Key Points
TLR4 and 7 agonists improve titers when coformulated with alum but not an emulsion formulation, but do not impact the titer half-lives.
Alum/TLR7 and pIC:LC are potent adjuvant formulations that improve the magnitude and quality of humoral and cellular responses to HIV Env.
Abstract
Adjuvants have a critical role for improving vaccine efficacy against many pathogens, including HIV. Here, using transcriptional RNA profiling and systems serology, we assessed how distinct innate pathways altered HIV-specific antibody responses in nonhuman primates (NHPs) using 8 clinically based adjuvants. NHPs were immunized with a glycoprotein 140 HIV envelope protein (Env) and insoluble aluminum salts (alum), MF59, or adjuvant nanoemulsion (ANE) coformulated with or without Toll-like receptor 4 (TLR4) and 7 agonists. These were compared with Env administered with polyinosinic-polycytidylic acid:poly-L-lysine, carboxymethylcellulose (pIC:LC) or immune-stimulating complexes. Addition of the TLR4 agonist to alum enhanced upregulation of a set of inflammatory genes, whereas the TLR7 agonist suppressed expression of alum-responsive inflammatory genes and enhanced upregulation of antiviral and interferon (IFN) genes. Moreover, coformulation of the TLR4 or 7 agonists with alum boosted Env-binding titers approximately threefold to 10-fold compared with alum alone, but remarkably did not alter gene expression or enhance antibody titers when formulated with ANE. The hierarchy of adjuvant potency was established after the second of 4 immunizations. In terms of antibody durability, antibody titers decreased ∼10-fold after the final immunization and then remained stable after 65 weeks for all adjuvants. Last, Env-specific Fc-domain glycan structures and a series of antibody effector functions were assessed by systems serology. Antiviral/IFN gene signatures correlated with Fc-receptor binding across all adjuvant groups. This study defines the potency and durability of 8 different clinically based adjuvants in NHPs and shows how specific innate pathways can alter qualitative aspects of Env antibody function.
Introduction
Development of a preventative HIV vaccine is a global health priority.1 Of the 6 clinical trials designed to show protection against HIV infection, only the RV144 trial (an ALVAC prime, AIDSVAX [protein/insoluble aluminum salts (alum)] boost regimen) demonstrated protection. The magnitude of non–immunoglobulin A (non-IgA) V1V2-directed antibody titers, in the absence of neutralization, the IgG3 isotype, and antibody-dependent cellular cytotoxicity (ADCC) activity all were associated with reduced risk of infection.2-4 Importantly, both protection and antibody titers were highest 6 months following immunization, at 60%, but dropped to 30% by 1 year. Together, these findings suggest that the magnitude, durability, and quality of HIV envelope (Env) antibody responses may play a role in protection from HIV. Thus, although the primary goal of a preventive HIV vaccine is to induce broadly neutralizing antibodies (bnAbs), it will also be critical to assess how adjuvants mediate these qualitative parameters.
Formulation of Env with immune adjuvants may be used to modulate the magnitude, durability, and also the quality of the antibody response through Fc-mediated effector functions.5 Alum are the most widely used adjuvants and provide a benchmark for comparing other adjuvants.6 Alum has intrinsic innate stimulatory effects through the inflammasome7,8 and can bind antigen (Ag) to enhance its uptake by Ag-presenting cells (APCs).9,10 Other commonly used adjuvants are oil-in-water emulsions, such as MF59, which enhances recruitment of mononuclear cells and neutrophils and increases Ag uptake.11-16
Alum and oil-in-water emulsions provide platforms for combining additional components to enhance their potency. For example, Toll-like receptor 4 (TLR4)17-19 and TLR7 agonists20-22 have been used. Improvements in antibody and T-cell immunity with these adjuvants are due to activation of distinct innate pathways in various APCs or direct activation of B cells. TLR4 agonists activate blood monocyte populations leading to activation of CD4 T cells23-25 whereas TLR7 stimulation of plasmacytoid dendritic cells (DCs) induces type I interferon (IFN).23,24,26-29 Moreover, direct TLR7 stimulation of B cells leads to their proliferation and differentiation.28,30 Additionally, polyinosinic:polycytidylic acid (polyI:C) signals through TLR3 in myeloid DCs to enhance Ag presentation, interleukin-12 (IL-12), and type I IFN production.31-36 Immune-stimulating complexes (ISCOMs), composed of saponins, cholesterol, and phospholipids, are a distinct class of adjuvant that act through both inflammatory and IFN pathways to enhance Ag uptake by DCs.37-41
Recent studies have used systems serology analyses to qualitatively assess antibody responses based on Fc-mediated effector functions,42-47 which have been found to play a role in protection against simian immunodeficiency virus (SIV)48 and HIV.4 To assess the mechanisms by which adjuvants and vaccines mediate such effector functions, transcriptional profiling has been used in mouse models,49-52 as well as in humans to define biomarkers to predict protection.53-55 However, mouse studies are limited because the tissue distribution of TLR expression is different than humans,56 and it is difficult to profile multiple vaccine adjuvants simultaneously in humans. Thus, nonhuman primates (NHPs) provide a unique opportunity to assess multiple adjuvants in parallel for translation to humans.23,28,57 Moreover, an in-depth analysis of humoral immunity has not been applied to a comparative adjuvant HIV study in NHPs nor integrated with innate transcriptional profiling.
In this report, NHPs were immunized with 8 different Env plus adjuvant formulations (alum or MF59 with or without TLR4 or 7 agonists, polyI:C, or ISCOMs). The animals were followed for nearly 2 years to assess the durability of Env antibody responses elicited by different adjuvant formulations. Finally, using transcriptional profiling and systems serology, correlations of specific innate pathways with antibody titer, effector functions, and CD4 T-cell cytokine responses were determined.
Methods
Study animals, immunizations, and sampling
Fifty-three female rhesus macaques of Indian origin were divided into 8 study groups of 6 and 1 group of 5 (supplemental Figure 1A). The average age of each group varied between 5.5 and 6.5 years. For vaccination, 100 μg of glycoprotein 140 (gp140) TV1ΔV2 protein (strain TV1c8.258 ) was adsorbed to alum and administered as such, or coadsorbed with 50 μg of TLR4 agonist (E6020, an monophosphoryl lipid derivative) or 100 μg of TLR7 agonist (a proprietary benzonaphthyridine of Novartis, Cambridge, MA). Env was also mixed with (250 μL) MF59 or with a similar adjuvant nanoemulsion (ANE) formulated with the TLR4 or 7 agonists (supplemental Methods; Novartis, Cambridge, MA59 ). Finally, Env was mixed with 1 mg of polyinosinic-polycytidylic acid:poly-L-lysine, carboxymethylcellulose (pIC:LC) (Hiltonol; Oncovir, Washington, DC) or Abisco100 ISCOMs (Isconova AB, Stockholm, Sweden). Animals were immunized intramuscularly in the quadriceps or deltoids in a homologous prime-boost fashion with Env alone, or with the adjuvant formulations at 0, 4, 12, 24, and 89 weeks. NHPs were housed at Bioqual, Inc (Rockville, MD) and cared for in accordance with American Association for Accreditation of Laboratory Animal Care standards in accredited facilities. Female BALB/c mice, age <8 months were obtained from The Jackson Laboratory (Bar Harbor, ME) and maintained at the Vaccine Research Center’s (VRC) Animal Care Facility (Bethesda, MD) under pathogen-free conditions. Vaccine doses consisted of 10 μg of gp140 TV1ΔV2 protein, 10 μg of TLR4, 25 μg of TLR7, 50 μg of pIC:LC in a 50-μL volume. Immunizations were given intramuscularly (quadriceps). All animal procedures were performed according to protocols approved by the Institutional Animal Care and Use Committees of the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health.
Peripheral blood mononuclear cell, lymph node cell, plasma, and serum separation
NHP blood processing was performed as previously reported,60 and is described in the supplemental Methods.
Flow cytometry phenotyping and intracellular cytokine staining
Peripheral blood mononuclear cells (PBMCs) were incubated with CD28, CD49d, brefeldin A, and 2 μg/mL clade C Env peptides, staphylococcal enterotoxin B (positive control), or media alone (negative control) at 37°C for 6 hours, then overnight at 4°C. Cells were stained for surface markers in phosphate-buffered saline plus serum, then permeabilized and stained intracellularly for cytokines (intracellular cytokine staining [ICS]). Antibodies are listed in supplemental Methods; positive cytokine signals were background subtracted from media-alone responses. Events were acquired on an LSR II cytometer (BD Biosciences) and fluorescence-activated cell sorter data were analyzed using FlowJo software (TreeStar), PESTLE (Mario Roederer, Vaccine Research Center, NIAID), and SPICE.61 Gating trees for cytokine positive populations are depicted in supplemental Figure 1B.
Cytokine ELISAs
Innate cytokines were measured from serum by the quantitative enzyme-linked immunosorbent assay (ELISA). IFN-α was measured using the VeriKine rhesus serum ELISA kit (PBL Interferon Source); IFN-γ–induced protein 10 (IP-10) was measured using the Quantikine human CXCL10 kit (R&D Systems); IL-6 and IL-1β were measured using a human proinflammatory 7-plex assay ultra-sensitive kit (Meso Scale Discovery). All samples were run in duplicate and according to manufacturer’s instructions.
T- and B-cell ELISpots
Enzyme-linked immunospot assays (ELISpots) were measured from freshly isolated PBMCs using ELISpotPlus for human IgG, ELISpotPro for human IL-4, and ELISpotPro for monkey IFN-γ (Mabtech) according to the manufacturer’s instructions. For IgG ELISpots, cells were plated in triplicate at 200 000 to 500 000 cells per well for Ag-specific responses and 25 000 cells per well for total IgG responses. Ag-specific spots were detected using biotinylated gp140 TV1ΔV2 protein at 0.4 μg/mL. For IL-4 and IFN-γ ELISpots, cells were plated in triplicate at 200 000 cells per well and incubated overnight with complete RPMI alone, with 1 to 2 μg/mL of a pool of overlapping 15-mer peptides spanning the clade C Env HIV-1 97/CN/54, or with 1.25 to 2.5 μg/mL Con A. After development, plates were allowed to dry overnight and then were read on an ImmunoSpot analyzer (Cellular Technology Ltd); spots were counted and quality controlled using ImmunoSpot software (CTL).
Titer and half-life analyses
ELISAs were performed as previously reported.60 Half-life analysis equations are described in the supplemental Methods.
Microarrays
Microarray analysis of whole-blood RNA (extraction protocol described in supplemental Methods) was conducted using an Agilent 8 sample × 60K implementation of the Agilent-026806 Macaca mulatta (Rhesus) Oligo Microarray v2. The probe sequence content is identical to Gene Expression Omnibus (GEO) platforms GPL17465 and GPL16026. Labeling was performed using the One-Color Microarray-Based Gene Expression Analysis (v.6.5 protocol; Agilent) as described previously.51 Microarray data for this study are available through GEO (accession number submission in process).
Microarray data analysis consisted of annotation, normalization, modularization, differential expression analysis, supervised clustering, correlation analysis, and enrichment analysis. Each step is described in detail in the supplemental Methods.
Systems serology assays
Systems serological analyses were performed as previously described,44,46,47 and are also detailed in the supplemental Methods.
Statistical analyses
Non–gene array data were graphed and analyzed with Prism software (GraphPad). A 1-way analysis of variance (ANOVA) Kruskal-Wallis test with a Dunn correction for multiple comparisons was used to compare between vaccine groups; a 2-way ANOVA with Bonferroni correction was used to compare vaccine groups over time. The “Env + alum” and prevaccination groups were set as the controls for comparison, unless otherwise noted.
Results
Study design
To determine the innate mechanisms by which adjuvants mediate their effects on HIV Env responses, NHPs were immunized with gp140 TV1ΔV2 Env protein alone, or with 8 different adjuvant formulations at 0, 4, 12, and 24 weeks (supplemental Figure 1A). This immunization schedule was based on the regimen of the RV144 and VAX003/004 vaccine trials.62-64 The TLR4 agonist, E6020,65 or a TLR7 agonist was adsorbed with Env to alum.22 Env was also administered with MF59, or an ANE modified to incorporate the TLR4 or 7 agonists.59 NHPs also received Env plus pIC:LC,66,67 or ISCOMs.38,68,69 No major local or systemic adverse events or changes in weight or temperature were observed with any of the adjuvants following the first immunization (supplemental Figure 2).
Adjuvants differentially enhance the magnitude and durability of antibody responses to Env
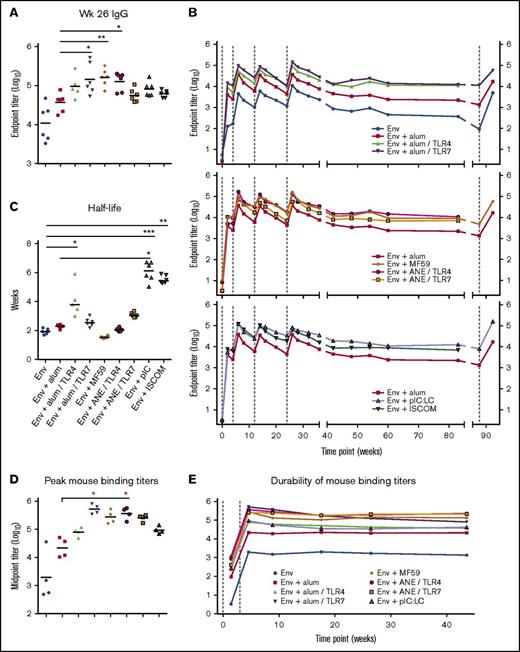
Antibody-binding titers were compared after the final immunization (week 26) (Figure 1A). Animals immunized with Env + alum/TLR7, MF59 or ANE/TLR4 had significantly higher peak Env IgG titers approximately threefold to fourfold over the benchmark formulation of Env + alum (P < .0001). Animals immunized with Env + alum/TLR4, pIC:LC and ISCOMs also had higher titers compared with Env + alum. Overall, the addition of TLR4 and 7 agonists had differential boosting effects on antibody titers when formulated with alum, but not ANE.
Adjuvants induce differences in the magnitude and durability of antibody responses. (A) Serum IgG-binding titers at week 26. (B) IgG-binding titers over time. Env and alum groups, top panel; emulsion groups, middle panel; pIC:LC and ISCOMs, bottom panel. Env + alum is shown as a benchmark; vaccine group geometric means are plotted; vertical dashed lines indicate immunizations. (C) Env IgG titer half-lives were calculated for each animal for the period between weeks 26 and 89. (D-E) Immunogenicity of clinically relevant adjuvant formulations in mice, vaccinated at weeks 0 and 3. Antibody titers are shown broken out at week 5 (D) or over a 44-week period (E). Horizontal bars indicate geometric means; *P < .05; **P < .01; ***P < .001 by the Kruskal-Wallis test in which the Env alone or Env + alum group (as indicated) were set as the control.
Adjuvants induce differences in the magnitude and durability of antibody responses. (A) Serum IgG-binding titers at week 26. (B) IgG-binding titers over time. Env and alum groups, top panel; emulsion groups, middle panel; pIC:LC and ISCOMs, bottom panel. Env + alum is shown as a benchmark; vaccine group geometric means are plotted; vertical dashed lines indicate immunizations. (C) Env IgG titer half-lives were calculated for each animal for the period between weeks 26 and 89. (D-E) Immunogenicity of clinically relevant adjuvant formulations in mice, vaccinated at weeks 0 and 3. Antibody titers are shown broken out at week 5 (D) or over a 44-week period (E). Horizontal bars indicate geometric means; *P < .05; **P < .01; ***P < .001 by the Kruskal-Wallis test in which the Env alone or Env + alum group (as indicated) were set as the control.
Durability of antibody titers was then determined over a period of 89 weeks (Figure 1B). Of note, binding titers were highest 2 weeks after the second immunization and then decreased approximately fivefold to 10-fold for all adjuvants. Antibody responses could be boosted after the third or fourth immunization to the level set after the second immunization. Importantly, for all adjuvants there was a ∼10-fold decrease in antibody titers after the final immunization, which remained constant by week 89. To determine whether a long interval would influence boosting, a subset of the groups was given a fifth immunization at week 89 (65 weeks after the fourth immunization). Titers were boosted to approximately the same peak response as after the second immunization. The effect of adjuvants on antibody half-life was done by modeling their decay from week 26 to 89. The median titer half-life elicited by each adjuvant was ∼2 to 6 weeks for the time period tested (Figure 1C). pIC:LC significantly (P < .05) increased the titer half-life compared with Env alone and Env + alum. ISCOMs also showed an increase in antibody half-life compared with unadjuvanted Env.
Similar studies were performed in mice. After 2 immunizations, antibody responses showed a similar hierarchy of potency of adjuvants for antibody titer (Figure 1D). However, in striking contrast to the results in NHPs showing a decrease in antibody titers over time, they remained constant in mice for ∼40 weeks (Figure 1E).
Qualitative antibody responses following NHP immunization
A clade C gp140 Env protein immunogen was used in this study, as it was being developed for a clinical trial in humans at the time the study was initiated. Based on the immunogen structure it was not likely to elicit bnAb responses. Although neutralization of tier 1 viruses was detected, which correlated with the binding titers, only low neutralization against the tier 2 TV1 strain was detected (supplemental Figure 3A). Alum/TLR7 and the MF59-based (ANE) formulations significantly increased avidity compared with Env only (supplemental Figure 3B).
To further explore other immune parameters that have been shown to have a role in preventive HIV vaccines,4,70,71 serum IgA and rectal mucosal IgG were assessed. NHPs immunized with alum/TLR7 and pIC:LC had significantly higher circulating IgA responses compared with alum alone (supplemental Figure 3C). PolyIC:LC and the emulsion-based formulations elicited statistically higher mucosal IgG titers compared with alum alone, with the ANE/TLR4 formulation inducing the highest titer (supplemental Figure 3D). Mucosal IgA responses were largely undetectable.
Effect of adjuvants on Env T-cell responses
We next assessed T helper cell 1 (TH1) and TH2 T-cell responses by enumerating IFN-γ and IL-4 ELISpots, the canonical cytokines for such responses, respectively (supplemental Figure 4A). Animals immunized with Env alone or Env + alum had low to undetectable IFN-γ or IL-4 responses. In contrast, alum/TLR7 or pIC:LC had a high frequency of TH1/IFN-γ responses ranging from ∼100 to ∼1000 ELISpots but low to undetectable TH2/IL-4 responses. Immunization with MF59, ANE formulations with TLR4 or 7, and ISCOMs resulted in mixed TH1/TH2 responses with comparable frequencies of IFN-γ– and IL-4–producing cells. Analysis of responses by flow cytometry showed that Env-specific CD4+ T cells producing IFN-γ was highest in animals immunized with pIC:LC (∼0.4%) (supplemental Figure 4B). NHPs immunized with alum/TLR7, pIC:LC and ISCOMs had the highest proportion of polyfunctional CD4+ T cells (supplemental Figure 4C). CD8+ T-cell responses were low (<0.1%) to undetectable in all vaccine groups.
Adjuvants induce differential innate transcriptional signatures
Based on the differences in antibody and T-cell immunity, whole-blood transcriptional profiling was performed at 0, 4, 24, 72 hours, and 14 days after the first vaccination (supplemental Tables 1 and 2). For all adjuvants, the most significant change in the magnitude of the transcriptional responses occurred at 24 hours and had resolved by 72 hours (supplemental Tables 3 and 4). The largest number of significantly upregulated genes at 24 hours was induced by pIC:LC, alum/TLR4 or 7, and ANE/TLR4 or 7, followed by ISCOMs and alum alone. MF59 alone induced the most limited blood transcriptional response.
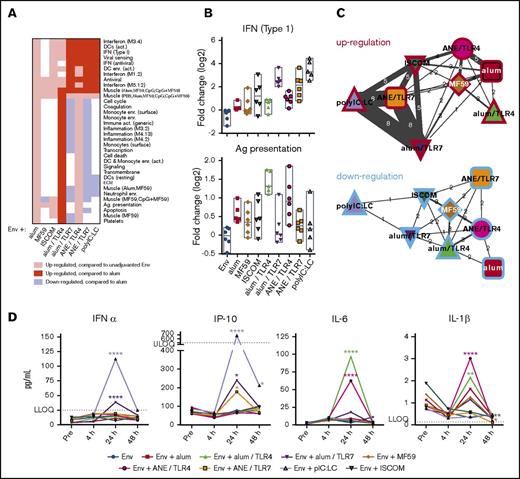
To determine the significance of the differential transcriptional responses, functionally associated gene coexpression modules were analyzed (supplemental Tables 5 and 6).49,72,73 Although each adjuvant had a unique pattern of gene module regulation, there were some common features across some of the adjuvant groups (Figure 2A-B; supplemental Tables 7 and 8). PolyIC:LC and TLR7-containing formulations induced IFN and antiviral modules whereas alum and TLR4-containing formulations induced inflammatory and myeloid-associated modules. Association analysis showed that responses to pIC:LC and TLR7 agonist-containing formulations were most similar to each other (Figure 2C; supplemental Tables 9 and 10).
Adjuvant-specific innate gene signatures. RNA transcripts were measured from peripheral blood sampling prevaccination and 4 hours, 24 hours, 72 hours, and 14 days after the initial immunization. (A) Heatmap showing upregulation and downregulation of innate gene modules by adjuvant at 24 hours. (B) Representative plots showing the expression level of the IFN (type 1) module (top panel) and Ag-presentation module (bottom panel). (C) Association networks depicting the number of modules regulated in common between each adjuvant. Similarities in upregulation (red borders) and downregulation (blue borders) are shown; width of connecting lines indicates the number of modules in common, also shown numerically. (D) Quantification of serum cytokines prevaccination, and 4 hours, 24 hours, or 48 hours following the first vaccination. All data points represent vaccine group medians. *P < .05; **P < .01; ****P < .0001 compared with the prevaccination time point by ANOVA. LLOQ, lower limit of quantitation; ULOQ, upper limit of quantitation.
Adjuvant-specific innate gene signatures. RNA transcripts were measured from peripheral blood sampling prevaccination and 4 hours, 24 hours, 72 hours, and 14 days after the initial immunization. (A) Heatmap showing upregulation and downregulation of innate gene modules by adjuvant at 24 hours. (B) Representative plots showing the expression level of the IFN (type 1) module (top panel) and Ag-presentation module (bottom panel). (C) Association networks depicting the number of modules regulated in common between each adjuvant. Similarities in upregulation (red borders) and downregulation (blue borders) are shown; width of connecting lines indicates the number of modules in common, also shown numerically. (D) Quantification of serum cytokines prevaccination, and 4 hours, 24 hours, or 48 hours following the first vaccination. All data points represent vaccine group medians. *P < .05; **P < .01; ****P < .0001 compared with the prevaccination time point by ANOVA. LLOQ, lower limit of quantitation; ULOQ, upper limit of quantitation.
To confirm that genes upregulated in the microarray analysis were transcribed into protein, we measured a subset of serum cytokines following the first immunization. Consistent with its IFN and antiviral microarray profile, pIC:LC induced transient but detectable IFN-α and IP-10 at 24 hours after vaccination, which resolved by 48 hours (Figure 2D). Similarly, alum/TLR7 or ANE/TLR7 induced transient but detectable production of IFN-α and IP-10. In contrast, the TLR4 agonist induced transient production of IL-6 and IL-1β, consistent with its inflammatory transcriptional profile.74-76
Differential transcriptional effects of TLR4 and TLR7 agonists in alum and MF59 formulations
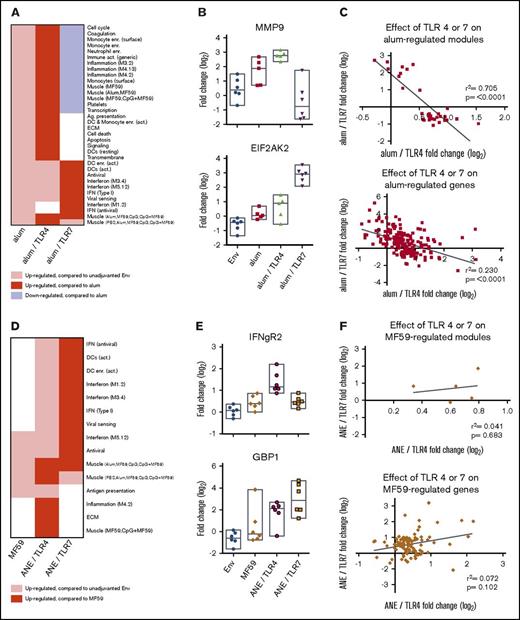
As alum and MF59 are the most widely used adjuvants, transcriptional analysis focused on changes with or without the TLR4 and 7 agonists. TLR4 and 7 agonists modulated the underlying alum-induced innate immune gene expression profile (Figure 3A-C; supplemental Tables 11 and 12). Addition of the TLR4 agonist to alum strongly enhanced myeloid cell and inflammation module induction but had no effect on antiviral and IFN modules. In contrast, the TLR7 agonist markedly suppressed the myeloid cell and inflammation module induction and enhanced the antiviral and IFN modules compared with alum. The expression profiles of MMP9 (a representative inflammatory gene from module 4.273 ) and EIF2AK2/protein kinase R (a representative antiviral IFN gene from module 3.473 ) illustrate the discordant effects of TLR4 and 7 agonists on the underlying alum-driven response (Figure 3B). In general, alum-induced transcriptional responses enhanced by TLR4 were suppressed by TLR7, but responses enhanced by TLR7 were not altered by TLR4 (Figure 3C).
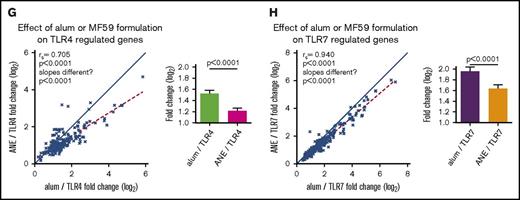
Genetic effects between TLR adjuvants and alum or MF59 formulations. (A) Heatmap depicting the effect of adding the TLR4 or 7 agonists to the alum formulation on gene module regulation. (B) Plots of representative genes showing the change in expression of MMP9 (top panel) and EIF2AK2 (bottom panel) for each alum-containing vaccine group compared with prevaccination. (C) Plots depicting the effect of TLR4 or 7 on the alum formulation. Symbols depict modules (top plot) and individual genes (bottom plot) found to be differentially regulated in the alum-only group, graphed by their change in expression found in the context of the TLR4 (x-axis) and TLR7 (y-axis) containing alum formulations. (D) Heatmap depicting the effect of coformulating ANE with the TLR4 or 7 agonists on gene module regulation. (E) Plots of representative genes showing the change in expression of IFNgR2 (top panel) and GBP1 (bottom panel) for each MF59 and ANE-based vaccine group compared with prevaccination. (F) Plots depicting the effect of ANE/TLR4 or 7 compared with the MF59 formulation. Symbols depict modules (top plot) and individual genes (bottom plot) found to be differentially regulated in the MF59-only group, graphed by their change in expression found in the context of the TLR4 (x-axis) and 7 (y-axis) containing ANE formulations. (G-H) Plots depicting the fold change of genes upregulated by TLR4 (G) or TLR7 (H) in the alum and ANE-based formulations. Blue lines (x = y) depict the hypothetical position of genes that have equal effects in both formulations; red dashed lines depict the linear regression of the actual data. The deviation in slope between both lines was evaluated by regression analysis; the correlation of each data set is indicated by the rs coefficient and P value. Bar graphs depict the mean fold change and standard error between the TLR-regulated genes in either the alum or ANE formulations; P values are derived from a pairwise comparisons of the means. MMP9, matrix metalloproteinase 9.
Genetic effects between TLR adjuvants and alum or MF59 formulations. (A) Heatmap depicting the effect of adding the TLR4 or 7 agonists to the alum formulation on gene module regulation. (B) Plots of representative genes showing the change in expression of MMP9 (top panel) and EIF2AK2 (bottom panel) for each alum-containing vaccine group compared with prevaccination. (C) Plots depicting the effect of TLR4 or 7 on the alum formulation. Symbols depict modules (top plot) and individual genes (bottom plot) found to be differentially regulated in the alum-only group, graphed by their change in expression found in the context of the TLR4 (x-axis) and TLR7 (y-axis) containing alum formulations. (D) Heatmap depicting the effect of coformulating ANE with the TLR4 or 7 agonists on gene module regulation. (E) Plots of representative genes showing the change in expression of IFNgR2 (top panel) and GBP1 (bottom panel) for each MF59 and ANE-based vaccine group compared with prevaccination. (F) Plots depicting the effect of ANE/TLR4 or 7 compared with the MF59 formulation. Symbols depict modules (top plot) and individual genes (bottom plot) found to be differentially regulated in the MF59-only group, graphed by their change in expression found in the context of the TLR4 (x-axis) and 7 (y-axis) containing ANE formulations. (G-H) Plots depicting the fold change of genes upregulated by TLR4 (G) or TLR7 (H) in the alum and ANE-based formulations. Blue lines (x = y) depict the hypothetical position of genes that have equal effects in both formulations; red dashed lines depict the linear regression of the actual data. The deviation in slope between both lines was evaluated by regression analysis; the correlation of each data set is indicated by the rs coefficient and P value. Bar graphs depict the mean fold change and standard error between the TLR-regulated genes in either the alum or ANE formulations; P values are derived from a pairwise comparisons of the means. MMP9, matrix metalloproteinase 9.
Despite reports demonstrating robust transcriptional responses to MF59 in mice at the muscle injection site,49 MF59 upregulated only 99 genes (Figure 2B; supplemental Tables 3 and 4) and 5 modules (Figure 3D; supplemental Tables 7 and 8) in blood of NHPs. Moreover, innate transcriptional responses with ANE/TLR4 or 7 agonists did not lead to discordant modulation of the MF59 response (Figure 3D; supplemental Tables 13 and 14) as observed with alum. Rather, coformulation of ANE with TLR4 and 7 agonists induced modules not already regulated by MF59 (such as Inflammation 4.2 represented by IFNGR2, or Interferon 3.4, represented by GBP1; Figure 3E). The effects of TLR4 or 7 agonists on the MF59 response were not negatively correlated (Figure 3F). Together, these results show that formulation (alum vs ANE) strongly affect how TLR4 and 7 agonists modulate innate immunity.
The overall analysis suggested similarities between formulations containing common TLR agonists (Figure 3A,D; supplemental Tables 9 and 10). Indeed, response magnitudes of genes commonly induced by TLR4 or 7 agonists when formulated with alum or ANE were highly correlated (P < .0001 for both TLR4 and 7 agonists; Figure 3G-H; supplemental Tables 15 and 16). This shows that the responses to TLR4 or 7 activation are dominant over the differences induced by alum and MF59 alone. Nevertheless, the magnitude of the fold changes were nearly universally lower in the ANE, compared with the alum formulations for both TLR4 and 7 (P < .0001 for both).
Assessment of antibody effector functions by systems serology
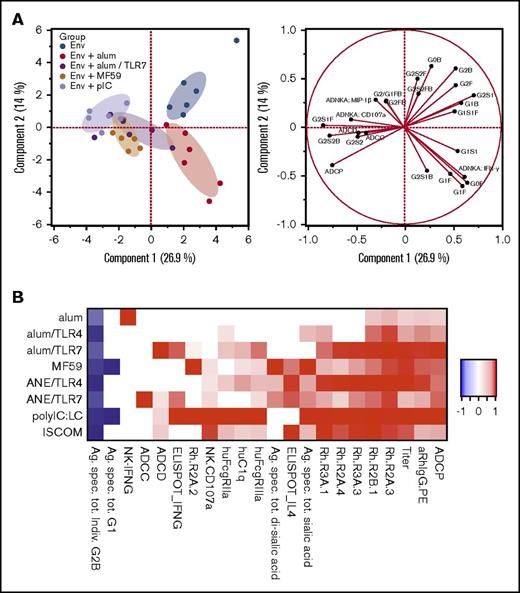
To study how adjuvants influence antibody quality, systems serology was used to cointerrogate immunoglobulin glycosylation structures, immunoglobulin Fc-receptor binding, and effector functions such as antibody-dependent cellular cytotoxicity (ADCC), cellular phagocytosis (ADCP), and complement deposition (ADCD), and Ig-mediated natural killer (NK)-cell activation (supplemental Figure 5; supplemental Table 26).77 A principal components analysis, encompassing all measured parameters, highlighted clear segregation of antibody Fc profiles induced by each of the adjuvants (Figure 4A).
Adjuvants influence immunoglobulin glycan composition and effector functions. Systems serology analyses were used to determine antibody glycan structures and effector functions from sera from study week 14. (A) Principal components analysis of antibody characteristics, differentiating animals within vaccines groups (left plot) by functional and glycan readouts (right plot); only a subset of vaccine groups is shown for simplicity. (B) Heatmap illustrating adaptive immune signature for each vaccine, based on antibody magnitude and effector functions, Fc-receptor binding, and T- and NK-cell cytokines. Heatmap colors indicate significant changes compared with the unadjuvanted group. Responses are scaled by the maximum observed absolute fold change, across all adjuvant groups. hu, human; Rh, rhesus macaque.
Adjuvants influence immunoglobulin glycan composition and effector functions. Systems serology analyses were used to determine antibody glycan structures and effector functions from sera from study week 14. (A) Principal components analysis of antibody characteristics, differentiating animals within vaccines groups (left plot) by functional and glycan readouts (right plot); only a subset of vaccine groups is shown for simplicity. (B) Heatmap illustrating adaptive immune signature for each vaccine, based on antibody magnitude and effector functions, Fc-receptor binding, and T- and NK-cell cytokines. Heatmap colors indicate significant changes compared with the unadjuvanted group. Responses are scaled by the maximum observed absolute fold change, across all adjuvant groups. hu, human; Rh, rhesus macaque.
To assess the potential mechanisms for the effect of adjuvants on FcR binding and antibody effector functions, Env-specific Fc-domain glycan structures were analyzed (supplemental Figure 6A). The adjuvants induced clear differences in the immunoglobulin glycan profiles, and specific glycoforms correlated with effector functions (supplemental Figure 6B). For example, G1 and bisecting GlcNAc glycans correlated with NK CD107α, IFN-γ, and MIP1β expression, and ADCP function; ADCD was better correlated with total sialic acid. Although certain glycan profiles were best correlated with each function, individual adjuvant groups showed unique associations between glycans and antibody function (supplemental Figure 6C), highlighting redundant glycan repertoires that can induce antibody effector functions.
The immunoglobulin glycosylation and effector function data with antibody binding titer and T-cell cytokine analysis were combined to present an adjuvant-specific adaptive immune signature for each formulation, showing both similarities and differences among adjuvants (Figure 4B; supplemental Tables 17-20). Of note, antibodies induced following vaccination with alum alone resulted in the highest in vitro NK IFN-γ production, those by alum/TLR7 resulted in the highest ADCD, whereas those by pIC:LC and ISCOM elicited the most polyfunctional profiles in vitro including more potent FCR binding.
Correlation between innate transcriptional signatures and adaptive immune responses
Based on differences in innate immunity induced by the adjuvants, we determined how blood transcriptional responses correlated with antibody and T-cell immunity. Because TLR4 and 7 agonists induced unique transcriptional responses that further differed in the alum- vs MF59- based (ANE) formulations, correlation analysis focused on those groups.
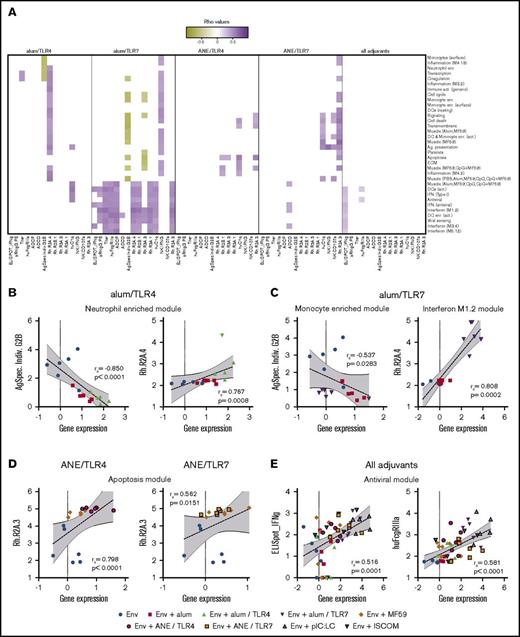
First, an overall adaptive immune response matrix was derived from antibody titer, ELISpot responses and the systems serology data (supplemental Table 17). This was compared with the matrix of significantly regulated genes and modules (supplemental Tables 6 and 8). As shown in Figure 5A and supplemental Table 21, correlations were found for both TLR4 and 7 agonists in each formulation and to a lesser extent among all adjuvant groups. Within each module, representative genes were identified that capture the overall correlation patterns exhibited by the parent modules (supplemental Figure 7; supplemental Tables 22 and 23). For example, Ag-specific individual G2B glycans negatively correlated with several gene modules in the alum/TLR4 and 7 formulations (Figure 5B-C), consistent with the anti-inflammatory properties of G2B. Similarly, binding to the pro-inflammatory rhesus FcR 2A.4 was correlated with many modules in the inflammatory and IFN pathways for the alum/TLR4 and 7 formulations, respectively (Figure 5B-C). Specifically, Bcl6 expression was positively correlated with 2A.4 binding for alum/TLR4, indicating a possible link between germinal center responses and improved antibody functionality. In the MF59-based formulations, binding to the less-characterized FcR 2A.3 was correlated with several modules, including those associated with apoptosis and survival (Figure 5D). Finally, correlations universal to all vaccine groups were found between IFN and antiviral modules and production of T-cell IFN-γ (Figure 5E), reinforcing the well-known mechanistic link between IFN signaling and TH1 responses.78 However, we also noted that several IFN and antiviral modules correlated with ADCD and C1q function in the alum/TLR4, alum/TLR7 and ANE/TLR7 formulations, suggesting a novel mechanistic link by which these pathways induce antibodies that can activate the complement system.
Correlations between innate gene modules and adaptive immune functional characteristics. (A) Expression data from genes that have been assigned to various innate modules were correlated with function characteristics including antibody titer and effector functions, Fc-receptor binding, and T- and NK-cell cytokines. The left 2 panels show correlations derived from the unadjuvanted group, with alum alone and alum/TLR4 or 7. The third and fourth panels show correlations derived from the unadjuvanted group, with MF59 alone and ANE/TLR4 or 7. Correlations that were common to all adjuvant groups are shown in the far-right panel. (B-E) Scatter plots show correlations for each group of vaccines between a representative gene module and either Ag-specific individual G2B glycan, rhesus Fc receptor 2A.4 binding, or 2A.3 binding. (B) Alum/TLR4; (C) alum/TLR7; (D) ANE formulations; (E) all adjuvant groups. For representative scatter plots, the correlation rs and P values are displayed numerically; dashed lines indicate linear regressions; gray shading highlights the 95% confidence interval (CI).
Correlations between innate gene modules and adaptive immune functional characteristics. (A) Expression data from genes that have been assigned to various innate modules were correlated with function characteristics including antibody titer and effector functions, Fc-receptor binding, and T- and NK-cell cytokines. The left 2 panels show correlations derived from the unadjuvanted group, with alum alone and alum/TLR4 or 7. The third and fourth panels show correlations derived from the unadjuvanted group, with MF59 alone and ANE/TLR4 or 7. Correlations that were common to all adjuvant groups are shown in the far-right panel. (B-E) Scatter plots show correlations for each group of vaccines between a representative gene module and either Ag-specific individual G2B glycan, rhesus Fc receptor 2A.4 binding, or 2A.3 binding. (B) Alum/TLR4; (C) alum/TLR7; (D) ANE formulations; (E) all adjuvant groups. For representative scatter plots, the correlation rs and P values are displayed numerically; dashed lines indicate linear regressions; gray shading highlights the 95% confidence interval (CI).
Discussion
Comparative adjuvant studies in NHPs facilitate downselection for translation to humans.23,79-82 This is the first NHP study to integrate transcriptional analysis, cellular profiling, and systems serology to assess the interactions between the innate and adaptive immune responses in the context of HIV Env protein vaccination.
A major focus of this study was to compare how coformulation of alum and ANE with TLR4 or 7 agonists influenced adaptive immunity.29,65 Consistent with prior studies, MF59 induced higher binding titers than alum alone.46,83-85 However, addition of the TLR4 and 7 agonists only improved antibody titers when formulated with alum and not ANE, which correlated with higher TLR-specific gene activation (Figure 3G-H). One explanation for this is that ANE incorporates the TLR agonists into the oil phase, which may be less effective retaining the TLR agonists compared with alum adsorption. Our findings are consistent with mouse studies in which the same TLR4 agonist did not improve titers to influenza hemagglutinin (HA) in an ANE formulation.59 Of note, a different oil-in-water stable emulsion (SE) elicits higher HA titers when formulated with the synthetic TLR4 agonist, glucopyranosyl lipid adjuvant (GLA).86 These data suggest that alum may provide a flexible platform for improving immunogenicity with these or other TLR agonists.
Although the goal for developing preventive HIV vaccines is to induce bnAbs, NHP vaccine studies48 and the RV-144 trial2,4 show that antibody Fc-mediated protection can be induced in the absence of bnAbs. The ongoing HVTN 702 phase 3 clinical trial is a follow-up to the RV144 trial, in which alum has been replaced with MF59 for the Env protein boost. This change was largely based on MF59 inducing higher antibody titers than alum. However, there are additional qualitative considerations for how adjuvants influence immunity and protection. A recent study by Vaccari et al administered alum or MF59 with SIV Env protein and compared the outcome in an NHP challenge model designed to mimic the RV144 trial.46 Although antibody titers were higher using MF59 compared with alum, enhanced protection was correlated with RAS signaling, which induces NK- and T-cell function, in the animals that received alum.46 Here, we substantiated in vitro that alum is a potent inducer of NK IFN-γ. Interestingly, addition of the TLR4 or 7 agonists suppressed this NK activation when coformulated with alum (Figure 4C; supplemental Figure 5). Given the potential importance of NK function, it was notable that pIC:LC and ISCOMs elicited antibodies capable of inducing the highest NK-cell degranulation, as measured in vitro by CD107a expression. These data highlight that replacing alum with MF59 or using additional TLR4 or 7 agonists with alum may enhance antibody titer but could potentially alter qualitative and functional responses that could play a role in protection against HIV.
Improving the durability of antibody responses remains a critical challenge for vaccine development for HIV and malaria.87-91 Here, antibody titers declined ∼10-fold over a 1-year period in NHPs but remain stable in mice (Figure 1), highlighting the importance of using NHPs for predicting adjuvant durability in humans, in which titers also decline. Interestingly, the hierarchy of potency between adjuvant groups was maintained throughout the follow-up period, suggesting that adjuvants should be used to induce the highest peak response, thus maintaining titers above a given threshold for the longest possible time as titers wane.82,92
In conclusion, in this large comparative adjuvant study, alum/TLR7 and pIC:LC were the most potent adjuvants across a variety of antibody and cellular assays. The alum/TLR7 formulation mediated its effects though innate antiviral and IFN pathways to induce strong TH1 T-cell responses, high binding titers, and a number of Fc-mediated effector functions. Similarly, pIC:LC mediated its effects through innate IFN pathways, leading to polyfunctional TH1 T cells, longer antibody half-lives, and NK-cell degranulation. These data support use of these adjuvants in humans for a variety of infections requiring antibodies and T cells.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank J. P. Todd, Carmelo Chiedi, Marlon Dillon, Kefale Wuddie, Will Williams, and Saran Bao for expert veterinary and technical assistance; Hongmei Gao and Kelli Greene for program management; Frank Liang and Karin Loré for helpful discussions; and Pamela Troisch from the Institute for Systems Biology Microarray Core for running the microarrays.
This work was supported in part by the Intramural Research Program of the Vaccine Research Center, NIAID, National Institutes of Health. Additional support was provided by the European Union FP7 (Advanced Immunization Technologies, 280873) and the Collaboration for AIDS Vaccine Discovery award (OPP1039775) from the Bill & Melinda Gates Foundation.
Authorship
Contribution: J.R.F., D.E.Z., G.A., and R.A.S. conceived the study and wrote the paper; E.S., N.M.V., P.M., E.D.G., S.W.B., P.S., M.S., S.J., A.S., and D.T.O. developed the vaccine formulations; J.R.F., C.L., N.L.Y., B.G., B.J.F., M. Ackerman, M. Alam, G.F., and G.D.T. designed and carried out experiments; C.J. and D.E.Z. performed the transcriptional analyses; M.J. performed half-life modeling; and A.A., D.E.Z., G.A. and R.A.S. gave conceptual advice and project oversight.
Conflict-of-interest disclosure: E.S., N.M.V., P.M., E.D.G., S.W.B., P.S., M.S., S.J., and D.T.O. were employees of Novartis Vaccines at this study’s conception. A.S. is the chief executive officer and chief science officer of and owns stock in Oncovir, Inc, the company that makes pIC:LC (Hiltonol). The remaining authors declare no competing financial interests.
Correspondence: Robert A. Seder, Cellular Immunology Section, Vaccine Research Center, National Institute of Allergy and Infectious Disease, National Institutes of Health, 40 Convent Dr, MSC 3025, Building 40, Room 3512, Bethesda, MD 20892; e-mail: rseder@mail.nih.gov.
References
Author notes
J.R.F. and D.E.Z. contributed equally to this work.