Key Points
TRAIL enhances receptor activator of NF-κB ligand–induced osteoclastogenesis and c-FLIP upregulation without osteoclast apoptosis induction.
TAK1 inhibition triggers TRAIL-induced apoptosis in osteoclasts, while potentiating TRAIL-induced myeloma cell death.
Abstract
Tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) agonists induce tumor-specific apoptosis indicating that they may be an attractive therapeutic strategy against cancers, including multiple myeloma (MM). Osteoclastogenesis is highly induced in MM, which in turn enhances MM growth, thereby forming a vicious cycle between MM tumor expansion and bone destruction. However, the effects of TRAIL on MM-enhanced osteoclastogenesis remain largely unknown. Here, we show that TRAIL induced apoptosis in MM cells, but not in osteoclasts (OCs), and that it rather facilitated receptor activator of NF-κB ligand–induced osteoclastogenesis along with upregulation of cellular FLICE inhibitory protein (c-FLIP). TRAIL did not induce death-inducing signaling complex formation in OCs, but formed secondary complex (complex II) with the phosphorylation of transforming growth factor β–activated kinase-1 (TAK1), and thus activated NF-κB signaling. c-FLIP knockdown abolished complex II formation, thus permitting TRAIL induction of OC cell death. The TAK1 inhibitor LLZ1640-2 abrogated the TRAIL-induced c-FLIP upregulation and NF-κB activation, and triggered TRAIL-induced caspase-8 activation and cell death in OCs. Interestingly, the TRAIL-induced caspase-8 activation caused enzymatic degradation of the transcription factor Sp1 to noticeably reduce c-FLIP expression, which further sensitized OCs to TRAIL-induced apoptosis. Furthermore, the TAK1 inhibition induced antiosteoclastogenic activity by TRAIL even in cocultures with MM cells while potentiating TRAIL’s anti-MM effects. These results demonstrated that osteoclastic lineage cells use TRAIL for their differentiation and activation through tilting caspase-8–dependent apoptosis toward NF-κB activation, and that TAK1 inhibition subverts TRAIL-mediated NF-κB activation to resume TRAIL-induced apoptosis in OCs while further enhancing MM cell death in combination with TRAIL.
Introduction
Implementation of novel agents and the availability of autologous stem-cell transplantation have revolutionized the treatment of multiple myeloma (MM); however, MM still remains incurable for the vast majority of patients. Because of the incurable nature of MM, clinical application of immunotherapies is ongoing and expected to open a new avenue for the MM treatment paradigm.
Tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) binds to its cognate death receptors (DRs) to activate caspase-8 and induce apoptosis in cancer cells.1-6 TRAIL-mediated immunotherapy is potentially an attractive therapeutic strategy against cancers, including MM.7-9 In addition, cytotoxic T cells and natural killer cells, major effectors in different types of immunotherapies, highly express TRAIL to induce tumor cell death. However, little information has been available on the effects of TRAIL on the tumor microenvironment.
Receptor activator of NF-κB ligand (RANKL), a critical mediator of osteoclastogenesis, is upregulated to extensively enhance osteoclastogenesis and bone resorption in MM.10-12 Thus, activated osteoclasts (OCs) in turn enhance MM growth, thereby forming a vicious cycle between MM tumor expansion and osteoclastic bone destruction.13,14 OCs are not merely bone resorbing cells, but rather facilitators for tumor growth; therefore, OCs should be targeted to improve treatment efficacy, especially in MM expanding in the bone marrow with enhanced bone resorption. However, the effects of TRAIL on osteoclastogenesis enhanced in MM remain largely unknown. The present study was therefore undertaken to clarify the impact of TRAIL on osteoclastogenesis and the MM-OC interaction.
We demonstrated here that TRAIL did not induce apoptosis, but rather facilitated RANKL-induced osteoclastogenesis along with upregulation of cellular FLICE inhibitory protein (c-FLIP), an endogenous inhibitor for caspase-8, in mouse RAW264.7 cells or bone marrow macrophages (BMMs). Although TRAIL induced death in MM cells through death-inducing signaling complex (DISC) formation and caspase-8 activation, TRAIL did not form the DISC in OCs, and instead facilitated complex II formation with the phosphorylation of transforming growth factor β–activated kinase-1 (TAK1) thereby activating NF-κB in OCs. However, TAK1 inhibition abrogated the TRAIL-induced NF-κB activation and c-FLIP induction to trigger apoptosis in OCs while potentiating TRAIL-induced apoptosis in MM cells. These observations provide a rationale for therapeutic strategies of TRAIL agonists in combination with TAK1 inhibition for cancers with osteoclastic bone destruction such as MM.
Materials and methods
Reagents
The following reagents were purchased from the indicated manufacturers: rabbit polyclonal anti-TAK1, mouse-specific caspase-8 antibody, rabbit monoclonal anti-c-FLIP, c-fos, RIP1, phosphorylated IκBα, cleaved caspase-8, mouse-specific cleaved caspase-8 antibody, mouse monoclonal anti-human caspase-8 antibody, horseradish peroxidase–anti-rabbit immunoglobulin G (IgG), and anti-mouse IgG from Cell Signaling Technology (Beverly, MA); rabbit polyclonal anti-phosphorylated TAK1, DR5 antibody, rabbit monoclonal anti-Pim-2, and TRAF2 antibody from Abcam (Cambridge, UK); mouse monoclonal anti-β-actin antibody and terameprocol (TMP) from Sigma-Aldrich (St. Louis, MO); mouse monoclonal antibodies against NFATc1, cathepsin K (CTSK), Sp1, and FADD, and the NF-κB inhibitor BAY 11-7082 from Santa Cruz Biotechnology Inc. (Santa Cruz, CA); TAK1 inhibitor, LLZ 1640-2 from BioAustralis (Smithfield, NSW, Australia); recombinant human TRAIL, human macrophage colony-stimulating factor (M-CSF) from Wako (Osaka, Japan); Z-lle-Glu(O-ME)-Thr-Asp(O-Me) uoromethyl ketone (Z-IETD-FMK) from Tonbo Biosciences (San Diego, CA); and human soluble RANKL (sRANKL) from Oriental Yeast Co. Ltd (Shiga, Japan).
MM cells and cell culture
The human MM cell lines RPMI 8226, U266-B1, and MM.1S and the murine preosteoclastic cell line RAW264.7 were obtained from the American Type Culture Collection (Rockville, MD). TSPC-1 was established in our laboratory.15 The human MM cell lines INA-6 were kindly provided by Renate Burger (University of Kiel, Kiel, Germany). The mouse MM cell line 5TGM1 was a kind gift from Gregory R. Mundy (Vanderbilt Center for Bone Biology, Vanderbilt University, Nashville, TN). Cells were cultured in RPMI 1640 medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham MA), penicillin G at 50 μg/mL and streptomycin at 50 μg/mL.
OC formation
The generation of OCs from RAW264.7 cells16 or mouse bone marrow cells17 was performed as previously described. The cells were cultured in Eagle's minimal essential medium α modification (Sigma-Aldrich) supplemented with 10% FBS, l-glutamine, and 50 μg/mL penicillin/streptomycin. RAW264.7 cells were cultured for 4 days in the presence of sRANKL at 25 to 50 ng/mL to generate mature OCs. Whole bone marrow cells were harvested from the femur of C57BL/6J mice (CLEA, Tokyo, Japan), and nonadherent cells were cultured with M-CSF (10 ng/mL) for 3 days to generate mouse BMMs. The BMMs were then cultured for 7 days with M-CSF (20 ng/mL) and sRANKL (50 ng/mL) to generate OCs. Human peripheral blood mononuclear cells (PBMCs) were isolated from 3 healthy donors as previously described15 and cultured for 14 days in the presence of M-CSF (10 ng/mL) and sRANKL (50 ng/mL) to generate OCs. Bone marrow mononuclear cells were isolated from fresh bone marrow aspirates of 2 patients with MM as previously described15 and cultured with M-CSF (10 ng/mL) for 3 days; the cells were then cultured for 14 days in the presence of M-CSF (10 ng/mL) and sRANKL (50 ng/mL) to generate OCs. To investigate the effects of TRAIL on osteoclatogenesis and bone resorption, we pretreated RAW264.7 cells with sRANKL (50 ng/mL) for 2 days, mouse BMMs with M-CSF (20 ng/mL) and sRANKL (50 ng/mL) for 2 days, and human PBMCs and patients’ bone marrow cells with M-CSF (10 ng/mL) and sRANKL (50 ng/mL) for 4 days. After the pretreatment, culture media were changed. The cells were then cultured in the presence or absence of TRAIL. Tartrate-resistant acid phosphatase (TRAP)–positive cells were detected with a Leukocyte Acid Phosphatase Assay kit (Sigma-Aldrich). TRAP-positive cells containing 3 or more nuclei were counted under a light microscope (BX50; Olympus, Tokyo, Japan). The cells were cultured in quadruplicate for each treatment on OC differentiation. All procedures involving human specimens were performed with written informed consent according to the Declaration of Helsinki and using a protocol approved by the Institutional Review Board for human protection.
Bone resorption assay
Bone resorption assay was performed using Corning Osteo-Assay Surface 96-well plates (Cat. No. 3989; Corning, Lowell, MA), as we described previously.16 The areas of dentin resorption were determined using image analysis techniques (National Institutes of Health ImageJ system; http://imagej.nih.gov/ij/), after attached cells were removed from the slides using 6% sodium hypochlorite.
Immunofluorescence staining
For immunofluorescence staining, RAW264.7 cells were cultured with sRANKL (50 ng/mL) for 3 days on glass bottom dishes. The cells were fixed for 10 minutes with 4% formaldehyde in phosphate-buffered saline (PBS) and permeabilized with 0.1% Triton X-100 in PBS. After blocking with 5% goat serum for 30 minutes, the cells were incubated with anti-DR5 antibody (1:100), anti-p65 and anti-CTSK antibody (1:100) overnight at 4°C. The cells were washed in PBS and incubated with Alexa Fluor 594-conjugated anti-rabbit IgG and Alexa Fluor 488–conjugated anti-mouse IgG secondary antibodies (1:100 in PBS; Thermo Fisher Scientific) for 1 hour. The F-actin ring was visualized with Alexa Fluor 488-conjugated phalloidin (Thermo Fisher Scientific). The nuclei were stained with Hoechst 33342 (Dojindo, Kumamoto, Japan) or 4’,6-diamino-2-phenylindole (Thermo Fisher Scientific). The wide field-of-view fluorescence images were examined using a fluorescence microscope (BX50, Olympus)
Reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was extracted using TRIZOL reagent (Gibco BRL, Rockville, MD). Two micrograms of total RNA was reverse-transcribed with Superscript II (Gibco) in a 20 μL reaction solution. The RT-PCR products were used for subsequent PCR analysis with 23 to 30 cycles of 95°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds. The following primer sequences were used: mouse DR5 F: AGTAGTGCTGCTGATTGGAG and R: CCTGTTTTCTGAGTCTTGCC, human DR4 F: TGGCACACAGCAATGGGAACATAG and R: GAAACACACCCTGTCCATGCACTT, DR5 F: TGAGTCTGCTCTGATCACCCAACA and R: ACTTCCGGCACATCTCAGGAGAAT, mouse c-FLIP F: AGTCCAGCCAAGGAGCAAGA and R: TGTAAGTCTCTTCACGGATG, human c-FLIP F: AATTCAAGGCTCACAAGCGA and R: GGCAGAAACTCTGCTGTTCC, and GAPDH F: TGTCTTCACCACCATGGAGAAGG and R: GTGGATGCAGGGATGATGTTCTG.
Real-time PCR
Each complementary DNA sample was amplified using SYBR Premix EX Taq II (Takara Bio Inc., Shiga, Japan) on the 7300 Real-time PCR System (Thermo Fisher Scientific). Briefly, the reaction conditions consisted of 2 μL of complementary DNA and 0.4 μM primers in a total volume of 20 μL. GAPDH was used as an endogenous control to normalize each sample.
Cell viability
Cell viability was determined by Cell Counting Kit-8 assay (Dojindo) according to the manufacturer’s instructions. Briefly, cells were plated on a 96-well plate and incubated with 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulphophenyl)-2H-tetrazolium monosodium salt (WST-8). After the incubation, the absorbance of each well was measured at 450 to 655 nm with an iMark microplate reader (Bio-Rad Laboratories, Hercules, CA).
TUNEL staining
To identify apoptotic cells, in situ cell death detection kit (Roche Diagnostics GmbH, Mannheim, Germany) was used. The cells were fixed with 4% paraformaldehyde for 1 hour and permeabilized with 0.1% Triton X-100 in PBS. The terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL)–positive multinucleated cells (MNCs) were counted.
Western blot analysis
Cells were collected and lysed in radioimmunoprecipitation assay (RIPA) buffer supplemented with a protease inhibitor cocktail (Santa Cruz). The cell lysates were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis on a 10% polyacrylamide gel and transferred to polyvinylidene difluoride membranes (Millpore, Billerica, MA). Membranes were blocked with 5% nonfat dry milk in tris(hydroxymethyl)aminomethane–buffered saline with 0.01% Tween 20 for 1 hour at room temperature and incubated for 16 hours at 4°C with primary antibodies. After washing, secondary horseradish peroxidase–conjugated antibody was added, and membranes were then developed, using the enhanced chemiluminescence plus western blotting detection system (GE Healthcare Life Sciences, Chicago, IL).
Immunoprecipitation
Briefly, cells were lysed in RIPA buffer supplemented with a protease inhibitor cocktail (Santa Cruz). For DR5, FADD immunoprecipitation, 1 mg of protein was incubated with 4 μg of anti-DR5 or FADD antibodies overnight at 4°C followed by the addition of 20 μL of protein A/G agarose beads (Santa Cruz) and then incubated for 1 hour at 4°C. The immune complexes were washed with RIPA buffer and subjected to western blotting.
Small interfering RNA (siRNA) transfection
RAW264.7 cells were seeded at a density of 2 × 105 cells per well on 6-well plates in Eagle's minimal essential medium α modification at 37°C with 5% CO2 overnight. c-FLIP siRNA was purchased from Santa Cruz and transfected into RAW264.7 cells using siRNA Transfection Reagent (Santa Cruz), following the manufacturer's protocol.
Statistical analysis
Statistical analysis was performed using the Student t test or 1-way analysis of variance. P ≤ .05 was considered as a significant difference. All statistics were performed using the Statistical Package for Social Sciences (SPSS 13.0 for Windows; Chicago, IL).
Results
Mature OCs express proapoptotic DRs
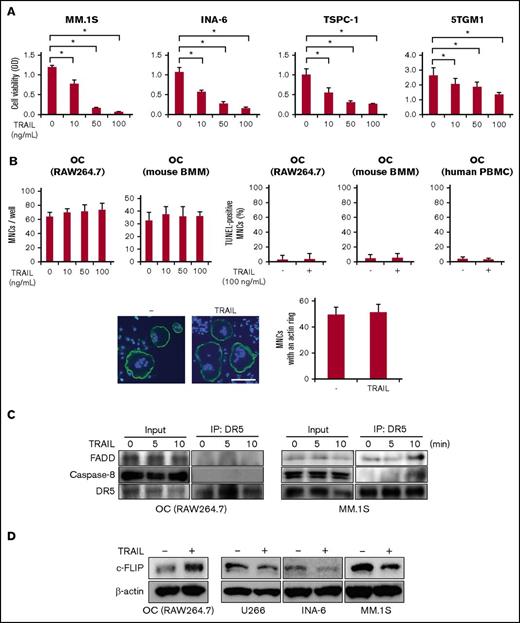
TRAIL binds to its cognate DRs to activate caspase-8 and induce apoptosis in cancer cells, which is intracellularly inhibited by c-FLIP.18 DR4 and DR5 are generally accepted as proapoptotic receptors for TRAIL in humans, whereas DR5 is regarded in mice as a single ortholog of human DR4 and DR5.19 To clarify the effects of TRAIL on OCs, we first examined the expression of DR5 in OCs in mouse bones. DR5 is expressed in CTSK-expressing OCs (Figure 1A).
Mature OCs express proapoptotic DRs. (A) CTSK and DR5 were immunostained in green and red, respectively, in a bone section. For staining OCs in bone, the tibiae of ICR-nu/nu mice (CLEA) were taken out, fixed for 2 days in 10% paraformaldehyde/PBS, and decalcified in 10% EDTA (pH 7.4) for 1 week. The tibiae specimens were embedded in paraffin, and then 5 μm thick serial sections were prepared. The deparaffinized sections were blocked with PBS containing 4.0% bovine serum albumin (PBS-BSA) for 1 hour at room temperature. To detect the localization of DR5 and CTSK in the tibia, the sections were incubated with anti-DR5 and anti-CTSK antibodies diluted 1:100 in PBS-BSA overnight at 4°C. After being washed 3 times with PBS, sections were reacted with Alexa Fluor 594–conjugated anti-rabbit IgG and Alexa Fluor 488–conjugated anti-mouse IgG secondary antibodies. The sections were observed under a fluorescence microscope (BX50, Olympus). Original magnification ×100. BM, bone marrow. (B) Murine RAW264.7 preosteoclastic cells were treated with sRANKL (50 ng/mL) for 72 hours, and DR5 mRNA expression was analyzed by RT-PCR (upper). Human monocytes from a healthy donor were treated with M-CSF (10 ng/mL) and sRANKL (50 ng/mL) for 5 days, and DR4 and DR5 mRNA expression was analyzed by RT-PCR (lower). GAPDH was used as an internal control. DR4 and DR5 mRNA expression was also determined using real-time PCR. GAPDH was used as an endogenous control to normalize each sample. Data were expressed as mean ± standard error. *P < .05. (C) Mature OCs derived from RAW264.7 cells were fixed and stained with anti-DR5 antibody (red, upper panels). F-actin and nuclei were stained with phalloidin (green) and Hoechst 33342 (Hoechst, red), respectively (lower panels). Samples were visualized with a fluorescence microscope (BX50, Olympus). Original magnification ×100. Bar represents 100 μm. (D) RAW264.7 cells (left) or mouse BMMs (right) were treated with sRANKL (50 ng/mL) for the indicated time periods, and then cell lysates were collected. The expression of DR5, c-FLIP, NFATc1, and CTSK was analyzed by western blotting. β-actin served as a loading control.
Mature OCs express proapoptotic DRs. (A) CTSK and DR5 were immunostained in green and red, respectively, in a bone section. For staining OCs in bone, the tibiae of ICR-nu/nu mice (CLEA) were taken out, fixed for 2 days in 10% paraformaldehyde/PBS, and decalcified in 10% EDTA (pH 7.4) for 1 week. The tibiae specimens were embedded in paraffin, and then 5 μm thick serial sections were prepared. The deparaffinized sections were blocked with PBS containing 4.0% bovine serum albumin (PBS-BSA) for 1 hour at room temperature. To detect the localization of DR5 and CTSK in the tibia, the sections were incubated with anti-DR5 and anti-CTSK antibodies diluted 1:100 in PBS-BSA overnight at 4°C. After being washed 3 times with PBS, sections were reacted with Alexa Fluor 594–conjugated anti-rabbit IgG and Alexa Fluor 488–conjugated anti-mouse IgG secondary antibodies. The sections were observed under a fluorescence microscope (BX50, Olympus). Original magnification ×100. BM, bone marrow. (B) Murine RAW264.7 preosteoclastic cells were treated with sRANKL (50 ng/mL) for 72 hours, and DR5 mRNA expression was analyzed by RT-PCR (upper). Human monocytes from a healthy donor were treated with M-CSF (10 ng/mL) and sRANKL (50 ng/mL) for 5 days, and DR4 and DR5 mRNA expression was analyzed by RT-PCR (lower). GAPDH was used as an internal control. DR4 and DR5 mRNA expression was also determined using real-time PCR. GAPDH was used as an endogenous control to normalize each sample. Data were expressed as mean ± standard error. *P < .05. (C) Mature OCs derived from RAW264.7 cells were fixed and stained with anti-DR5 antibody (red, upper panels). F-actin and nuclei were stained with phalloidin (green) and Hoechst 33342 (Hoechst, red), respectively (lower panels). Samples were visualized with a fluorescence microscope (BX50, Olympus). Original magnification ×100. Bar represents 100 μm. (D) RAW264.7 cells (left) or mouse BMMs (right) were treated with sRANKL (50 ng/mL) for the indicated time periods, and then cell lysates were collected. The expression of DR5, c-FLIP, NFATc1, and CTSK was analyzed by western blotting. β-actin served as a loading control.
RAW264.7 cells, an authentic murine preosteoclastic cell line, and human monocytes obtained from peripheral blood of a normal subject only weakly expressed DR5, and both DR4 and DR5 at messenger RNA (mRNA) levels, respectively (Figure 1B). However, treatment with RANKL upregulated DR5 expression in OCs derived from RAW264.7 cells. DR4 and DR5 expression was increased in OCs induced from human monocytes upon treatment with M-CSF and RANKL. Furthermore, DR5 was clearly expressed at the periphery of mature OCs with F-actin ring formation derived from RAW264.7 cells (Figure 1C). The upregulation of DR5 is consistent with the previous observation by Colucci et al.20 DR5 protein levels were increased over time in RAW264.7 cells and mouse BMMs upon treatment with RANKL in parallel with induction of NFATc1 and CTSK, markers of osteoclastic differentiation (Figure 1D). Interestingly, c-FLIP, an endogenous caspase-8 inhibitor, was also upregulated during osteoclastogenesis in parallel with the expression of DR5. These results demonstrated that the expression of DRs and the endogenous caspase-8 inhibitor c-FLIP is upregulated in OCs.
TRAIL induces cell death in MM cells but not in OCs
As previously reported,8,21 TRAIL dose dependently induced cell death in MM cell lines, MM.1S, INA-6, and TSPC-1, and murine MM cell line 5TGM1 at 10 ng/mL and more (Figure 2A). In sharp contrast, although mature OCs express DRs (Figure 1A), TRAIL did not induce cell death in OCs derived from RAW264.7 cells, murine BMMs, or human PBMCs even at 100 ng/mL (Figure 2B, upper), nor did it affect F-actin ring formation in OCs even at 500 ng/mL (Figure 2B, lower), indicating the resistance of OCs to TRAIL-induced apoptosis.
TRAIL induces cell death in MM cells but not in OCs. (A) Human MM cell lines, MM.1S, INA-6, and TSPC-1, and murine 5TGM1 MM cell line were treated in triplicate with the indicated concentrations of TRAIL for 48 hours. Cell viability was determined by WST-8 assay. Data were expressed as mean ± standard deviation (SD). *P < .05. (B) To generate mature OCs, RAW264.7 cells were cultured for 4 days with sRANKL at 50 ng/mL, BMMs for 7 days with a combination of M-CSF (20 ng/mL) and sRANKL (50 ng/mL), and human PBMCs for 14 days in the presence of M-CSF (10 ng/mL) and sRANKL (50 ng/mL). Mature OCs from RAW264.7, mouse BMMs, and human PBMCs were treated in triplicate with the indicated concentrations of TRAIL for 48 hours. Representative results are shown. The numbers of TRAP-positive MNCs were counted (upper, left). Cell viability was also determined by TUNEL assay (upper, right). Data were expressed as mean ± SD. The experiment was performed by 3 independent experiments with triplicate. Mature OCs from RAW264.7 cells were treated with TRAIL (500 ng/mL) for 48 hours, and F-actin and nuclei were stained with phalloidin (green) and 4’,6-diamino-2-phenylindole (blue), respectively (lower, left). Samples were visualized with a fluorescence microscope (BX50, Olympus). Original magnification ×100. Bar represents 100 μm. MNCs with an intact actin ring were counted (lower, right). (C) Mature OCs from RAW264.7 cells and MM.1S were starved with 1% FBS for 24 hours, and then incubated with TRAIL at 100 ng/mL for the indicated periods. For immunoprecipitation, cell lysates were collected and an equal amount of each protein lysate was incubated with anti-DR5 antibody. DISC formation was analyzed by western blotting with anti-FADD, anti-caspase-8, and anti-DR5 antibodies. (D) Mature OCs from RAW264.7 cells and the indicated MM cell lines were treated with TRAIL at 100 ng/mL for 48 hours. The protein levels of c-FLIP were analyzed by western blotting. β-actin was used as a protein loading control.
TRAIL induces cell death in MM cells but not in OCs. (A) Human MM cell lines, MM.1S, INA-6, and TSPC-1, and murine 5TGM1 MM cell line were treated in triplicate with the indicated concentrations of TRAIL for 48 hours. Cell viability was determined by WST-8 assay. Data were expressed as mean ± standard deviation (SD). *P < .05. (B) To generate mature OCs, RAW264.7 cells were cultured for 4 days with sRANKL at 50 ng/mL, BMMs for 7 days with a combination of M-CSF (20 ng/mL) and sRANKL (50 ng/mL), and human PBMCs for 14 days in the presence of M-CSF (10 ng/mL) and sRANKL (50 ng/mL). Mature OCs from RAW264.7, mouse BMMs, and human PBMCs were treated in triplicate with the indicated concentrations of TRAIL for 48 hours. Representative results are shown. The numbers of TRAP-positive MNCs were counted (upper, left). Cell viability was also determined by TUNEL assay (upper, right). Data were expressed as mean ± SD. The experiment was performed by 3 independent experiments with triplicate. Mature OCs from RAW264.7 cells were treated with TRAIL (500 ng/mL) for 48 hours, and F-actin and nuclei were stained with phalloidin (green) and 4’,6-diamino-2-phenylindole (blue), respectively (lower, left). Samples were visualized with a fluorescence microscope (BX50, Olympus). Original magnification ×100. Bar represents 100 μm. MNCs with an intact actin ring were counted (lower, right). (C) Mature OCs from RAW264.7 cells and MM.1S were starved with 1% FBS for 24 hours, and then incubated with TRAIL at 100 ng/mL for the indicated periods. For immunoprecipitation, cell lysates were collected and an equal amount of each protein lysate was incubated with anti-DR5 antibody. DISC formation was analyzed by western blotting with anti-FADD, anti-caspase-8, and anti-DR5 antibodies. (D) Mature OCs from RAW264.7 cells and the indicated MM cell lines were treated with TRAIL at 100 ng/mL for 48 hours. The protein levels of c-FLIP were analyzed by western blotting. β-actin was used as a protein loading control.
It is intriguing that TRAIL induces cell death in MM cells but not in mature OCs, although both types of cells express proapoptotic DRs. c-FLIP is generally accepted as one of the most important factors to prevent caspase-8 activation and thereby inhibit apoptosis.22 DR-bound FADD forms a DISC with caspase-8.23 c-FLIP competes with caspase-8 for binding to FADD and interferes with caspase recruitment to the DISC, thereby antagonizing DISC formation to inhibit caspase-8 activation after DR-mediated stimulation.24 c-FLIP, an endogenous caspase-8 inhibitor, was induced during osteoclastogenesis in parallel with the expression of DR5 (Figure 1D), suggesting that upregulated c-FLIP in mature OCs plays a key role in their resistance to TRAIL-induced apoptosis. Therefore, we next looked at the formation of a DISC in mature OCs derived from RAW264.7 cells and MM cells. Immunoprecipitation analyses with an anti-DR5 antibody demonstrated that TRAIL did not induce the formation of a DISC, consisting of DR5, FADD, and caspase-8, in OCs, but the DISC was formed in MM.1S cells at 10 minutes after addition of TRAIL (Figure 2C). Interestingly, c-FLIP protein levels were substantially increased in the OCs, but reduced in MM cells after the TRAIL treatment (Figure 2D). These results suggest a reverse correlation between TRAIL-induced cell death and c-FLIP levels.
c-FLIP knockdown impairs osteoclastogenesis and resumes TRAIL-induced apoptosis in mature OCs
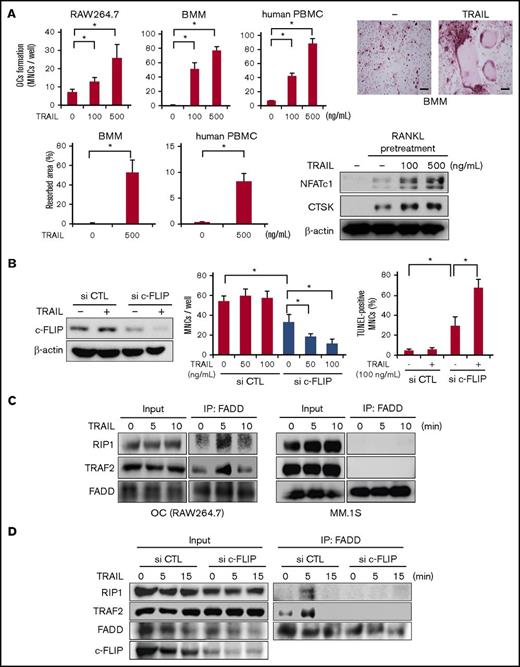
As DR5 expression was upregulated during osteoclastogenesis (Figure 1D) and because TRAIL does not induce cell death in OCs (Figure 2B), we examined whether TRAIL affects osteoclastogenesis by itself. RAW264.7 cells or mouse BMMs were treated with RANKL for 2 days to upregulate their DR5 expression, and thereafter deprived of RANKL and cultured with TRAIL. TRAIL was able to dose dependently increase the formation of TRAP-positive MNCs, namely mature OCs, in the absence of RANKL (Figure 3A). Similar results were obtained in osteoclastogenesis from PBMCs from normal human donors (Figure 3A) and bone marrow mononuclear cells from patients with MM (supplemental Figure 1A). Bone resorption activity was also increased in a similar fashion to the OC formation by TRAIL (Figure 3A). Emergence of NFATc1 and CTSK expression in the mouse BMMs also indicated the induction of osteoclastogenesis upon treatment with TRAIL (Figure 3A). These results demonstrated that osteoclastic lineage cells use TRAIL to facilitate their osteoclastogenesis. In addition, TRAIL was able to stimulate mature OCs to slightly enhance bone resorptive activity (supplemental Figure 1B, left). When we compared the distribution of nuclei per cell for OCs generated by TRAIL and sRANKL at 100 ng/mL, sRANKL and TRAIL similarly enhanced the formation of TRAP-positive MNCs with 3 to 5 nuclei as well as those with >5 (supplemental Figure 1B, right).
c-FLIP knockdown impairs osteoclastogenesis and resumes TRAIL-induced apoptosis in mature OCs. (A) RAW264.7 cells were pretreated with sRANKL (50 ng/mL) for 48 hours, mouse BMMs were pretreated with M-CSF (20 ng/mL) and sRANKL (50 ng/mL) for 48 hours, and human PBMCs were pretreated in quadruplicate with M-CSF (10 ng/mL) and sRANKL (50 ng/mL) for 4 days. After the pretreatment, the media were changed, and TRAIL was added at the indicated concentrations. RAW264.7 cells, mouse BMMs, and human PBMCs were further cultured for 4, 7, and 14 days, respectively. The numbers of TRAP-positive MNCs were counted. Data were expressed as mean ± SD. *P < .05. The representative photos of TRAP staining are shown. Bars represent 100 μm. Original magnification ×100. Bone resorption activity was also analyzed using Osteo assay plates. Resorption area was visualized by von Kossa staining, and observed using a light microscope (BX50, Olympus). The results were expressed as % area of bone resorption. The cell lysates were collected, and NFATc1 and CTSK were analyzed by western blotting. β-actin served as a loading control. (B) c-FLIP siRNA or control siRNA was transfected into RAW264.7 cells. After the transfection, the cells were treated with sRANKL (50 ng/mL) for 4 days. The cells were then washed and treated with the indicated concentrations of TRAIL for 48 hours. The protein levels of c-FLIP were analyzed by western blotting (left). β-actin was used as a protein loading control. The numbers of TRAP-positive MNCs were counted. To identify apoptotic cells, the cells were stained with TUNEL staining. The TUNEL-positive multinuclear cells were counted. Data were expressed as mean ± SD. *P < .05. (C) Mature OCs from RAW264.7 cells and MM.1S cells were starved with 1% FBS for 24 hours and then incubated with TRAIL at 100 ng/mL for the indicated time periods. The cell lysates were collected and an equal amount of each protein lysate was incubated with anti-FADD antibody. Complex II formation was analyzed by western blotting with anti-RIP1, anti-TRAF2, and anti-FADD antibodies. (D) RAW264.7 cells transfected either c-FLIP or control siRNA were treated with sRANKL (50 ng/mL) for 48 hours. The cells were then washed and treated with at 100 ng/mL for the indicated time periods. The complex II formation was analyzed.
c-FLIP knockdown impairs osteoclastogenesis and resumes TRAIL-induced apoptosis in mature OCs. (A) RAW264.7 cells were pretreated with sRANKL (50 ng/mL) for 48 hours, mouse BMMs were pretreated with M-CSF (20 ng/mL) and sRANKL (50 ng/mL) for 48 hours, and human PBMCs were pretreated in quadruplicate with M-CSF (10 ng/mL) and sRANKL (50 ng/mL) for 4 days. After the pretreatment, the media were changed, and TRAIL was added at the indicated concentrations. RAW264.7 cells, mouse BMMs, and human PBMCs were further cultured for 4, 7, and 14 days, respectively. The numbers of TRAP-positive MNCs were counted. Data were expressed as mean ± SD. *P < .05. The representative photos of TRAP staining are shown. Bars represent 100 μm. Original magnification ×100. Bone resorption activity was also analyzed using Osteo assay plates. Resorption area was visualized by von Kossa staining, and observed using a light microscope (BX50, Olympus). The results were expressed as % area of bone resorption. The cell lysates were collected, and NFATc1 and CTSK were analyzed by western blotting. β-actin served as a loading control. (B) c-FLIP siRNA or control siRNA was transfected into RAW264.7 cells. After the transfection, the cells were treated with sRANKL (50 ng/mL) for 4 days. The cells were then washed and treated with the indicated concentrations of TRAIL for 48 hours. The protein levels of c-FLIP were analyzed by western blotting (left). β-actin was used as a protein loading control. The numbers of TRAP-positive MNCs were counted. To identify apoptotic cells, the cells were stained with TUNEL staining. The TUNEL-positive multinuclear cells were counted. Data were expressed as mean ± SD. *P < .05. (C) Mature OCs from RAW264.7 cells and MM.1S cells were starved with 1% FBS for 24 hours and then incubated with TRAIL at 100 ng/mL for the indicated time periods. The cell lysates were collected and an equal amount of each protein lysate was incubated with anti-FADD antibody. Complex II formation was analyzed by western blotting with anti-RIP1, anti-TRAF2, and anti-FADD antibodies. (D) RAW264.7 cells transfected either c-FLIP or control siRNA were treated with sRANKL (50 ng/mL) for 48 hours. The cells were then washed and treated with at 100 ng/mL for the indicated time periods. The complex II formation was analyzed.
As c-FLIP expression is induced during osteoclastogenesis in parallel with the expression of DR5 (Figure 1D), c-FLIP is suggested to be involved in osteoclastogenesis and resistance to TRAIL-induced cell death in OCs. To clarify the role of c-FLIP in osteoclastogenesis and OC survival, we examined the effects of c-FLIP knockdown on RANKL-induced osteoclastogenesis and the susceptibility of OCs to TRAIL. c-FLIP siRNA was transfected into RAW264.7 cells, and then the cells were cultured with RANKL to give rise to OCs. The c-FLIP knockdown substantially reduced c-FLIP protein levels and abolished TRAIL-induced c-FLIP upregulation in the OCs (Figure 3B, left). The c-FLIP knockdown alone reduced osteoclastogenesis by RANKL (Figure 3B, middle). Addition of TRAIL was able to dose dependently induce cell death in the OCs differentiated from RAW264.7 cells with c-FLIP knockdown (Figure 3B, middle and right). These results suggest the critical role of c-FLIP in OC differentiation and maturation by RANKL, as well as the resistance of OCs to TRAIL-induced apoptosis.
FADD has been demonstrated to interact with TRAF2 and RIP1 to form a secondary complex (complex II), but not DISC, as well as to initiate canonical NF-κB activation in a portion of transformed cells in a context-dependent manner.25 Therefore, we investigated the effects of TRAIL on complex II formation in OCs. Immunoprecipitation analyses with an anti-FADD antibody revealed that upon TRAIL treatment, FADD formed a complex with TRAF2 and RIP1, namely complex II, in OCs at 5 minutes but not in MM cells (Figure 3C). However, TRAIL did not induce complex II formation in OCs differentiated from RAW264.7 cells with c-FLIP knockdown, indicating the critical role of c-FLIP in induction of complex II formation by TRAIL in OCs (Figure 3D). These results collectively suggest that the upregulation of c-FLIP facilitates osteoclastogenesis and confers resistance to TRAIL-induced cell death in OCs.
TRAIL activates the TAK1/NF-κB signaling in OCs
TAK1 plays a critical role in activation of the canonical NF-κB and MAPK pathways downstream of complex II.26 Therefore, we next looked at the effects of TRAIL on the activation of TAK1 and thereby NF-κB signaling in RAW264.7 cell-derived OCs with DR5 expression. Consistent with the formation of complex II (Figure 3C), treatment with TRAIL induced the phosphorylation of TAK1 in OCs but not in MM cells (Figure 4A). The induction of TAK1 phosphorylation by TRAIL was followed by the phosphorylation of IκBα in OCs (Figure 4B, left). Nuclear localization of p65, a subunit of NF-κB, was also confirmed in OCs upon the TRAIL treatment (Figure 4B, right). However, the TAK1 inhibitor LLZ1640-2 abolished the TRAIL-induced phosphorylation of IκBα (Figure 4B, left) and the nuclear localization of p65 (Figure 4B, right) in OCs.
TRAIL activates TAK1/NF-κB signaling in OCs. (A) Mature OCs from RAW264.7 cells and human MM cell lines, MM.1S and INA-6, were starved with 1% FBS for 24 hours and then incubated with TRAIL at 100 ng/mL for the indicated time periods. The expression of phosphorylated TAK1 (p-TAK1) and TAK1 was detected by western blotting. β-actin was used as a protein loading control. (B) Mature OCs from RAW264.7 cells were starved with 1% FBS for 24 hours and then incubated with or without the TAK1 inhibitor LLZ1640-2 (LLZ) at 300 nM, followed by the addition of TRAIL at 100 ng/mL for the indicated time periods. The expression of p-TAK1, TAK1, and phosphorylated IκBα (p-IκBα) was detected by western blotting (left). β-actin was used as a protein loading control. To observe p65 localization, the cells were fixed and stained with anti-p65 antibody (green), and their nuclei were stained with Hoechst 33342 (red). These images are taken by wide field-of-view fluorescence microscope (BX50, Olympus). Bar represents 100 μm. (C) Mature OCs from RAW264.7 cells were cultured in the presence or absence of LLZ at 300 nM for 48 hours. TRAIL was added as indicated. The protein levels of Pim-2 were analyzed by western blotting. β-actin was used as a protein loading control. (D) Mature OCs from RAW264.7 cells were cultured in the presence or absence of TRAIL (100 ng/mL) for 6 and 24 hours to perform RT-PCR and western blotting, respectively. LLZ at 300 nM and BAY11-7085 at 20 nM (upper), and the Sp1 inhibitor TMP at 100 μM (lower) were added as indicated. c-FLIP expression was analyzed. c-FLIP mRNA expression was also analyzed by real-time PCR. The results are expressed using Δ-Δ Ct method. GAPDH and β-actin served as loading controls, respectively.
TRAIL activates TAK1/NF-κB signaling in OCs. (A) Mature OCs from RAW264.7 cells and human MM cell lines, MM.1S and INA-6, were starved with 1% FBS for 24 hours and then incubated with TRAIL at 100 ng/mL for the indicated time periods. The expression of phosphorylated TAK1 (p-TAK1) and TAK1 was detected by western blotting. β-actin was used as a protein loading control. (B) Mature OCs from RAW264.7 cells were starved with 1% FBS for 24 hours and then incubated with or without the TAK1 inhibitor LLZ1640-2 (LLZ) at 300 nM, followed by the addition of TRAIL at 100 ng/mL for the indicated time periods. The expression of p-TAK1, TAK1, and phosphorylated IκBα (p-IκBα) was detected by western blotting (left). β-actin was used as a protein loading control. To observe p65 localization, the cells were fixed and stained with anti-p65 antibody (green), and their nuclei were stained with Hoechst 33342 (red). These images are taken by wide field-of-view fluorescence microscope (BX50, Olympus). Bar represents 100 μm. (C) Mature OCs from RAW264.7 cells were cultured in the presence or absence of LLZ at 300 nM for 48 hours. TRAIL was added as indicated. The protein levels of Pim-2 were analyzed by western blotting. β-actin was used as a protein loading control. (D) Mature OCs from RAW264.7 cells were cultured in the presence or absence of TRAIL (100 ng/mL) for 6 and 24 hours to perform RT-PCR and western blotting, respectively. LLZ at 300 nM and BAY11-7085 at 20 nM (upper), and the Sp1 inhibitor TMP at 100 μM (lower) were added as indicated. c-FLIP expression was analyzed. c-FLIP mRNA expression was also analyzed by real-time PCR. The results are expressed using Δ-Δ Ct method. GAPDH and β-actin served as loading controls, respectively.
We and others recently reported that Pim-2 is upregulated not only in MM cells but also in OCs in MM bone lesions.16,27-29 RANKL activates the NF-κB signaling and thereby transcriptionally induces Pim-2 in osteoclastic lineage cells during their osteoclastogenesis; and Pim-2 inhibition abolishes RANKL-mediated osteoclastogenesis, indicating Pim-2 as a critical mediator of osteoclastogenesis.16 Consistent with Pim-2 upregulation by NF-κB activation, treatment with TRAIL upregulated Pim-2 expression in OCs (Figure 4C). However, LLZ1640-2 abrogated the Pim-2 upregulation by TRAIL, suggesting involvement of the TRAIL/TAK1/NF-κB/Pim-2 pathway in osteoclastogenesis.
TRAIL enhanced c-FLIP expression (Figure 2D) and activated the NF-κB signaling in OCs (Figure 4B). c-FLIP has been demonstrated to be upregulated in different types of cells through NF-κB activation.18 Sp1 is known to be a transcription factor downstream of NF-κB signaling30 and to be responsible for c-FLIP gene expression.18 Consistent with these observations, the NF-κB inhibitor BAY11-7085 as well as LLZ1640-2 suppressed c-FLIP induction in OCs by TRAIL (Figure 4D, upper). Furthermore, c-FLIP expression almost completely disappeared by treatment with TMP, an inhibitor of Sp1 binding to DNA (Figure 4D, lower), suggesting a critical role of Sp1 in c-FLIP expression in OCs. These results demonstrated that TRAIL induces the TAK1/NF-κB/Sp1/c-FLIP induction axis besides upregulating the osteoclastogenic mediator Pim-2 in OCs and suggest that TRAIL activates TAK1/NF-κB signaling in OCs while mitigating DR-induced caspase-8 activation through c-FLIP induction.
TAK1 inhibition triggers TRAIL-induced death in OCs and potentiates it in MM cells
As TRAIL appears to activate the complex II/TAK1-mediated signaling pathway in OCs, we next looked at the role of TRAIL-induced TAK1 activation in the survival of OCs. As shown in Figure 2B, TRAIL alone did not affect the viability of OCs; however, TRAIL triggered and dose dependently reduced OC numbers and increased TUNEL-positive OCs in the presence of the TAK1 inhibitor LLZ1640-2 (Figure 5Ai-ii). Treatment with TRAIL upregulated c-FLIP protein levels without activation of caspase-8 in OCs (Figure 5Aiii). However, the TAK1 inhibition substantially reduced c-FLIP expression and triggered activation of caspase-8 in OCs. TRAIL substantially reduced Sp1 and c-FLIP protein levels in parallel with caspase-8 activation in the presence of the TAK1 inhibitor LLZ1640-2, which were restored by the caspase-8 inhibitor Z-IETD-FMK (Figure 5B), indicating enzymatic degradation of Sp1 and thereby c-FLIP reduction. This is consistent with our recent observation in MM cells of caspase-8-driven degradation of Sp1 protein and thereby reduction of Sp1-target gene expression.31
TAK1 inhibition triggers TRAIL-induced death in OCs and potentiates it in MM cells. (A) Mature OCs from RAW264.7 cells and mouse BMMs were cultured in the presence or absence of LLZ1640-2 (LLZ) at 300 nM for 48 hours. TRAIL was added at the indicated concentrations. (Ai) The numbers of TRAP-positive MNCs were counted. (Aii) Percent distribution of TUNEL-positive cells was also estimated within OCs derived from mouse BMMs and human PBMCs. Data were expressed as mean ± SD. *P < .05. (Aiii) The protein levels of c-FLIP, caspase-8, and cleaved caspase-8 were analyzed by western blotting. β-actin was used as a protein loading control. (B) Mature OCs from RAW264.7 cells were cultured in the presence or absence of LLZ at 150 nM for 48 hours. TRAIL at 100 ng/mL and the caspase-8 inhibitor Z-IETD-FMK at 100 μM were added as indicated. The protein levels of caspase-8, cleaved caspase-8, Sp1, and c-FLIP were analyzed by western blotting. β-actin was used as a protein loading control. (C) MM cell lines, RPMI 8226, MM.1S, and 5TGM1, were cultured in the presence or absence of LLZ at 300 nM for 48 hours. TRAIL was added at the indicated concentrations. The cell viability was measured by WST-8 cell proliferation assay. Results were expressed as the mean ± SD. *P < .05. (D) The indicated MM cell lines were cultured in the presence or absence of LLZ at 300 nM for 6 and 24 hours to perform RT-PCR and western blotting, respectively. TRAIL was added at 100 ng/mL as indicated. The protein levels of caspase-8, cleaved caspase-8, Sp1, and c-FLIP were analyzed by western blotting (left). β-actin was used as a protein loading control. Z-IETD-FMK was added at 100 μM as indicated. c-FLIP mRNA expression was analyzed by RT-PCR. GAPDH served as an internal control.
TAK1 inhibition triggers TRAIL-induced death in OCs and potentiates it in MM cells. (A) Mature OCs from RAW264.7 cells and mouse BMMs were cultured in the presence or absence of LLZ1640-2 (LLZ) at 300 nM for 48 hours. TRAIL was added at the indicated concentrations. (Ai) The numbers of TRAP-positive MNCs were counted. (Aii) Percent distribution of TUNEL-positive cells was also estimated within OCs derived from mouse BMMs and human PBMCs. Data were expressed as mean ± SD. *P < .05. (Aiii) The protein levels of c-FLIP, caspase-8, and cleaved caspase-8 were analyzed by western blotting. β-actin was used as a protein loading control. (B) Mature OCs from RAW264.7 cells were cultured in the presence or absence of LLZ at 150 nM for 48 hours. TRAIL at 100 ng/mL and the caspase-8 inhibitor Z-IETD-FMK at 100 μM were added as indicated. The protein levels of caspase-8, cleaved caspase-8, Sp1, and c-FLIP were analyzed by western blotting. β-actin was used as a protein loading control. (C) MM cell lines, RPMI 8226, MM.1S, and 5TGM1, were cultured in the presence or absence of LLZ at 300 nM for 48 hours. TRAIL was added at the indicated concentrations. The cell viability was measured by WST-8 cell proliferation assay. Results were expressed as the mean ± SD. *P < .05. (D) The indicated MM cell lines were cultured in the presence or absence of LLZ at 300 nM for 6 and 24 hours to perform RT-PCR and western blotting, respectively. TRAIL was added at 100 ng/mL as indicated. The protein levels of caspase-8, cleaved caspase-8, Sp1, and c-FLIP were analyzed by western blotting (left). β-actin was used as a protein loading control. Z-IETD-FMK was added at 100 μM as indicated. c-FLIP mRNA expression was analyzed by RT-PCR. GAPDH served as an internal control.
In contrast, TRAIL dose dependently induced death in MM cells, which was further augmented in the presence of LLZ1640-2 (Figure 5C). TRAIL was able to activate caspase-8, and substantially reduced Sp1 and c-FLIP protein levels in MM cells, which was further enhanced by the TAK1 inhibition (Figure 5D, left). TRAIL potently reduced c-FLIP mRNA expression in MM cells; however, the reduction of c-FLIP mRNA by TRAIL was abolished under the caspase-8 inhibition (Figure 5D, right; supplemental Figure 1C), indicating caspase-8–dependent reduction of c-FLIP in MM cells by TRAIL. From these results, TAK1 inhibition permits TRAIL-induced apoptosis in OCs while further enhancing TRAIL-induced MM cell death.
TRAIL disrupts MM cell–OC interaction under TAK1 inhibition
MM cells enhance osteoclastogenesis and OC activation through upregulation of RANKL expression in MM bone lesions. Cocultures with MM cells enhanced the formation of TRAP-positive MNCs, namely mature OCs, from bone marrow cells (Figure 6A). Treatment with TRAIL further enhanced osteoclastogenesis in the cocultures; however, TAK1 inhibition abolished the enhancement of osteoclastogenesis by TRAIL in the presence of MM cells.
TRAIL disrupts MM cell-OC interaction under TAK1 inhibition. (A) Murine bone marrow mononuclear cells were plated onto 24-well culture plates (1 × 106 cells per well). The cells were treated with M-CSF for 3 days, followed by sRANKL (25 ng/mL) for 2 days to generate adherent preosteoclastic cells. After washing, the adherent preosteoclastic cells were cocultured for 3 days with murine 5TGM1 MM cells at 1 × 104 cells per well. TRAIL at 100 ng/mL and LLZ at 300 nM were added as indicated. TRAP-positive MNCs (5 or more nuclei) were counted as OCs. Data were expressed as mean ± standard error. *P < .05. Microscopic images of TRAP staining in representative cultures are shown (lower panels). Original magnification ×100. Bar represents 100 μm. (B) Human OCs were generated in 24-well culture plates with M-CSF and sRANKL from PBMCs from a healthy donor. The OCs were tightly attached on the culture plates. MM cell lines, INA-6, RPMI 8226, and MM.1S, were cultured alone or cocultured with the OCs for 48 hours. TRAIL at 100 ng/mL and LLZ at 300 nM were added as indicated. MM cells were collected, and their viability was measured by WST-8 assay. Data were expressed as mean ± SD. *P < .05.
TRAIL disrupts MM cell-OC interaction under TAK1 inhibition. (A) Murine bone marrow mononuclear cells were plated onto 24-well culture plates (1 × 106 cells per well). The cells were treated with M-CSF for 3 days, followed by sRANKL (25 ng/mL) for 2 days to generate adherent preosteoclastic cells. After washing, the adherent preosteoclastic cells were cocultured for 3 days with murine 5TGM1 MM cells at 1 × 104 cells per well. TRAIL at 100 ng/mL and LLZ at 300 nM were added as indicated. TRAP-positive MNCs (5 or more nuclei) were counted as OCs. Data were expressed as mean ± standard error. *P < .05. Microscopic images of TRAP staining in representative cultures are shown (lower panels). Original magnification ×100. Bar represents 100 μm. (B) Human OCs were generated in 24-well culture plates with M-CSF and sRANKL from PBMCs from a healthy donor. The OCs were tightly attached on the culture plates. MM cell lines, INA-6, RPMI 8226, and MM.1S, were cultured alone or cocultured with the OCs for 48 hours. TRAIL at 100 ng/mL and LLZ at 300 nM were added as indicated. MM cells were collected, and their viability was measured by WST-8 assay. Data were expressed as mean ± SD. *P < .05.
We and others demonstrated that MM cells and OCs interact with each other to augment their growth and activity, thereby forming a vicious cycle that leads to extensive bone destruction and MM expansion.13,14,32 Therefore, we next examined the impact of TRAIL in combination with TAK1 inhibition on the viability of MM cells in cocultures with OCs. Cocultures with OCs lowered TRAIL-induced MM cell death (Figure 6B). However, addition of the TAK1 inhibitor LLZ1640-2 substantially reduced MM cell viability in combination with TRAIL in cocultures with OCs, although there was still protection by OCs from the MM cell killing observed when both TRAIL and LLZ1640-2 are present. These results suggest that TAK1 inhibition may disrupt MM cell-OC interaction and potentiate TRAIL’s antitumor effects while also converting TRAIL to an anti–bone resorptive agent.
Discussion
MM has a unique propensity to develop and expand almost exclusively in the bone marrow and generates destructive bone disease. MM cells enhance osteoclastogenesis33,34 while suppressing osteoblastogenesis,35-39 leading to devastating bone destruction with rapid loss of bone. Along with the progression of bone disease, the bone marrow microenvironment is skewed by MM cells, which underlies the unique pathophysiology of MM and confers aggressiveness and drug resistance in MM cells.40,41 Although MM cells perturb bone metabolism to develop bone destruction, cross talk between MM cells and the microenvironment in bone lesions leads to a progressive vicious cycle phase of tumor growth and bone destruction.13,14,32 The present study demonstrated that osteoclastic lineage cells use TRAIL for their differentiation into OCs through tilting TRAIL-mediated caspase-8–dependent apoptosis into activation of the NF-κB signaling. Therefore, use of TRAIL agonists to treat patients with MM and other cancers with osteolytic bone metastases may result in enhanced progression of osteolytic bone destruction.
There have been conflicting data in the literature as to whether TRAIL increases the formation of human and murine OCs from their precursors and induction of apoptosis in them. Some reported that TRAIL induces apoptosis in OCs and suppresses osteoclastogenesis.20,42,43 Others demonstrated that TRAIL induces osteoclastogenesis.44-46 We obtained the results showing facilitation of RANKL-primed osteoclastogenesis without induction of OC death by TRAIL. According to our present results, RANKL activates TAK1 and induces c-FLIP expression in OCs and their precursors, which is able to protect them from TRAIL-induced apoptosis. In such a condition, TRAIL further upregulates c-FLIP expression. Further, TRAIL was also found to facilitate osteoclastogenesis even in the presence of RANKL (supplemental Figure 1D). However, because antagonism between TRAIL and RANKL has been demonstrated,47 we need to dissect the molecular mechanisms of the combinatory effects of TRAIL and RANKL. Checking the activation status of TAK1 and thereby upregulation of c-FLIP in each individual experimental condition may help to explain the conflicting results of TRAIL effects on osteoclastogenesis and the viability of OCs.
As RANKL induces the expression of DRs and c-FLIP in osteoclastic lineage cells along with osteoclastogenesis (Figure 1D), an anti-RANKL antibody may ameliorate TRAIL’s osteoclastogenic actions. Therefore, combinatory treatment with an anti-RANKL antibody warrants further study. However, in the present study, we demonstrated that TAK1 inhibition subverts TRAIL-mediated NF-κB activation to regain TRAIL-induced apoptosis in OCs while further enhancing MM cell death in combination with TRAIL. Combination of TAK1 inhibitors with TRAIL agonists may become an effective therapeutic option yielding an improved clinical outcome for MM.
DRs can mediate diverse signals in a cell type–specific and context-dependent manner.48 Here, we dissected the mechanisms of resistance to TRAIL-induced apoptosis in OCs. TRAIL was demonstrated to induce the formation of FADD-driven complex II in OCs to activate TAK1 and its downstream signaling pathways, including the NF-κB pathway. During osteoclastogenesis, the expression of c-FLIP, an endogenous inhibitor of DISC formation, was substantially increased in parallel with DR expression. The c-FLIP upregulation appears to block DISC formation to allow FADD-driven complex II formation. Therefore, c-FLIP upregulation is suggested to primarily play an important role in rendering OCs activated and refractory to TRAIL-induced apoptosis.
The activation of the TRAIL/TAK1/NF-κB pathway by TRAIL not only stimulates osteoclastogenesis and OC survival but also enhances c-FLIP expression in OCs, which further suppresses TRAIL-induced apoptosis. Intriguingly, MM cells tend to undergo apoptosis along with reducing c-FLIP in response to TRAIL in sharp contrast to OCs.
We recently reported that caspase-8 activation enzymatically degrades the transcription factor Sp1 and thereby reduces its target gene expression in MM cells.31 We demonstrated here that this c-FLIP reduction in MM cells is largely because of caspase-8–mediated enzymatic degradation of Sp1, a transcription factor responsible for c-FLIP gene expression, because treatment with a caspase-8 inhibitor almost completely restored the c-FLIP levels reduced in MM cells by TRAIL (Figure 5D). Therefore, TRAIL-induced activation of caspase-8 is suggested to lead to further potentiation of the DR/caspase-8–mediated apoptotic pathway in MM cells. Of note, TRAIL was able to reduce c-FLIP levels and activate caspase-8 in OCs under TAK1 inhibition (Figure 5A). Although c-FLIP production is transcriptionally upregulated to inhibit DISC formation and thereby apoptosis in OCs by TRAIL, TAK1 inhibition is able to suppress c-FLIP production through disrupting complex II–mediated NF-κB signaling and triggering the caspase-8–mediated degradation of Sp1, which causes induction of apoptosis in OCs.
Although c-FLIP expression was increased along with RANKL-mediated OC differentiation, c-FLIP knockdown significantly impaired RANKL-mediated osteoclastogenesis. Therefore, c-FLIP is suggested to play a critical role in osteoclastogenesis other than inhibition of the DISC formation in osteoclastic lineage cells. c-FLIP has been reported to interact with intracellular signaling components, such as TRAF2 and RIP1, to perform cellular functions in different types of cells,25 which warrants for further study on the functional role of c-FLIP in osteoclastogenesis.
In conclusion, TAK1 inhibition subverts TRAIL-mediated NF-κB activation to restore TRAIL-induced apoptosis in OCs while further enhancing MM cell death in combination with TRAIL, providing a rationale for therapeutic strategies of TRAIL agonists in combination with TAK1 inhibition for cancers with osteoclastic bone destruction such as MM.
The full-text version of this article contains a data supplement.
Acknowledgments
This work was supported in part by Grants-in-Aid for Young Scientists (B) (15K19551 [H.M.], 15K20385 [R.A.], and 16K18420 [S.N.]); Grants-in-Aid for Scientific Research (C) (16K11504 [J.T.] and 26461422 [M.A.]); and a Grant-in-Aid for Cancer Research (17-16) (M.A.) from the Ministry of Health, Labor and Welfare of Japan.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authorship
Contribution: H.T., J.T., and M.A. designed the research and conceived the project; H.T., J.T., A.O., R.A., M.H., A.B.-E., K.W., M.I., T.H., S.F., K.K., K.S., S.N., H.M., K.K., S.Y., K.A., and I.E. performed the experiments; H.T., J.T., E.T., T.M., and M.A. analyzed and interpreted the data; and H.T., J.T., and M.A. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Masahiro Abe, Department of Hematology, Endocrinology and Metabolism, Tokushima University Graduate School, 3-18-15 Kuramoto, Tokushima 770-8503, Japan; e-mail: masabe@tokushima-u.ac.jp.