Key Points
WES can be applied for precise RH genotyping, detection of new or uncommon variants, and determination of RHD zygosity.
An altered RH genotype is a risk factor for Rh alloimmunization in patients with sickle cell anemia.
Abstract
RH genes are highly polymorphic and encode the most complex of the 35 human blood group systems. This genetic diversity contributes to Rh alloimmunization in patients with sickle cell anemia (SCA) and is not avoided by serologic Rh-matched red cell transfusions. Standard serologic testing does not distinguish variant Rh antigens. Single nucleotide polymorphism (SNP)–based DNA arrays detect many RHD and RHCE variants, but the number of alleles tested is limited. We explored a next-generation sequencing (NGS) approach using whole-exome sequencing (WES) in 27 Rh alloimmunized and 27 matched non-alloimmunized patients with SCA who received chronic red cell transfusions and were enrolled in a multicenter study. We demonstrate that WES provides a comprehensive RH genotype, identifies SNPs not interrogated by DNA array, and accurately determines RHD zygosity. Among this multicenter cohort, we demonstrate an association between an altered RH genotype and Rh alloimmunization: 52% of Rh immunized vs 19% of non-immunized patients expressed variant Rh without co-expression of the conventional protein. Our findings suggest that RH allele variation in patients with SCA is clinically relevant, and NGS technology can offer a comprehensive alternative to targeted SNP-based testing. This is particularly relevant as NGS data becomes more widely available and could provide the means for reducing Rh alloimmunization in children with SCA.
Introduction
Transfusion management of patients with sickle cell anemia (SCA) remains a significant challenge in clinical transfusion medicine.1,2 Most comprehensive sickle cell programs aim to reduce alloimmunization by providing Rh D, C, E, and K phenotype-matched red blood cells, meaning that units selected for transfusion are antigen-negative when the patient lacks the antigen.3 Less often, extended phenotype matching is included for Duffy (Fya/b), Kidd (Jka/b), and MNS (Ss) blood groups.4 Despite these strategies, alloimmunization to Rh antigens remains problematic even for patients transfused with phenotypic Rh antigen–matched red cells from African American donors who have similar blood group antigen profiles.5 The frequency of RH variant alleles in this patient population contributes to the persistence of Rh alloimmunization through expression of partial or novel Rh antigen expression.5,6
The RH locus comprises 2 homologous genes, RHD and RHCE, which encode the D antigen and the CcEe antigens in various combinations (ce, cE, Ce, CE), respectively. The RH genes are highly polymorphic, particularly in individuals of African ancestry. Standard serologic Rh typing does not detect the many Rh antigenic variations commonly expressed on erythrocytes from this population. These include the absence of common Rh epitopes termed “high-frequency antigens” (eg, hrB, hrS, HrB)7 and/or expression of novel Rh epitopes called “low-frequency antigens” (eg, V, VS, Goa) on erythrocytes.8 Altered RH alleles encode weak and/or partial expression of D, C, e and, less often, c and E antigens. The term “partial” describes red cells that lack some common epitopes associated with expression of an antigen. As a result, patients with SCA can produce antibodies to foreign Rh epitopes they lack as well as to Rh epitopes that differ from those expressed on their own cells. It is often difficult to define the precise Rh epitope specificities of the antibodies in the plasma of immunized patients with altered RH alleles, because the reactivity pattern in laboratory testing can mirror that seen with an autoantibody, yet can cause destruction of transfused cells.
We previously reported that nearly 90% of patients with SCA inherit at least 1 RH allele that differs from those commonly found in Europeans.5 Standard serologic testing does not detect altered Rh antigen expression. Currently available DNA array and laboratory-developed assays identify the most common polymorphic RH alleles but cannot detect all RH variation. Next-generation sequencing (NGS) with either whole-genome sequencing (WGS) or whole-exome sequencing (WES) is rapidly moving toward routine practice for patients with chronic diseases and should allow comprehensive analysis of RH genetic variation. It was recently shown that NGS-based mapping of conventional complementary DNA–annotated erythrocyte and platelet antigens enables accurate prediction from WGS data for most blood group systems.9,10 Alignment of NGS sequence reads is more difficult in the duplicated and homologous gene families, including RHD and RHCE. Application of NGS using a non-targeted WES approach for RH genotyping of a cohort of patients with SCA who are known to have increased RH genetic diversity has not previously been investigated.
The overarching goal of this study was to investigate the utility of WES for comprehensive RH genotyping in children with SCA. We analyzed WES data from 54 children with SCA (27 Rh alloimmunized and 27 non-alloimmunized) randomly assigned in the SWiTCH (Stroke With Transfusions Changing to Hydroxyurea; NCT 00122980) study.11 Specific goals were to compare the accuracy of a WES approach to current single nucleotide polymorphism (SNP)–based testing and to determine associations of Rh antibody production with the RH genotype.
Methods
Study population, phenotyping, and DNA extraction
SWiTCH had an average age at enrollment of 13 years with approximately 7 years of transfusion duration.11 A total of 161 patients were enrolled from 2006 to 2010, all with at least 18 months of chronic transfusions to prevent secondary stroke. Most participating institutions provided prophylactic C-, E-, K-matched transfusions. All patients had their transfusion history and antibody records reviewed, and data were recorded at study entry. We identified 27 patients in the randomized cohort (n = 134) with known Rh antibodies at enrollment. We paired a matched control for each alloimmunized patient with an unrelated patient of similar age who had a similar number of years of transfusion exposure and the same sex when possible but with no alloimmunization (Table 1). Patients with RH and RHAG genotypes were compared with patients from Children’s Hospital of Philadelphia who received prophylactic C-, E-, K-matched transfusions from minority donors. Patient enrollment, clinical data collection, and sample analysis were conducted with approval by the institutional review boards at Baylor College of Medicine and CHP. Genomic DNA was extracted from whole blood by using standard methods and was stored at −20°C until analysis.
Patient characteristics
| Characteristic . | Overall SWiTCH . | Rh immunized . | Controls . | P . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean ± SD . | No. . | Female (%) . | Mean ± SD . | No. . | Female (%) . | Mean ± SD . | No. . | Female (%) . | ||
| No. of patients | 134 | 46 | 27 | 67 | 27 | 56 | NA | |||
| Age, y | ||||||||||
| At enrollment | 13.2 ± 3.9 | 14.9 ± 3.5 | 14.2 ± 3.1 | .44 | ||||||
| At first stroke | 5.9 ± 2.9 | 7.2 ± 3.9 | 6.6 ± 3.0 | .53 | ||||||
| Duration of transfusions, y | 7.2 ± 3.7 | 7.6 ± 4.6 | 7.6 ± 4.2 | 1.0 | ||||||
| Characteristic . | Overall SWiTCH . | Rh immunized . | Controls . | P . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean ± SD . | No. . | Female (%) . | Mean ± SD . | No. . | Female (%) . | Mean ± SD . | No. . | Female (%) . | ||
| No. of patients | 134 | 46 | 27 | 67 | 27 | 56 | NA | |||
| Age, y | ||||||||||
| At enrollment | 13.2 ± 3.9 | 14.9 ± 3.5 | 14.2 ± 3.1 | .44 | ||||||
| At first stroke | 5.9 ± 2.9 | 7.2 ± 3.9 | 6.6 ± 3.0 | .53 | ||||||
| Duration of transfusions, y | 7.2 ± 3.7 | 7.6 ± 4.6 | 7.6 ± 4.2 | 1.0 | ||||||
Among all 134 randomly assigned SWiTCH study participants, 27 children had documented Rh immunization at study entry. Controls were identified from nonimmunized SWiTCH participants and matched first for duration of transfusions (mean difference, 0.0 y; median difference, 0.0 y) and then for age at first stroke (mean difference, 0.6 y; median difference, 0.0 y). Sex was matched when possible.
NA, not applicable; SD, standard deviation.
RH genotyping
DNA samples were tested with RHD and RHCE BeadChip DNA arrays (Bioarray, Warren, NJ) and polymerase chain reaction (PCR)–based assays, as described previously.5,12 Briefly, the DNA array targets 35 RHD and 25 RHCE single nucleotide changes or insertions. PCR- restriction fragment length polymorphism–based analysis was performed for RHD exon 8 (c.1136C>T) and RHCE exon 2 (c.254C>G), exon 4 (c.577A>G), and exon 6 (c.907 del C). RHD exons 2 and 7 were amplified, and Sanger sequencing was performed to distinguish some alleles. Discordant results between SNP-based assays and WES were investigated with Sanger sequencing of implicated exons that were amplified by using RHCE- and/or RHD-specific primers as previously described.13 RHD zygosity was determined by direct amplification of a 1507-bp fragment from the region associated with deletion of RHD,14 and for some, RHD zygosity was also determined by PstI-restriction fragment length polymorphism.15 RHD and RHCE alleles were assigned on the basis of either a single or a combination of genetic markers and haplotype associations based on reported alleles.5,16,17
WES
WES data were generated by using NimbleGen VCRome 2.1 capture reagents, followed by sequencing on an Illumina HiSeq platform. Individual sequence reads were aligned to the reference human genome, and variants were annotated. All samples passed WES quality control parameters with an average of 92% of all exonic regions sequenced at greater than 20× coverage per sample. A project-level variant call format was generated for each sample and included all RHD and RHCE gene variations present in at least 1 sample.
Sequencing reads were mapped to the GRCh37/hg19 reference genome by using the Burrows-Wheeler algorithm.18 Sample-level genome variants were identified and annotated by using an integrated Mercury pipeline19 which includes quality score recalibration, genome variant identification by AtlasSNP,20 and annotation using Cassandra software. All reagents and data processing are standard methods used for WES and were not adjusted to focus analysis on RHD and RHCE.
RHAG (V270I) genotyping
We used Applied Biosystems StepOne (Applied Biosystems, Foster City, CA) PCR systems to perform a TaqMan assay for the RHAG SNP c.808G>A (rs16879498) associated with p.Val270Ile. After 40 amplification cycles, threshold cycle values were automatically calculated, and the individual SNP genotypes were determined by StepOne v2.0 software (Applied Biosystems). Samples from 54 individuals from the SWiTCH cohort and 488 individuals with SCA from CHP were tested.
Statistical analysis
We performed genetic case-control comparison by using Fisher’s exact test or Pearson’s χ2 test. Phenotypes (groups) were classified as Rh alloimmunized or non-alloimmunized. An additive genetic model was used for testing, with adjustment for age, sex, and population stratification when possible. Any variant with an association having a P value below .05 was considered significant, although none of the P values remained statistically significant when adjusted for the number of multiple comparisons. For the WES data analysis of candidate genes, quality control filtering steps included minimum sequence coverage of 10×, SNP missingness check, and application of an excess heterozygosity filter.
Results
Rh immunization in SWiTCH study participants
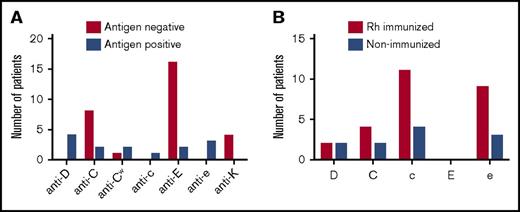
Among 134 randomly assigned SWiTCH study participants, 27 patients (20.1%) had a history of antibodies to Rh antigens at study entry. Controls were identified from non-alloimmunized SWiTCH participants (Table 1) and matched first for duration of transfusions (mean difference, 0.0 years; median difference, 0.0 years) and then for age at first stroke (mean difference, 0.6 years; median difference, 0.0 years). The mean duration of transfusions was 7.6 years for both groups, and the mean age at first stroke was 7.2 years for the Rh immunized and 6.6 years for the control group (P = .53; Table 1). There were more females in the Rh alloimmunized group compared with the overall SWiTCH cohort (P < .04). From antibody data reported at study entry, there were 4 anti-D, 10 anti-C, 3 anti-Cw, 1 anti-c, 18 anti-E, and 3 anti-e antibodies among the 27 Rh-immunized patients (Figure 1A; supplemental Table 1). Fourteen (35.8%) of these 39 Rh antibodies were unexpected in that they were present in individuals who typed positive by serology for that antigen. The remaining 25 Rh antibodies (8 anti-C, 1 anti-Cw, and 16 anti-E) were found in patients who were antigen-negative by serologic testing. The finding of anti-K is consistent with lack of antigen matching and was identified in 4 Rh-alloimmunized individuals (Figure 1A).
Anti-Rh and anti-K in SWiTCH patients at trial enrollment. (A) The number and specificity of antibodies identified in 27 patients who are either negative or positive for the corresponding antigen by serologic testing. (B) The number of patients with partial Rh antigens (and no corresponding conventional allele) determined by high-resolution RH genotyping and at risk of alloantibody (n = 27 Rh alloimmunized and 27 non-alloimmunized at enrollment).
Anti-Rh and anti-K in SWiTCH patients at trial enrollment. (A) The number and specificity of antibodies identified in 27 patients who are either negative or positive for the corresponding antigen by serologic testing. (B) The number of patients with partial Rh antigens (and no corresponding conventional allele) determined by high-resolution RH genotyping and at risk of alloantibody (n = 27 Rh alloimmunized and 27 non-alloimmunized at enrollment).
RH genotyping by SNP assay
Samples were tested by a combination of DNA array and PCR-based assays targeting markers of RHD (supplemental Table 2) and RHCE (supplemental Table 3) gene variation. Overall, 9 different RHD and 13 RHCE alleles were identified by either single or combinations of markers among this cohort, of which 5 RHD and 8 RHCE are reported to encode partial Rh antigens defined as lacking antigenic epitopes (Table 2). Two alleles that differ from the conventional alleles, RHD*DAU0 and RHCE*ce48C, encode single amino acid changes and are common in African populations. These alleles have not been reported to encode proteins lacking epitopes or to encode novel epitopes (ie, partial antigens). Exceptions may exist, but patients with these alleles seem to be at less risk for Rh alloimmunization than patients with other altered alleles (our unpublished observations).
Comparison of allele frequencies between participants in the SWiTCH trial (n = 54) and children with SCA from CHP (n = 488)
| Rh genotype . | Rh phenotype . | Diagnostic nucleotide changes (cDNA) . | SWiTCH cohort . | CHP cohort . | P . |
|---|---|---|---|---|---|
| RHD* | |||||
| Deleted D | D– | 0.1666 | 0.1445 | .5678 | |
| Inactive Dψ | D– | In3-19 (37 bp), 609G>A, 654G>C, 667T>G, 674C>T, 807T>G | 0.0185 | 0.0297 | .7616 |
| DIIIa-CE(4-7)-D | D–, partial C+ | 186G>T, 410C>T, 455A>C | 0.0648 | 0.0277 | .0721 |
| D | D+ | 0.5648 | 0.5410 | .7601 | |
| DAU0 | D+ (unknown) | 1138C>T | 0.1019 | 0.1619 | .1239 |
| DAU3 | Partial D+ | 835G>A, 1138C>T | 0.0278 | 0.0113 | .1588 |
| DAR | Partial D+ | 602C>G, 667T>G, 1025C>T | 0.0093 | 0.0041 | .4116 |
| D 4.0 | Partial D+ | 602C>G, 667T>G | 0.0278 | 0.0256 | .7553 |
| DIIIa | Partial D+ | 186G>T, 410C>T, 455A>C, 602C>G, 667T>G | 0.0185 | 0.0123 | .6435 |
| Others | NA | 0.0419 | NA | ||
| RHCE* | |||||
| ce | c+, e+ | 0.2407 | 0.2490 | .9068 | |
| Ce | C+, e+ | 48G>C, 150T>C, 178C>A, 201A>G, 203A>G, 307C>T | 0.0926 | 0.1199 | .4360 |
| cE | c+, E+ | 676G>C | 0.0463 | 0.0871 | .1969 |
| ce48C | c+, e+ (unknown) | 48G>C | 0.2130 | 0.1926 | .6118 |
| ce48C 733G | Partial c+, partial e+ | 48G>C, 733C>G | 0.0556 | 0.0686 | .6911 |
| ce733G | Partial c+, partial e+ | 733C>G | 0.1852 | 0.1455 | .3200 |
| ce254G | Partial c+, partial e+ | 254C>G | 0.0370 | 0.0471 | .8107 |
| ce1025T | Partial c+, partial e+ | 1025C>T | 0.0093 | 0 | .1005 |
| ceJAL | Partial c+, partial e+ | 340C>T, 733C>G | 0.0093 | 0.0010 | .1909 |
| ceCF | Partial c+, partial e+ | 48G>C, 697C>G, 733C>G | 0.0093 | 0.0020 | .2724 |
| ceS | Partial c+, partial e+ | 48G>C, 733C>G, 1006G>T | 0.0833 | 0.0379 | .0408 |
| ceAR | Partial c+, partial e+ | 48G>C, 712A>G, 733C>G, 787A>G, 800T>A, 916A>G | 0.0093 | 0.0031 | .3457 |
| cE48C | c+, E+ (unknown) | 676G>C, 48G>C | 0.0093 | 0.0010 | .1909 |
| Rh genotype . | Rh phenotype . | Diagnostic nucleotide changes (cDNA) . | SWiTCH cohort . | CHP cohort . | P . |
|---|---|---|---|---|---|
| RHD* | |||||
| Deleted D | D– | 0.1666 | 0.1445 | .5678 | |
| Inactive Dψ | D– | In3-19 (37 bp), 609G>A, 654G>C, 667T>G, 674C>T, 807T>G | 0.0185 | 0.0297 | .7616 |
| DIIIa-CE(4-7)-D | D–, partial C+ | 186G>T, 410C>T, 455A>C | 0.0648 | 0.0277 | .0721 |
| D | D+ | 0.5648 | 0.5410 | .7601 | |
| DAU0 | D+ (unknown) | 1138C>T | 0.1019 | 0.1619 | .1239 |
| DAU3 | Partial D+ | 835G>A, 1138C>T | 0.0278 | 0.0113 | .1588 |
| DAR | Partial D+ | 602C>G, 667T>G, 1025C>T | 0.0093 | 0.0041 | .4116 |
| D 4.0 | Partial D+ | 602C>G, 667T>G | 0.0278 | 0.0256 | .7553 |
| DIIIa | Partial D+ | 186G>T, 410C>T, 455A>C, 602C>G, 667T>G | 0.0185 | 0.0123 | .6435 |
| Others | NA | 0.0419 | NA | ||
| RHCE* | |||||
| ce | c+, e+ | 0.2407 | 0.2490 | .9068 | |
| Ce | C+, e+ | 48G>C, 150T>C, 178C>A, 201A>G, 203A>G, 307C>T | 0.0926 | 0.1199 | .4360 |
| cE | c+, E+ | 676G>C | 0.0463 | 0.0871 | .1969 |
| ce48C | c+, e+ (unknown) | 48G>C | 0.2130 | 0.1926 | .6118 |
| ce48C 733G | Partial c+, partial e+ | 48G>C, 733C>G | 0.0556 | 0.0686 | .6911 |
| ce733G | Partial c+, partial e+ | 733C>G | 0.1852 | 0.1455 | .3200 |
| ce254G | Partial c+, partial e+ | 254C>G | 0.0370 | 0.0471 | .8107 |
| ce1025T | Partial c+, partial e+ | 1025C>T | 0.0093 | 0 | .1005 |
| ceJAL | Partial c+, partial e+ | 340C>T, 733C>G | 0.0093 | 0.0010 | .1909 |
| ceCF | Partial c+, partial e+ | 48G>C, 697C>G, 733C>G | 0.0093 | 0.0020 | .2724 |
| ceS | Partial c+, partial e+ | 48G>C, 733C>G, 1006G>T | 0.0833 | 0.0379 | .0408 |
| ceAR | Partial c+, partial e+ | 48G>C, 712A>G, 733C>G, 787A>G, 800T>A, 916A>G | 0.0093 | 0.0031 | .3457 |
| cE48C | c+, E+ (unknown) | 676G>C, 48G>C | 0.0093 | 0.0010 | .1909 |
The allele frequency was similar (two-sided Fisher’s exact test). Only the specific alleles shared by both groups are shown. The alleles in the SWiTCH cohort comprised 95.8% of RHD and 95.5% of RHCE alleles also present in the larger CHP cohort.
cDNA, complementary DNA.
Overall, RH allele frequencies were similar between Rh alloimmunized and non-immunized individuals (P > .05; data not shown). RH allele frequencies also did not differ significantly from 488 patients from the CHP cohort, some of whom have been previously reported, with the exception that RHCE*ceS was more frequent in the CHP cohort (P = .0408).5 Analysis of the comprehensive RH genotypes for each individual revealed that 14 Rh alloimmunized patients were predicted to express known altered or partial Rh antigens without co-expression of conventional protein compared with 5 non-immunized individuals (51.9% vs 18.5%; P = .0103) (supplemental Table 1). Variant e and/or c (ie, RHCE*ce) were the most common variants in both the Rh alloimmunized and the non-immunized children (Figure 1B). Correlation of unexpected Rh antibodies (present in individuals who typed positive for the antigen; n = 14) revealed 1 anti-D, 1 anti-C, and 1 anti-e in 3 patients with altered alleles and absence of the conventional allele (supplemental Table 1). Anti-Cw in 2 C+ patients can likely be explained by exposure to altered C antigen (Cw) present in 2% of white donors. Nine Rh specificities (3 anti-D, 1 anti-C, 1 anti-c, 2 anti-E, 2 anti-e) in 8 patients remain unexplained (antibody present in patient with conventional allele). However, with 1 exception (unique patient identifier [UPID] 75 whose anti-e would be classified as an autoantibody; supplemental Table 1), 7 of these individuals have altered RH genotypes.
RH genotype by WES
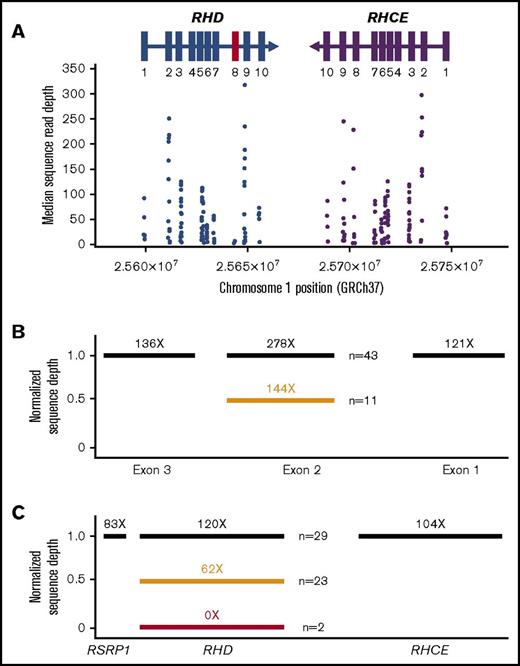
We then examined whether analysis of WES data covering RHD and RHCE gene loci agreed with results from RH genotyping by SNP-based methods. The publicly available NGS analysis pipeline Mercury19 was used to integrate multiple sequence analysis components across many computational steps and to provide a fully annotated list of variants (Figure 2). We assessed concordance for detection of SNPs between the 2 methods and performed Sanger re-sequencing for discordant samples. Good-coverage exome data were generated for 9 of 10 RHD exons (median coverage, 19×-317×) and all RHCE exons (39×-297×; supplemental Tables 2 and 3). RHD exon 8 had low sequence coverage (median coverage, <6×; Figure 3A), and therefore WES did not reliably detect the c.1138C>T change (p.Thr379Met) in exon 8 that has an allele frequency of 0.1019 to 0.1710 in African individuals (Table 2).5,16 WES also did not detect the RHD 37-bp duplication that inactivates D antigen expression (D– phenotype) referred to as the RHD pseudogene (supplemental Table 2). However, the RHD pseudogene has the stop codon c.807T>G (p.Try269Stop) in exon 6 that was identified by WES. Finally, as expected for exon-only interrogation, WES cannot detect the RHCE 109-bp intron 2 insertion in RHCE*Ce associated with a C+ phenotype (supplemental Table 3).
Mercury analysis pipeline. (A) Raw data from the sequencing instrument is passed to primary analysis software to generate sequence reads and base call confidence values (qualities). (B) Reads and qualities are passed along to a mapping tool Burrows-Wheeler algorithm (BWA) for comparison with a reference genome. The placement of reads on the reference genome produces a Binary-format Sequence Alignment Map (BAM) file and individual event BAMs were merged to make a single sample-level BAM file. (C) AtlasSNP and Genome Analysis Toolkit (GATK) are used to identify variants and produce annotated variant call files (VCFs). (D) In this study, we specifically interrogated WES data for the RHD and RHCE genes compared with conventional targeted assays.
Mercury analysis pipeline. (A) Raw data from the sequencing instrument is passed to primary analysis software to generate sequence reads and base call confidence values (qualities). (B) Reads and qualities are passed along to a mapping tool Burrows-Wheeler algorithm (BWA) for comparison with a reference genome. The placement of reads on the reference genome produces a Binary-format Sequence Alignment Map (BAM) file and individual event BAMs were merged to make a single sample-level BAM file. (C) AtlasSNP and Genome Analysis Toolkit (GATK) are used to identify variants and produce annotated variant call files (VCFs). (D) In this study, we specifically interrogated WES data for the RHD and RHCE genes compared with conventional targeted assays.
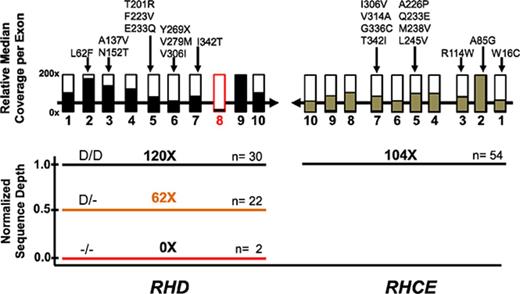
WES coverage for RHD and RHCE genes. (A) The median number of individual sequence reads are given for each polymorphism identified by exome sequencing. The sequence reads were aligned to the human reference sequence GRCh37, and median coverage was calculated for the entire SWiTCH cohort (n = 134). All exons (rectangles) had greater than 10× median coverage except RHD exon 8 (marked in red). (B) Normalized sequence read depth for RHCE exons 1, 2, and 3 (n = 54). Individuals with RHCE*Ce have a reduced ratio for exon 2 compared with exons 1 and 3 (orange bar). (C) Normalized sequence read depth for RHD, RHCE, and neighboring genes (n = 54). Genes with 2 copies are indicated as black bars, 1 copy as orange bars, and no copies as red bars.
WES coverage for RHD and RHCE genes. (A) The median number of individual sequence reads are given for each polymorphism identified by exome sequencing. The sequence reads were aligned to the human reference sequence GRCh37, and median coverage was calculated for the entire SWiTCH cohort (n = 134). All exons (rectangles) had greater than 10× median coverage except RHD exon 8 (marked in red). (B) Normalized sequence read depth for RHCE exons 1, 2, and 3 (n = 54). Individuals with RHCE*Ce have a reduced ratio for exon 2 compared with exons 1 and 3 (orange bar). (C) Normalized sequence read depth for RHD, RHCE, and neighboring genes (n = 54). Genes with 2 copies are indicated as black bars, 1 copy as orange bars, and no copies as red bars.
Determining the C+ phenotype by using WES data requires an alternative approach. Exon 2 of RHD and RHCE*Ce share identical sequences, and RHCE*Ce exon 2 WES sequence reads will map to RHD exon 2. To overcome this, we calculated read depth ratios of each sample for RHCE exons 1, 2, and 3 and compared the average value for all samples. Patients with RHCE*Ce have a reduced ratio for exon 2 (fewer reads) compared with exons 1 and 3 (Figure 3B). By using this approach, all 10 C+ individuals with RHCE*Ce were identified. One individual (UPID 19) was identified as having an RHCE*Ce allele with WES, whereas SNP array demonstrated RHCE*cE/ce.
WES and SNP concordance
Comparison of WES and DNA array SNP genotypes demonstrated differences in 19 samples involving 10 nucleotide positions (Table 3). Sanger re-sequencing was performed for each discordant result. Alignment of sequence reads to the homologous RH gene was the primary reason for discordant WES assignments. Four hemizygous and/or homozygous G nucleotide positions in RHD were identified as heterozygous by WES: c.186G>T in exon 2, c.602C>G in exon 4, and c.667T>G and c.697G>C in exon 5. For a heterozygous call, a sequence ratio of 50:50 is expected. For each of these calls, 11% to 13% of sequence reads were RHCE sequences that were assigned to RHD. The integrated Mercury pipeline used to process the WES data interpreted any polymorphism with reference-to-variant sequence reads greater than 90% of total as being homozygous. A slight relaxation of this threshold to 85% of total calls would have resulted in all of these RHD markers called as homozygous.
Summary of discordance between WES and SNP methods designated by nucleotide position in cDNA and RH exon that were resolved by Sanger re-sequencing
| Reason . | cDNA position . | Exon . | WES . | SNP array . | Sanger re-sequence . | Sample UPIDs . |
|---|---|---|---|---|---|---|
| RHCE nucleotide assigned to RHD by WES | RHD c.186G>T | 2 | G/T | G/G | G/G | 10 |
| RHD c.602C>G | 4 | C/G | G/G | G/G | 60 | |
| RHD c.667T>G | 5 | T/G | G/G | G/G | 60 | |
| RHD c.697G>C | 5 | G/C | G/G | G/G | 84 | |
| RHD nucleotide assigned to RHCE by WES | RHCE c.410C>T | 3 | C/T | C/C | C/C | 7, 50, 82 |
| RHCE c.654G>C | 5 | G/C | G/G | G/G | 39 | |
| RHCE c.916A>G | 6 | A/G | A/A | A/A | 67, 104 | |
| Misassignment as a result of reference sequence | RHCE c.48G>C | 1 | G/C | G/G | G/G | 33, 81, 124 |
| RHCE c.48G>C | 1 | C/C | G/C | G/C | 38, 85 | |
| WES detects changes not included in SNP testing | RHD c.916G>A | 6 | G/A | NT | G/A | 28, 81 |
| RHCE c.941T>C | 7 | T/C | NT | T/C | 89, 102 |
| Reason . | cDNA position . | Exon . | WES . | SNP array . | Sanger re-sequence . | Sample UPIDs . |
|---|---|---|---|---|---|---|
| RHCE nucleotide assigned to RHD by WES | RHD c.186G>T | 2 | G/T | G/G | G/G | 10 |
| RHD c.602C>G | 4 | C/G | G/G | G/G | 60 | |
| RHD c.667T>G | 5 | T/G | G/G | G/G | 60 | |
| RHD c.697G>C | 5 | G/C | G/G | G/G | 84 | |
| RHD nucleotide assigned to RHCE by WES | RHCE c.410C>T | 3 | C/T | C/C | C/C | 7, 50, 82 |
| RHCE c.654G>C | 5 | G/C | G/G | G/G | 39 | |
| RHCE c.916A>G | 6 | A/G | A/A | A/A | 67, 104 | |
| Misassignment as a result of reference sequence | RHCE c.48G>C | 1 | G/C | G/G | G/G | 33, 81, 124 |
| RHCE c.48G>C | 1 | C/C | G/C | G/C | 38, 85 | |
| WES detects changes not included in SNP testing | RHD c.916G>A | 6 | G/A | NT | G/A | 28, 81 |
| RHCE c.941T>C | 7 | T/C | NT | T/C | 89, 102 |
NT, not tested.
The converse, alignment of RHD sequence reads to RHCE by WES, occurred in 6 samples (Table 3). The c.410C>T change in exon 3 from the RHD locus hybrid RHD*DIIIa-CE(4-7)-D was assigned to RHCE in 3 samples, as was the c.654G>C change in exon 5 from the RHD pseudogene in 1 sample and the c.916A>G in exon 6 in 2 samples. As with RHD, relaxation of the Mercury pipeline reference:variant ratio threshold would have resulted in concordant calls for RHCE c.410C/C, c.654G/G, and c.916A/A. Five samples were discordant for RHCE c.48G>C. Exon 1 of the RH genes has only this 1 coding polymorphic nucleotide position between RHD and some but not all RHCE alleles, but the human genome reference sequence for RHCE lists c.48C as wild-type, which also complicates allelic assignment by NGS. Relaxation of the reference:variant ratio threshold would have resulted in 1 of these calls being concordant with DNA array SNP testing.
Four samples had changes identified by WES that were not identified by SNP assays. WES detected an RHD exon 6 change c.916G>A in 2 samples (UPID 28 and 81) associated with RHD*weak D type 66, and the exon 7 RHCE change c.941T>C in 2 samples (UPID 89 and 102). These polymorphisms have been previously reported but are not interrogated by SNP assay. Overall, the concordance between the genotyping methods ranged from 91.7% to 100% for RHD and RHCE polymorphisms (supplemental Tables 2 and 3).
Determination of RHD zygosity
RHD zygosity can be determined by analysis of RHD exon copy number compared with RHCE exon copy number. With very rare exceptions, 2 RHCE alleles are present in all samples, and in most populations, the D– phenotype results from deletion of RHD.15 To determine RHD copy number, we generated an average read depth ratio between the entire RHD and RHCE genes (Figure 3C). For patients with homozygous deletion (absence) of RHD (n = 2), the sequence read depth was zero for RHD but reached an average of 104 ± 56 reads for RHCE (1.0 normalized read depth ratio) representing 2 copies of RHCE. By using a calculated read depth ratio, 23 patients had 1 copy (0.62 ± 0.09 read depth ratio), and 29 patients had 2 copies of RHD (1.20 ± 0.11 read depth ratio) (supplemental Table 4). We compared these results to RHD zygosity determined by PCR methods. Fifty-three (98%) of 54 were concordant. One sample by WES analysis was RHD hemizygous but gave conflicting results (D−/− and D+/+) by 2 different PCR-based methods.
Search for genetic associations with Rh alloimmunization
RhD and RhCE proteins physically associate with Rh-associated glycoprotein (RhAG), CD47, LW, and glycophorin B in a larger Rh complex in the erythrocyte membrane. We hypothesized that mutations in these proteins may alter Rh epitopes or topology and contribute to recognition of the Rh complex as foreign, with subsequent antibody production. WES data were therefore also examined for polymorphisms in RHAG, CD47, LW, and GYPB. Among the SWiTCH cohort, no variant in CD47, LW, GYB, or RHAG was significantly different between the Rh alloimmunized group compared with the non-immunized group. Eight (14.8%) of 54 SWiTCH participants were heterozygous for the V270I variant, of which 5 were Rh-immunized and 3 were non-immunized (18.5% vs 11.1%; P = .70). RhAG is important for targeting RhD and RhCE to the erythrocyte membrane, and gene-inactivating mutations in RHAG are responsible for loss of Rh antigen expression (Rh-null).21 The RHAG V270I (c.808G>A change) variant was previously reported to encode the RHAG4 antigen in a familial case with maternal sensitization and hemolytic disease in her newborn.22 To ensure that an association of RHAG V270I was not detected because of the small sample size of the SWiTCH cohort, we further investigated possible significance in a larger cohort of 488 individuals with SCA from CHP. The overall frequency of the RHAG V270I variant was similar: 14.6% (n = 71) were heterozygous and 0.6% were homozygous (n = 3). Among patients with at least 10 unit exposures (n = 157; median exposure, 178 units; mean, 341 units), there was no difference in the frequency of the RHAG V270I variant in the Rh alloimmunized (9 of 60) compared with non-alloimmunized (15 of 97) individuals (15.0% vs 15.5%; P = 1.0).
Discussion
RH genetic diversity contributes to the prevalence of Rh antibodies encountered after red cell transfusion in patients with SCA.5,6,23 Importantly, Rh antibodies continue to occur in patients who receive donor units that are antigen-matched by serology for Rh D, C, and E antigens because of RH variation at the genetic level. We previously reported that individuals who express only altered or partial Rh antigen (and no corresponding conventional protein) are at increased risk of alloimmunization.5
We RH genotyped a matched cohort of SWiTCH trial participants to extend our studies on RH variation and to assess whether Rh antibody formation correlated with specific RH alleles in a multicenter cohort. Genotyping revealed that RH diversity in the SWiTCH participants parallels that seen in our previously reported CHP cohort.5 At enrollment, 20.1% of SWiTCH participants were Rh alloimmunized and, consistent with our previous observations, 36% of these Rh antibodies were unexpected in that patients were positive for the corresponding antigen. Comparison of 27 Rh alloimmunized with 27 non-alloimmunized individuals matched for age and transfusion years indicated that Rh alloimmunization was correlated to inheritance of altered RH alleles (P = .0103). Although only 3 of the antibodies (1 anti-D, 1 anti-C, 1 anti-e) could be directly explained by inheritance of the corresponding partial antigen and absence of conventional protein, 11 additional Rh-immunized patients expressed at least 1 partial Rh antigen, further supporting our previous suggestion that the presence of any partial Rh may disrupt the Rh complex and contribute to Rh alloimmunization.5
The Rh antibodies found in this population challenge convention in that patients whose erythrocytes are positive for an antigen, or patients who were never exposed to the antigen, are not expected to have the corresponding antibody identified by the laboratory as present in their plasma. Thus, RH genotyping has a potentially important role in transfusion support for patients with SCA to resolve confusing serologic antibody reactivity and to guide selection of units for transfusion. Currently available DNA arrays identify the most common polymorphic RH alleles but cannot detect all RH variation, require expertise to interpret, may be cost prohibitive for some, and are not widely available. Alternatives would advance patient care, especially if they involve translation of data that may already be available. WGS and WES are moving toward routine practice for patients with many chronic diseases and should allow comprehensive analysis of RH genetic variation in patients with SCA. Examining whether WES data can be accurately aligned and interpreted, given the challenges of duplicated genes with significant homology, provides proof of concept for the development of targeted NGS panels for RHD and RHCE.
In this study, WES data using a standard non-targeted approach with modified analysis pipelines to identify RH variants and determine RHD zygosity demonstrated a high concordance rate with SNP typing. Comprehensive RH genotypes for 49 (90.7%) of 54 SWiTCH participants in this study were concordant with SNP-based assays. The 4 discordant RHCE exon 1 c.48C alignment challenges would not have an impact on risk for clinically significant antibody production or donor choice, because RHCE*ce48C has not been associated with partial antigen expression. Alignments for RHCE vs RHD in exon 1, which differ only by 1 nucleotide (c.48) in some but not all RHCE alleles, require greater read depth with algorithm adjustment. The other limitation was the very low sequence coverage for RHD exon 8 (median coverage, <6×), which was supported by examination of the ExaC database, which shows poor coverage of RHD exon 8 in more than 60 000 individuals with WES data. A recent study that designed targeted exome sequencing for blood group systems encountered similar limitations in coverage and sequence alignment for exons 1 and 8 of RHD and RHCE.24
Approaches used here to improve accuracy included a slight relaxation of the Mercury pipeline reference:variant ratio threshold, consideration of RHCE exon 2 copy number to detect the C+ phenotype associated with RHCE*Ce, and recognition of the common hybrid allele RHD*DIIIa-CE(4-7)-D as an RHD locus variant (Table 4). In the future, standardization of the human genome reference sequence and increased coverage of exon 1 for RHCE*ce48C and exon 8 for RHD*1138T detection would further enhance accuracy (Table 4).
Summary of RH alleles that require modification of data analysis or algorithm for assignment
| Genotype . | RBC phenotype . | cDNA location . | Genomic coordinates . | Consideration for correct assignment . |
|---|---|---|---|---|
| RHD*DAU0 | D+ | Exon 8, c.1138C>T (p.Thr379Met) | NC_000001.11:g.25317062T>C | Reference sequence and increased genomic coverage |
| RHCE*ce48C | c+e+ | Exon 1, c.48G>C (p.Trp16Cys) | NC_000001.11:g.25420739G | Reference sequence and increased genomic coverage |
| RHD*Ce | C+ | 109-bp intron 2 insertion | NC_000001.11:g.25732088-25732107 | Exon copy number calculation |
| RHD*Dpsi | D– | 37-bp insertion | NC_000001.11:g.25627431-25627454dup | Use of stop codon to detect |
| RHD*DIIIa-CE(4-7)-D | C+ partial, D– | Exons 4-7 | NA | Detection of novel insert for assignment to RHD rather than RHCE |
| Genotype . | RBC phenotype . | cDNA location . | Genomic coordinates . | Consideration for correct assignment . |
|---|---|---|---|---|
| RHD*DAU0 | D+ | Exon 8, c.1138C>T (p.Thr379Met) | NC_000001.11:g.25317062T>C | Reference sequence and increased genomic coverage |
| RHCE*ce48C | c+e+ | Exon 1, c.48G>C (p.Trp16Cys) | NC_000001.11:g.25420739G | Reference sequence and increased genomic coverage |
| RHD*Ce | C+ | 109-bp intron 2 insertion | NC_000001.11:g.25732088-25732107 | Exon copy number calculation |
| RHD*Dpsi | D– | 37-bp insertion | NC_000001.11:g.25627431-25627454dup | Use of stop codon to detect |
| RHD*DIIIa-CE(4-7)-D | C+ partial, D– | Exons 4-7 | NA | Detection of novel insert for assignment to RHD rather than RHCE |
NT, not tested; RBC, red blood cell.
Analysis of NGS data for 36 blood group systems from the 1000 Genomes Project demonstrated extensive variation in a multiethnic cohort.25 Of the 1241 nonsynonymous variants identified in the coding regions, only 241 were known blood group polymorphisms. Therefore, performing NGS-based RH predictions on a diverse population of serologically and molecularly typed individuals will be necessary to further refine interpretation algorithms. The development of sequencing technology that allows longer read lengths, targeted NGS panels for the RH loci, and automated algorithms for interpretation will further improve accuracy, availability, and affordability.
Interrogation of our WES data to identify polymorphisms in other membrane proteins known to interact physically with RhD and RhCE was unrevealing. Although the RHAG V270I variant was identified in 15% of the SWiTCH and CHP cohorts, it was not associated with expression of RhAG4 antigen (not shown) and was not associated with a higher risk for Rh immunization. Similar to prior studies looking for genetic polymorphisms associated with alloimmunization risk, our relatively small sample size (n = 54 and n = 488) is a limitation.
Recognition of RH variants can refine antibody identification, improve red blood cell matching, and potentially minimize alloimmunization in patients with SCA. We demonstrate that WES data can be used for accurate prediction of Rh antigen expression in patients with SCA who have tremendous RH genetic variation, and adjustments will increase accuracy. In the future, improved RH genotyping technology and accessibility may allow consideration of both recipient and donor RH genotypes and ultimately reduce Rh alloimmunization in individuals with SCA.
Acknowledgments
The authors thank the patients and families who enrolled in the studies, members of the Immunohematology and Genomics Laboratory at the New York Blood Center, the Human Genome Sequencing Centre at Baylor College of Medicine for assistance in generating the whole-exome sequencing data, and Perry Evans for statistical support.
This work was supported by the Doris Duke Innovations in Clinical Research Award grants 2013151 (S.T.C., C.M.W., and R.E.W.), 2011097, and 2015133 (S.T.C. and C.M.W.); by National Institutes of Health (NIH) National Human Genome Research Institute award U54-HGOO3273 (R.E.W.); by NIH National Heart, Lung, and Blood Institute award HL-078787 for the SWiTCH study (R.E.W.); and in part by a donation from the DiGaetano family (S.T.C.).
Authorship
Contribution: S.T.C., J.M.F., R.E.W., and C.M.W. designed the study, analyzed results, and wrote the manuscript; S.V. conducted research and analyzed results; N.L.C.L. was the SWiTCH transfusion consultant; R.C.B. enrolled patients and collected data for the SWiTCH study; and N.L.C.L. and R.C.B. edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stella T. Chou, The Children’s Hospital of Philadelphia, 3615 Civic Center Blvd, Abramson Research Center, Room 316D, Philadelphia, PA 19104; e-mail: chous@email.chop.edu.
References
Author notes
S.T.C. and J.M.F. contributed equally to this work.
The full-text version of this article contains a data supplement.