar concentration to the FVa) that is cleaved from FV when it is activated by FXa.6 However, micromolar concentrations of LIKTKRKRKK are required to inhibit prothrombinase containing FV810,22 similar to prothrombinase containing FXa-activated FVa, suggesting that nanomolar concentrations of the FV B-domain basic peptide have little effect on the inhibitory reaction.12 In this study, 2 additional interactions were identified that greatly increase the efficiency of prothrombinase inhibition, allowing for inhibition at low nanomolar concentrations of TFPIα: (1) the TFPIα K2 domain binds the FXa active site, and (2) residues Leu252-Thr255 of the TFPIα C-terminus bind the FVa heavy chain.
TFPIα directly inhibits FXa by binding to the FXa active site through its K2 domain.23 We previously showed that inhibition of prothrombinase assembled with FXa-activated FVa by TFPIα occurs at an IC50 2 orders of magnitude lower than inhibition by a 26-residue peptide mimicking the TFPIα C-terminus and proposed that inhibition requires interactions with both FVa and FXa.12 Here, the requirement for FXa active site binding was established using 2 altered forms of TFPIα: TFPIΔK2, which has an altered K2 domain that cannot bind FXa, and K3C, which lacks the K2 domain entirely. These proteins were 20- to 30-fold weaker inhibitors of prothrombinase than TFPIα. Therefore, binding to the active site of FXa increases the efficiency of prothrombinase inhibition by TFPIα.
The TFPIα and FV basic regions contain conserved basic and uncharged residues. The importance of the basic residues is evident from the ability of negatively charged polymers, such as heparin, polyphosphate, and fucoidan, to block prothrombinase inhibition.12,17-20,39 However, the basic residues are not sufficient for inhibitory activity, because poly-L-lysine does not block prothrombinase assembly or function.21 Here, we establish that the uncharged residues of the TFPIα basic region (Leu252, Ile253, Thr255) are required for efficient prothrombinase inhibitory activity in experiments using recombinant TFPIα with these amino acids substituted with Ala and several basic region peptides with the 3 amino acids either all or individually substituted with Ala. Interestingly, even though the uncharged residues are highly conserved, they did not contribute to the high-affinity interaction with the FVa acidic region, but they were required for prothrombinase inhibition.
The requirement of the uncharged amino acids for prothrombinase inhibition suggested that these amino acids may interact with the FVa heavy chain in a manner that blocks prothrombinase assembly by blocking the binding of FXa to FVa, as proposed by Bunce et al.21 Studies with FVL were used to further pursue this possibility. FVL is the most common genetic risk factor for thrombosis in whites and consists of the substitution of Arg506 in the heavy chain with Gln.22 FVL is resistant to proteolysis by activated protein C, providing 1 biochemical mechanism for the increased prevalence of thrombosis.40-42 Our laboratory recently found that TFPIα is a 1.7-fold weaker inhibitor of prothrombinase containing FVL810 compared with FV810.22 Similarly, the prothrombinase inhibitory activity of LIKTKRKRKK was decreased 2.1-fold22 and that of TFPI-R107A was reduced 1.6-fold when using FVL810. These small shifts in inhibitory kinetics become physiologically meaningful under conditions of reduced TFPIα concentration. For example, the combination of FVL and TFPI haploinsufficiency is lethal in mice, which die as a result of perinatal thrombosis.43 Because FXa binds FVa near Arg506,44-47 it was plausible that the uncharged amino acids within the TFPIα basic region also bind near Arg506, perhaps explaining the decreased inhibitory activity toward FVL. Therefore, experiments were performed to examine the ability of the different altered forms of TFPIα to inhibit prothrombinase assembled with FVL. Unlike TFPIα, the inhibitory activity of TFPI-AAKA was not affected by FVL, a finding consistent with the uncharged amino acids interacting with the FVa heavy chain. The small shift in kinetics suggests that this interaction is near, but may not directly involve, Arg506.
Collectively, these findings can be described using a model where TFPIα inhibits prothrombinase via at least 3 biochemical interactions: (1) the TFPIα basic region binds the FVa acidic region in a charge-dependent interaction that is mediated by the respective basic and acidic residues; (2) amino acids Leu252-Thr255 interact with the FVa heavy chain in the vicinity of Arg506, either preventing FXa from binding FVa or causing it to bind in a manner that does not promote thrombin generation; and (3) the K2 domain of TFPIα binds the FXa active site. These interactions might occur in any order, although it seems most plausible that TFPIα binds to the FVa acidic region before the heavy chain, because the latter is a low-affinity interaction. This model explains the discrepancy between the high affinity and low inhibitory activity of the TFPIα and FV basic region peptides. Although these peptides are able to bind the FVa acidic region and heavy chain and block prothrombinase assembly, they do not block the FXa active site. High concentrations of peptide are required to push the equilibrium such that no FXa can bind and thus no activity can be measured. The model also explains why poly-L-lysine has no inhibitory activity,21 because it does not contain the Leu, Ile, and Thr residues and therefore would not interact with the FVa heavy chain and displace FXa.
This model is limited because it does not include other described mechanisms for how TFPIα may alter prothrombinase activity. It was recently reported that TFPIα, as well as a peptide mimicking the TFPIα C-terminus, inhibits thrombin-mediated cleavage of FV at Arg1545.48 This inhibition is proposed to be the result of steric hindrance (ie, that the acidic region is close enough to Arg1545 such that when TFPIα or the peptide is bound to the B-domain, thrombin cannot access the cleavage site). Thus, TFPIα not only reduces thrombin generation through inhibition of prothrombinase assembly and activity, but also reduces the feedback activity of thrombin by blocking further FVa processing. Similarly, TFPIα may alter cleavage of FVa at Arg506 by activated protein C49 or at Arg510 by factor XIa,50 because the TFPIα basic region binds near these sites.
In summary, TFPIα inhibits prothrombinase activity through the combination of disrupting prothrombinase assembly and blocking the FXa active site. This inhibition is mediated by the C-terminal basic region and the K2 domain. The basic and uncharged amino acids of the C-terminal region serve distinct roles in the inhibitory biochemistry. Each of these 3 interactions increases the efficiency of prothrombinase inhibition by TFPIα. Therefore, all 3 are likely required for prothrombinase inhibition under physiological conditions.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank D. Jensen (Medical College of Wisconsin) and T. Holyst (BloodCenter of Wisconsin) for technical assistance.
This work was supported by National Institutes of Health, National Heart, Lung, and Blood Institute grants HL068835 (A.E.M.) and HL129193 (J.P.W.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Authorship
Contribution: J.P.W. performed experiments, analyzed results, and wrote the manuscript; H.H.P., B.Y., X.W., and I.H. produced the TFPI variant proteins and wrote the manuscript; and A.E.M. designed the research, analyzed results, and wrote the manuscript.
Conflict-of-interest disclosure: A.E.M. receives grant support from Novo Nordisk. H.H.P., B.Y., X.W., and I.H. are employees of Novo Nordisk A/S. J.P.W. declares no competing financial interests.
The current affiliation for J.P.W. is Division of Cardiovascular Medicine, Gill Heart and Vascular Institute, University of Kentucky, Lexington, KY.
Correspondence: Alan E. Mast, Blood Research Institute, BloodCenter of Wisconsin, PO Box 2178, Milwaukee, WI 53201; e-mail: alan.mast@bcw.edu.